Abstract
Since the first draft of the human genome was published 10 years ago, scientists have tried to develop new treatment strategies for various types of diseases based on individual genomes. It is called personalized (or individualized) medicine and is expected to increase efficacy and reduce adverse reactions of drugs. Much progress has been made with newly developed technologies, though individualized pain medicine is still far from realization. Efforts on the integrative genomic analyses along with understandings of interactions between other related factors such as environment will eventually translate complex genomic information into individualized pain medicine.
Keywords: individualized medicine, pharmacogenomics, genetic polymorphism
Variability of bio-behavioral traits among humans is tremendous, especially for complex traits such as pain which is influenced by an individual s genetic composition, prior learning, current physiological status, expectations and sociocultural environment.[1] Not only do personal experiences such as natural child birth or traumatic injury influence an individual s response to pain but also the sociocultural experiences of a specific population alter pain reports. For example, males usually report less pain in response to an unpleasant stimuli compared to females, a phenomenon that can partially be explained by gender role expectation.[2,3] It has also been reported that people in economically or socially weak groups tend to report pain more frequently.[4]
Even though multiple non-physiologic factors influence nociception significantly, there are also actual biological differences in pain perception and processing between sexes / races as well as between pain inducing modalities. Animal studies, which are less affected by gender role expectation, show trends similar to human studies as males show less nociceptive responses to pain stimuli.[5,6] Nociceptive responses to painful stimuli vary also by inbred strains as well as by the type of pain producing modality[7], also suggesting actual biological difference exist between sexes / races. These underlying biological differences may also induce variable responses to drugs, with some drugs having differential efficacy or toxicity that varies by race. This concept has already resulted in the first FDA approved “race-specific” drug, BiDil, to the market in 2005. BiDil is specifically recommended only for African Americans who suffer from hypertension but not for Caucasians. It demonstrates that genetically determined demographics such as sex or race can be used as a criterion for selecting drugs. This may be the initial step of the individualized medicine in a broad sense,[8,9] though skepticism still exists.[10]
Genomic studies provide clues for better individualized pain management
While it is estimated that any two humans are 99.9% identical in their DNA sequence,[11] the small differences of ~0.1% of the genome and factors influencing the expression of individual genes are fundamental to variability between humans. The initial process inducing variability in response to drugs starts with genetic variations at the DNA level. Investigation of what and how genetic variations are associated with pain and drug responses, therefore, is the beginning point of individualized medicine for analgesia.
Single nucleotide polymorphisms (SNPs) are considered to be an appropriate genetic marker for identification of phenotype related loci (Figure 1), despite that more than 90% of SNPs are in introns or intergenic regions so that little difference in protein structure is expected, regardless of potential profound functional consequences. Relatively even distribution of SNPs across the human genome along with their high frequency make SNPs serve as superior genetic markers. Some SNPs in exons induce amino acid change from the encoding proteins and affect their biological features. SNPs in the promoter regions alter the level of gene expression. It is also possible that any portion of DNA can function as an RNA interference element. Any SNP can be meaningful because of the complicated dynamics of DNA structure and gene expression.[12]
Figure 1.
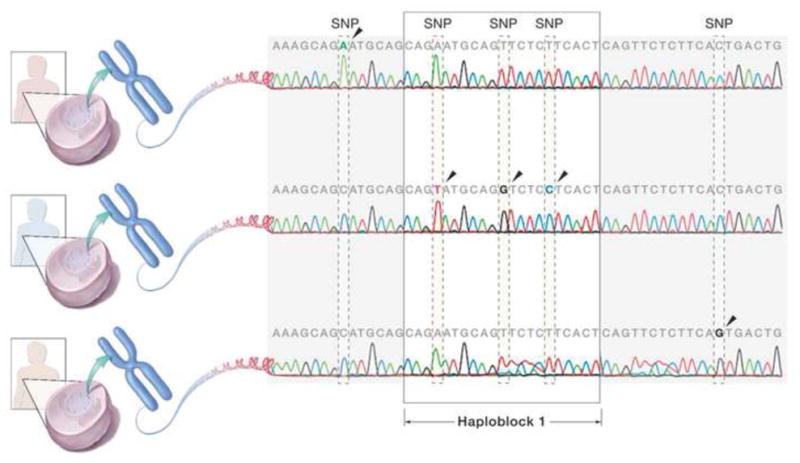
Single nucleotide polymorphism (SNP) and Haplotype. SNPs are the marker of choice for the association studies. They are as frequent as more than one per kb on average. Haplotype based on linkage disequilibrium can also be used for the association studies, though still debatable.
Candidate gene association study (CGAS) focusing on polymorphisms in a small set of markers or genes suspected to be related to the phenotypes have been performed so far mostly for pain responses. However, accumulated knowledge based on CGAS is not enough for routine clinical application yet. Most CGASs of pain investigate variations of genes in pain pathways rather than genes related to analgesic drugs. Information from these studies may be effective for finding new targets for pain control and developing novel analgesic drugs. For example, based on the role of tetrahydrobiopterin (BH4) as an intrinsic regulator of pain sensitivity and chronicity, variations of GTP cyclohydrolase gene have been investigated and suggested as a marker of pain traits. These studies may eventually be extented to develop BH4 as a new target for analgesic drugs.[13,14] However, almost all new analgesics approved during the last 25 years are modifications of opioids and aspirin- like drugs.[15] There are still only a few analgesic drug classes available, regardless of the large accumulated knowledge on the mechanisms of pain and analgesia. Considering the enormous resources needed for developing new drugs, targets for novel analgesic drugs might be more successfully identified by casting a genome-wide net rather than continuing to focus on traditional targets.
Furthermore, associations found in CGAS for pain are usually weak and often fail to replicate.[14,16–18] Dissection of the genetic influence on pain sensitivity has been difficult due to its physiologic complexity. Most variance in pain sensitivity likely result from multiple genes with small effects and environmental factors, interacting to produce pain traits rather than from one single gene with a major effect. Considering the size of the human genome along with the complexity of pain pathways, subtle effects of these individual genetic influences can easily be masked or sometimes even exaggerated due to other factor such as population stratification. Unfortunately, the most common causes of a failure to replicate genetic-association findings are insufficient statistical power, population stratification, or various forms of between-study heterogeneity or environmental influences. Though disparate findings could reflect biological variability across various types of painful conditions, a lack of methodological consistency among pain researchers may also contribute to non-replicable associations.[19]
In order to overcome some limitations of CGAS, genome-wide association studies (GWAS) based on high-throughput genotyping technology has been suggested.[20] Several platforms using arrays allow genotyping of more than 1 million SNPs at a single run. Large scale GWAS for rheumatoid arthritis (RA) revealed novel insights into the pathogenesis of RA for a significantly associated SNP that is in linkage disequilibrium with a gene encoding the tumor necrosis factor (TNF) receptor-associated factor.[21] Independent studies of the pharmacogenetics of anti-TNF treatment for RA patients have reported the influence of genetic variations in genes encoding TNF, TNF receptors and other TNF-relevant genetic loci on treatment efficacy and potential application of pharmacogenetic information in clinics.[22–26] However, GWAS with meaningful sample size in pain and analgesia is even rarer than CGAS. Recently developed technologies, so-called next generation sequencing, even make it possible to sequence one person s entire genome within a few weeks.[27–30] The technology of sequencing is no longer a limiting factor but clinical studies using next generation sequencing have not been reported yet.
Promising findings from pharmacogenomic studies
For faster application of genomic information to clinical practice, genetic association studies evaluating responses to currently used analgesic drugs may be an alternative. As a phenotype, responses to analgesic drugs may be comparatively more objective (i.e., did the drug produce analgesia?) and practical (i.e., is the drug safe?) target for genetic association studies. All currently available analgesics have safety issues. Consequently, patients suffering from pain related conditions frequently reduce dosage of analgesic drugs due to the adverse reactions. In this situation, the choice of drug and dose for an individual becomes critical. Lacking the analgesic magic bullet , individualizing analgesic therapy for a specific patient based on pharmacogenomics may improve analgesic efficacy and safety at less expense than traditional 'one size fits all' pain management.
Regardless of this potential immediate effect on clinical practice, pharmacogenomic genetic association studies have not been widely reported yet. The most extensively studied pharmacogenetic mechanism is the influence of genetic variations in the isoforms of cytochrome P450: CYP2D6, CYP2C9 and CYP2C19, which collectively account for the metabolism of 40% of marketed drugs. The genes encoding the isoforms likely possess a large number of functionally significant polymorphisms. Consequently, some people very effectively metabolize drugs (so-called ultra metabolizers) while some are very poor metabolizers. It is estimated that 6% of Caucasians have inactive CYP2D6 and do not benefit from codeine administration. The major metabolic pathway of codeine consists of glucuronidation; CYP2D6 mediated O-methylation produces morphine which is 200 to 3,000 times more affinitive to opioid receptor. Individuals with ultra-metabolizing CYP2D6 produce greater amount of morphine from codeine and have significantly increased analgesic efficacy and even toxicity, while poor metabolizing forms of CYP2D6 have reduced efficacy or even treatment failure. On the contrary, for drugs inactivated by CYP2D6, poor metabolizers are exposed to increased risk of adverse reactions whereas ultra metabolizers have decreased efficacy.[31–33] In January 2005, a genetic test kit for the rapid genotyping of CYP2D6 and CYP2C19 variants for diagnostic use was approved by FDA. While this kit, called AmpliChip CYP450 Array, has been available in the market since June 2005, the actual testing with this kit is still rarely performed in clinical practice.[34,35]
Other genes and their polymorphisms have been reported with responses to opioid drugs. The polymorphism inducing amino acid change from asparagine to aspartate at codon 40 in the mu opioid receptor gene increases morphine requirements in patients with pain caused by malignant diseases.[36] Interestingly, genetic variability in genes relevant to non-opioid systems, such as catechol O-methyltransferase gene (COMT) also shows an association with the clinical efficacy of morphine when administered to cancer pain patients. Cancer patients with Met/Met genotype at codon 158 of COMT need lower doses of morphine to relieve pain than patients with Val/Val genotype. The investigators expanded these results to explore interactions between OPRM1 and COMT to confirm the joint effect of the above SNPs on opioid analgesic efficacy.[37,38] Though intriguing, independent studies of this association have reported conflicting results. An initial report which inspired the above studies reported the opposite influence of the same SNP in COMT, as significantly increased experimental pain responses in Val/Val genotype subjects.[17] Some studies have failed to find any association of OPRM1 and COMT SNPs with pain.[39]
There are also some reports of the genetic influence of COX-2 encoding gene (PTGS2) on the response to frequently used over-the-counter, COX inhibitors. One genetic variation in PTGS2 showed clinical impact on non-steroidal antiinflammatory drugs (NSAIDs) responses in an in vivo model and suggests promise for individualized therapy.[40] However, other investigators reported no impact of this genetic variation based on an ex vivo model.[41] Because of its cardiovascular effect, the pharmacogenomic understanding of aspirin will also help to enhance individualized approaches that maximize analgesic and antithrombotic benefit while minimizing bleeding risk. However, it is still not known which specific variants contribute to phenotype or through what mechanisms. GWAS regarding NSAIDs is very rare and only one preliminary result has been reported so far.[42] Findings from genetic association studies are not readily applicable to clinical pain and analgesia until enough knowledge is accumulated based on studies following principles of genetic research.
Genes function through transcription to messenger RNA and final production of its encoding protein. The expression profiles of entire genes across the genome can be another tool for individualized analgesic drug selection.[43] Analysis of gene expression profiles based on microarray platforms in a peripheral blood sample may provide meaningful information for evaluating gene expression patterns in specific organs, where direct tissue collection is not easy, so that it can potentially be used as a surrogate.[44] Even though only a few clinical trials have been initiated for chemotherapy in the oncology area so far,[45,46] the same strategy of using gene expression profiles may be applicable for individualized selection of analgesic drugs.
Challenges to individualizing pain therapeutics
Increasing evidence suggests that patients in painful conditions are molecularly unique, although the final phenotype of pain may appear similar.[47] The pathogenesis of most pain inducing diseases is polygenic in nature,[48–51] as multiple molecular and cellular pathways operate in parallel in both the peripheral and central nervous system to produce different forms of pains. It is also likely that most analgesic drugs exert their actions through multiple mechanisms mediated by many different molecular genetic pathways.[52,53] It is not likely, therefore, that a universal analgesic can reduce different forms of pain in heterogenous patients. Even for individual patients with a single painful condition, molecular genetic mechanism underlying pain can be changed over time. For example, gene expression pattern of COX-2 initially increase in the immediate post-injury phase, but then decrease dramatically once the initial stage of acute inflammation is over by 48 hours. When COX-2 inhibitors are administered, however, COX-2 gene expression increases further at 48 hours after the injury.[40] Therefore, individualized analgesic medicine may not only apply to specific individual patients suffering from the same disease, but also for a specific conditions causing pain or for a specific stage of disease in the same patient. In addition, the strength of a genetic effect can vary by age and may require age-varying medication for the same patient with the same disease.[54] (This is why the term "individualized medicine" is preferred to "personalized medicine")
For making individualized pain medicine a reality, many barriers still remain which require persistent and enormous effort. The first step should be careful phenotype data collection. Because of the multi-factorial nature of pain, simple and objective phenotypes measuring analgesic response must be collected from a large sample of homogenous subjects. Then genetic association studies (GWAS or CGAS) or expression profiles of the entire genome following administration of specific drugs can provide initial clues for targets. Dense genotyping or real time PCR of selected candidate genomic regions may strengthen the findings and narrow the candidates. However, the significance of associations is just a statistical relationship, not necessarily a proof of causality. Functional genomic studies combined with related areas such as proteomics and bioinformatics should follow to reveal the underlying mechanisms. Considering significant findings of GWAS are mostly located in non-annotated region, follow up studies are critical. With the current technologies, all of these studies are feasible even with a very small amount of molecules, such as genomic DNA from a single cell. However, analyses of each result separately may not be consistent with the final outcome as a phenotype in humans. Integrative genomic analysis which analyzes all genomic information together collectively at the same time is eventually necessary before it will be practical for individualized selection of analgesic drugs in the future.
After all those integrative genomic analyses are accomplished, the entire process should be repeated for different analgesic drugs and for other specific diseases. Integrative genomic analysis may be combined with epigenetic factors, environmental factors, and psychological factors to understand the whole process of pain medicine in human biology. In addition to the individual's genetic makeup, complex interactions between genes, proteins, environment and drugs also influence drug responses. Other physiological and cultural factors such as age, gender, ethnicity, smoking, and financial situation are other determinants of variable drug responses. Realization of complete individualized pain medicine will not be achieved until all of the factors can be collectively analyzed together. Unfortunately, methods for integrative genomic analyses are not fully available yet, mainly due to limitations of computing power as well as lack of appropriate algorithms that need to be developed. Because more than 1011 interaction models are possible for all pairs of SNPs only from ~500K SNP arrays, it requires enormous computing times. Therefore new, but arguable approaches for multiple interactions at the genome wide level have been proposed.[55–57] When these methods successfully apply to investigations of molecular-genetic mechanisms of pain, it may soon translate complex genomic information into individualized pain medicine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kim H, et al. Genetic influence on variability in human pain sensitivity associated with gender, ethnicity and psychological temperament. Pain. 2004;109:488–496. doi: 10.1016/j.pain.2004.02.027. [DOI] [PubMed] [Google Scholar]
- 2.Kupers R. Sex differences in pain: And now for something completely different. Behav Brain Sci. 1997;20 (3):455–456. [Google Scholar]
- 3.Robinson ME, et al. Altering gender role expectations: Effects on pain tolerance, pain threshold, and pain ratings. Journal of Pain. 2003;4 (5):284–288. doi: 10.1016/s1526-5900(03)00559-5. [DOI] [PubMed] [Google Scholar]
- 4.Green CR, et al. Race, age, and gender influences among clusters of African American and white patients with chronic pain. J Pain. 2004;5 (3):171–182. doi: 10.1016/j.jpain.2004.02.227. [DOI] [PubMed] [Google Scholar]
- 5.Hamann SR, et al. Interactions of "ultra-low" doses of naltrexone and morphine in mature and young male and female rats. Receptors Channels. 2004;10 (2):73–81. doi: 10.1080/10606820490464334. [DOI] [PubMed] [Google Scholar]
- 6.Wilson MA, et al. Sex differences in delta opioid receptor immunoreactivity in rat medial amygdala. Neurosci Lett. 2002;328 (2):160–164. doi: 10.1016/s0304-3940(02)00481-0. [DOI] [PubMed] [Google Scholar]
- 7.Lariviere WR, et al. Heritability of nociception. III. Genetic relationships among commonly used assays of nociception and hypersensitivity. Pain. 2002;97 (1–2):75–86. doi: 10.1016/s0304-3959(01)00492-4. [DOI] [PubMed] [Google Scholar]
- 8.Lee S. Personalized medicine and pharmacogenomics: ethical and social challenges. Personalized Medicine. 2005;2 (1):29–35. doi: 10.1517/17410541.2.1.29. [DOI] [PubMed] [Google Scholar]
- 9.Daar AS, Singer PA. Pharmacogenetics and geographical ancestry: implications for drug development and global health. Nat Rev Genet. 2005;6 (3):241–246. doi: 10.1038/nrg1559. [DOI] [PubMed] [Google Scholar]
- 10.Kahn J. Misreading race and genomics after BiDil. Nat Genet. 2005;37 (7):655–656. doi: 10.1038/ng0705-655. [DOI] [PubMed] [Google Scholar]
- 11.Gibbs RA, et al. The International HapMap Project. Nature. 2003;426 (6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 12.Bentley DR. Genomes for medicine. Nature. 2004;429 (6990):440–445. doi: 10.1038/nature02622. [DOI] [PubMed] [Google Scholar]
- 13.Tegeder I, et al. Reduced hyperalgesia in homozygous carriers of a GTP cyclohydrolase 1 haplotype. Eur J Pain. 2008;12 (8):1069–1077. doi: 10.1016/j.ejpain.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Tegeder I, et al. GTP cyclohydrolase and tetrahydrobiopterin regulate pain sensitivity and persistence. Nat Med. 2006;12 (11):1269–1277. doi: 10.1038/nm1490. [DOI] [PubMed] [Google Scholar]
- 15.Woodcock J, et al. Stimulating the development of mechanism–based, individualized pain therapies. Nat Rev Drug Discov. 2007;6 (9):703–710. doi: 10.1038/nrd2335. [DOI] [PubMed] [Google Scholar]
- 16.Kim H, Dionne RA. Lack of influence of GTP cyclohydrolase gene (GCH1) variations on pain sensitivity in humans. Mol Pain. 2007;3:6. doi: 10.1186/1744-8069-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zubieta JK, et al. COMT val158met genotype affects mu-opioid neurotransmitter responses to a pain stressor. Science. 2003;299 (5610):1240–1243. doi: 10.1126/science.1078546. [DOI] [PubMed] [Google Scholar]
- 18.Diatchenko L, et al. Genetic basis for individual variations in pain perception and the development of a chronic pain condition. Hum Mol Genet. 2005;14 (1):135–143. doi: 10.1093/hmg/ddi013. [DOI] [PubMed] [Google Scholar]
- 19.Kim H, et al. Genetic contributions to clinical pain and analgesia: Avoiding Pitfalls in Genetic Research. J Pain. 2009;10 (7):663–693. doi: 10.1016/j.jpain.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirschhorn JN, Daly M. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 21.Plenge RM, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis--a genomewide study. N Engl J Med. 2007;357 (12):1199–1209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu C, et al. Genome-wide association scan identifies candidate polymorphisms associated with differential response to anti-TNF treatment in rheumatoid arthritis. Mol Med. 2008;14 (9–10):575–581. doi: 10.2119/2008-00056.Liu. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seitz M, et al. The -308 tumour necrosis factor-alpha gene polymorphism predicts therapeutic response to TNFalpha-blockers in rheumatoid arthritis and spondyloarthritis patients. Rheumatology (Oxford) 2007;46 (1):93–96. doi: 10.1093/rheumatology/kel175. [DOI] [PubMed] [Google Scholar]
- 24.Guis S, et al. Influence of -308 A/G polymorphism in the tumor necrosis factor alpha gene on etanercept treatment in rheumatoid arthritis. Arthritis Rheum. 2007;57 (8):1426–1430. doi: 10.1002/art.23092. [DOI] [PubMed] [Google Scholar]
- 25.Criswell LA, et al. The influence of genetic variation in the HLA-DRB1 and LTA-TNF regions on the response to treatment of early rheumatoid arthritis with methotrexate or etanercept. Arthritis Rheum. 2004;50 (9):2750–2756. doi: 10.1002/art.20469. [DOI] [PubMed] [Google Scholar]
- 26.Danila MI, et al. Pharmacogenetics of etanercept in rheumatoid arthritis. Pharmacogenomics. 2008;9 (8):1011–1015. doi: 10.2217/14622416.9.8.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hashimoto S, et al. High-resolution analysis of the 5'-end transcriptome using a next generation DNA sequencer. PLoS ONE. 2009;4 (1):e4108. doi: 10.1371/journal.pone.0004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rothberg JM, Leamon JH. The development and impact of 454 sequencing. Nat Biotechnol. 2008;26 (10):1117–1124. doi: 10.1038/nbt1485. [DOI] [PubMed] [Google Scholar]
- 29.Quinn NL, et al. Assessing the feasibility of GS FLX Pyrosequencing for sequencing the Atlantic salmon genome. BMC Genomics. 2008;9:404. doi: 10.1186/1471-2164-9-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta PK. Single-molecule DNA sequencing technologies for future genomics research. Trends Biotechnol. 2008;26 (11):602–611. doi: 10.1016/j.tibtech.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 31.Gardiner SJ, Begg EJ. Pharmacogenetic testing for drug metabolizing enzymes: is it happening in practice? Pharmacogenet Genomics. 2005;15 (5):365–369. doi: 10.1097/01213011-200505000-00013. [DOI] [PubMed] [Google Scholar]
- 32.Samer CF, et al. Individualizing analgesic prescription Part I: pharmacogenetics of opioid analgesics. Personalized Medicine. 2006;3 (3):239–269. doi: 10.2217/17410541.3.3.239. [DOI] [PubMed] [Google Scholar]
- 33.Samer CF, et al. Individualizing analgesic prescription. Part II:pharmacogenetics of anti-inflammatory analgesics and co-analgesics. Personalized Medicine. 2006;3 (3):271–297. doi: 10.2217/17410541.3.3.271. [DOI] [PubMed] [Google Scholar]
- 34.Xie H, Frueh FW. Pharmacogenomics step toward personalized medicine. Personalized Medicine. 2005;2 (4):325–337. doi: 10.2217/17410541.2.4.325. [DOI] [PubMed] [Google Scholar]
- 35.de Leon J, et al. Clinical guidelines for psychiatrists for the use of pharmacogenetic testing for CYP450 2D6 and CYP450 2C19. Psychosomatics. 2006;47 (1):75–85. doi: 10.1176/appi.psy.47.1.75. [DOI] [PubMed] [Google Scholar]
- 36.Beyer A, et al. Effect of the A118G polymorphism on binding affinity, potency and agonist-mediated endocytosis, desensitization, and resensitization of the human mu-opioid receptor. J Neurochem. 2004;89 (3):553–560. doi: 10.1111/j.1471-4159.2004.02340.x. [DOI] [PubMed] [Google Scholar]
- 37.Rakvag TT, et al. The Val158Met polymorphism of the human catechol-O-methyltransferase (COMT) gene may influence morphine requirements in cancer pain patients. Pain. 2005;116 (1–2):73–78. doi: 10.1016/j.pain.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 38.Reyes-Gibby CC, et al. Exploring joint effects of genes and the clinical efficacy of morphine for cancer pain: OPRM1 and COMT gene. Pain. 2007;130 (1–2):25–30. doi: 10.1016/j.pain.2006.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim H, et al. Genetic polymorphisms in monoamine neurotransmitter systems show only weak association with acute post-surgical pain in humans. Mol Pain. 2006;2 (1):24. doi: 10.1186/1744-8069-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee Y, et al. Genetically mediated inter-individual variation in analgesic responses to COX inhibitory drugs. Clin Pharmacol Ther. 2006;79 (5):407–418. doi: 10.1016/j.clpt.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 41.Skarke C, et al. The cyclooxygenase 2 genetic variant -765G>C does not modulate the effects of celecoxib on prostaglandin E2 production. Clin Pharmacol Ther. 2006;80 (6):621–632. doi: 10.1016/j.clpt.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 42.Kim H, et al. Genome-wide association study of acute post-surgical pain in humans. Pharmacogenomics. 2009;10 (2):171–179. doi: 10.2217/14622416.10.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stranger BE, et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1 (6):e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharp FR, et al. The future of genomic profiling of neurological diseases using blood. Arch Neurol. 2006;63 (11):1529–1536. doi: 10.1001/archneur.63.11.1529. [DOI] [PubMed] [Google Scholar]
- 45.Dobbe E, et al. Gene-expression assays: new tools to individualize treatment of early-stage breast cancer. Am J Health Syst Pharm. 2008;65 (1):23–28. doi: 10.2146/ajhp060352. [DOI] [PubMed] [Google Scholar]
- 46.L'Esperance S, et al. Global gene expression analysis of early response to chemotherapy treatment in ovarian cancer spheroids. BMC Genomics. 2008;9:99. doi: 10.1186/1471-2164-9-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scholz J, Woolf CJ. Can we conquer pain? Nat Neurosci Suppl. 2002;5:1062–1067. doi: 10.1038/nn942. [DOI] [PubMed] [Google Scholar]
- 48.Ligthart L, et al. A genome-wide linkage scan provides evidence for both new and previously reported loci influencing common migraine. Am J Med Genet B Neuropsychiatr Genet. 2008;147B (7):1186–1195. doi: 10.1002/ajmg.b.30749. [DOI] [PubMed] [Google Scholar]
- 49.Buskila D, et al. The genetics of fibromyalgia syndrome. Pharmacogenomics. 2007;8 (1):67–74. doi: 10.2217/14622416.8.1.67. [DOI] [PubMed] [Google Scholar]
- 50.Diatchenko L, et al. Genetic architecture of human pain perception. Trends Genet. 2007;23 (12):605–613. doi: 10.1016/j.tig.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 51.Wang XM, et al. Upregulation of IL-6, IL-8 and CCL2 gene expression after acute inflammation: Correlation to clinical pain. Pain. 2009;142 (3):275–283. doi: 10.1016/j.pain.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang XM, et al. Rofecoxib modulates multiple gene expression pathways in a clinical model of acute inflammatory pain. Pain. 2007;128 (1–2):136–147. doi: 10.1016/j.pain.2006.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang XM, et al. Rofecoxib regulates the expression of genes related to the matrix metalloproteinase pathway in humans: implication for the adverse effects of cyclooxygenase-2 inhibitors. Clin Pharmacol Ther. 2006;79 (4):303–315. doi: 10.1016/j.clpt.2005.12.306. [DOI] [PubMed] [Google Scholar]
- 54.Lasky-Su J, et al. On the replication of genetic associations: timing can be everything! Am J Hum Genet. 2008;82(4):849–858. doi: 10.1016/j.ajhg.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park J, et al. Genome-wide analysis of haplotype interaction for the data from the North American Rheumatoid Arthritis Consortium. BMC Proc. 2009;3(Suppl 7):S34. doi: 10.1186/1753-6561-3-s7-s34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kovac IP, Dube MP. Different models and single-nucleotide polymorphisms signal the simulated weak gene-gene interaction for a quantitative trait using haplotype-based and mixed models testing. BMC Proc. 2009;3(Suppl 7):S77. doi: 10.1186/1753-6561-3-s7-s77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jung J, et al. Allelic-based gene-gene interaction associated with quantitative traits. Genet Epidemiol. 2009;33 (4):332–343. doi: 10.1002/gepi.20385. [DOI] [PMC free article] [PubMed] [Google Scholar]


