Abstract
In the molecular structure of the title hydrazide derivative, C27H24ClN3OS, the acetohydrazide group is approximately planar, with a maximum deviation of 0.017 (3) Å. The dihedral angle between the naphthylene system and the phenyl ring is 78.91 (18)°. The crystal structure is stabilized by one weak intermolecular C—H⋯O hydrogen bond and two aliphatic C—H⋯π hydrogen-bonding associations.
Related literature
For the applications and bioactivity of hydrazide derivatives, see: Feng et al. (2006 ▶); Yang et al. (2007 ▶); Kamal et al. (2007 ▶); Masunari & Tavares (2007 ▶); Rando et al. (2002 ▶). For bond-length data, see: Demir et al. (2006 ▶).
Experimental
Crystal data
C27H24ClN3OS
M r = 474.00
Monoclinic,

a = 7.498 (5) Å
b = 12.823 (5) Å
c = 24.924 (5) Å
β = 92.185 (5)°
V = 2394.6 (19) Å3
Z = 4
Mo Kα radiation
μ = 0.27 mm−1
T = 296 K
0.36 × 0.22 × 0.12 mm
Data collection
Stoe IPDS 2 CCD diffractometer
Absorption correction: integration (X-RED32; Stoe & Cie, 2002 ▶) T min = 0.797, T max = 0.961
12524 measured reflections
4206 independent reflections
1375 reflections with I > 2σ(I)
R int = 0.135
Refinement
R[F 2 > 2σ(F 2)] = 0.069
wR(F 2) = 0.156
S = 0.90
4206 reflections
298 parameters
H-atom parameters constrained
Δρmax = 0.14 e Å−3
Δρmin = −0.16 e Å−3
Data collection: X-AREA (Stoe & Cie, 2002 ▶); cell refinement: X-AREA; data reduction: X-RED32 (Stoe & Cie, 2002 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶) and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536811000183/zs2087sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811000183/zs2087Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg1 and Cg2 are the centroids of the C12/C13/S1/C14/N1 and C22–C27 rings, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C13—H13⋯O1i | 0.93 | 2.41 | 3.243 (7) | 148 |
| C8—H7A⋯Cg1ii | 0.96 | 2.94 | 3.748 (3) | 143 |
| C16—H16B⋯Cg2iii | 0.97 | 2.93 | 3.740 (8) | 142 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Acknowledgments
This study was supported financially by the Research Center of Ondokuz Mayıs University (Project No. F-461).
supplementary crystallographic information
Comment
Hydrazide derivatives of various compounds are very important units in host–guest chemistry due to their special arrangement of donor-acceptor functional groups (Feng et al., 2006; Yang et al., 2007). Hydrazine derivatives have also been associated with remarkable anticancer (Kamal et al., 2007), antibacterial (Masunari & Tavares, 2007) and tuberculostatic (Rando et al., 2002) activities. The title compound, the hydrazide derivative C27H24N3OClS (I) has been synthesized and its crystal structure is reported here.
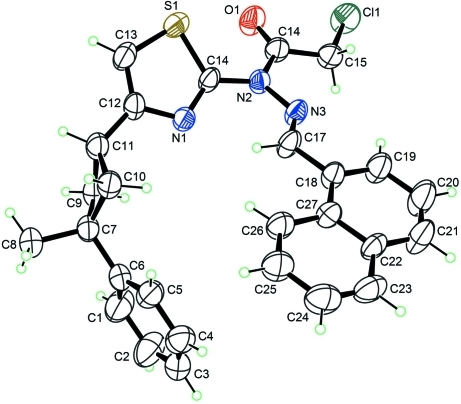
In the structure of (I) (Fig. 1) the phenyl and thiazole rings are cis-related with respect to the cyclobutane ring. The dihedral angle between the naphthylene fragment with the thiazole and phenyl rings are 35.76 (17)° and 78.91 (18)°, respectively. The cyclobutane ring is puckered, with a dihedral angle of 25.20 (5)° between the two three-membered halves of the ring. The dihedral angle between the acetohydrazide group and the thiazole ring is 32.28 (38)°. The C═O bond distance is 1.190 (6) Å comparing with a literature value of 1.187 (16) Å (Demir et al., 2006).
The crystal packing involves a weak intermolecular thiazole C13—H···O1 hydrogen bond (Table 1, Fig. 2) and intermoleculer C8—H···π (thiazole ring C12/C13/S1/C14/N1), C16—H···π (ring C22—C27) hydrogen-bonding associations (Fig. 3).
Experimental
The synthesis of the title compound was simply carried out in the following reaction (Fig. 4). A solution of 0.3975 gram (1 mmol) of N-[4-(3-methyl-3-phenyl-cyclobutyl)-thiazol-2-yl]-N-naphthalen -1-ylmethylenehydrazine was dissolved in 20 ml of dioxane containing 1 mmol triethylamine. To this solution, 90 µL (1 mmol) of chloroacetyl chloride solution in 20 ml 1,4-dioxane was added dropwise over a two hour period at room temperature with stirring. Mixture was stirred two hours more and then neutralized with 5% aqueous ammonia. The compound thus precipitated was filtered, washed with copious water and crystallized from ethanol, giving brown crystals (yield, 93%).
Refinement
The data was poor because of the weakly diffracting crystals which were not of good quality. Although a long exposure time (5 minute) was applied, the reflections were quite weak, resulting in a too low observed/unique reflection ratio. H atoms were positioned geometrically and treated using a riding model, fixing the bond lengths at 0.96, 0.97, 0.98 and 0.93 Å for CH3, CH2, CH and CH(aromatic), respectively. The displacement parameters of the H atoms were constrained with Uiso(H) = 1.2Ueq (aromatic, methylene or methine C) or 1.5Ueq (methyl C).
Figures
Fig. 1.
An ORTEP-3 (Farrugia, 1997) drawing of (I), showing the atomic numbering scheme. Displacement ellipsoids are drawn at the 30% probability level.
Fig. 2.
Part of the crystal structure of the title compound, showing the C—H···O hydrogen bonding. For clarity, only H atoms involved in hydrogen bonding have been included. For symmetry codes, see Table 1.
Fig. 3.
Part of the crystal structure of the title compound, showing the C—H···π interactions. For clarity, only H atoms involved in hydrogen bonding have been included. For symmetry codes, see Table 1.
Fig. 4.
Reaction scheme for the title compound.
Crystal data
| C27H24ClN3OS | F(000) = 992 |
| Mr = 474.00 | Dx = 1.315 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71069 Å |
| Hall symbol: -P 2ybc | Cell parameters from 9611 reflections |
| a = 7.498 (5) Å | θ = 1.6–27.3° |
| b = 12.823 (5) Å | µ = 0.27 mm−1 |
| c = 24.924 (5) Å | T = 296 K |
| β = 92.185 (5)° | Prism, pale brown |
| V = 2394.6 (19) Å3 | 0.36 × 0.22 × 0.12 mm |
| Z = 4 |
Data collection
| Stoe IPDS 2 CCD diffractometer | 4206 independent reflections |
| Radiation source: fine-focus sealed tube | 1375 reflections with I > 2σ(I) |
| plane graphite | Rint = 0.135 |
| rotation method scans | θmax = 25.0°, θmin = 1.6° |
| Absorption correction: integration (X-RED32; Stoe & Cie, 2002) | h = −8→7 |
| Tmin = 0.797, Tmax = 0.961 | k = −15→15 |
| 12524 measured reflections | l = −29→29 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.069 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.156 | H-atom parameters constrained |
| S = 0.90 | w = 1/[σ2(Fo2) + (0.0426P)2] where P = (Fo2 + 2Fc2)/3 |
| 4206 reflections | (Δ/σ)max < 0.001 |
| 298 parameters | Δρmax = 0.14 e Å−3 |
| 0 restraints | Δρmin = −0.16 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.1715 (3) | 0.89358 (11) | 0.38214 (7) | 0.1256 (8) | |
| S1 | 0.0165 (3) | 0.51892 (13) | 0.27709 (7) | 0.1101 (7) | |
| N1 | 0.2484 (6) | 0.4174 (3) | 0.33380 (17) | 0.0726 (14) | |
| N2 | 0.1837 (6) | 0.5845 (3) | 0.37084 (17) | 0.0721 (14) | |
| N3 | 0.2157 (6) | 0.5688 (3) | 0.42568 (17) | 0.0760 (15) | |
| O1 | 0.1257 (7) | 0.7154 (3) | 0.31249 (16) | 0.1143 (18) | |
| C1 | 0.7867 (12) | 0.1538 (6) | 0.3653 (3) | 0.127 (3) | |
| H1 | 0.8339 | 0.1724 | 0.3327 | 0.153* | |
| C2 | 0.8984 (11) | 0.1442 (7) | 0.4126 (4) | 0.140 (3) | |
| H2 | 1.0198 | 0.1577 | 0.4106 | 0.168* | |
| C3 | 0.8345 (13) | 0.1166 (6) | 0.4591 (4) | 0.110 (3) | |
| H3 | 0.9116 | 0.1100 | 0.4891 | 0.132* | |
| C4 | 0.6583 (13) | 0.0977 (5) | 0.4638 (3) | 0.103 (2) | |
| H4 | 0.6128 | 0.0789 | 0.4966 | 0.123* | |
| C5 | 0.5461 (9) | 0.1073 (4) | 0.4178 (3) | 0.091 (2) | |
| H5 | 0.4248 | 0.0943 | 0.4208 | 0.109* | |
| C6 | 0.6064 (11) | 0.1348 (4) | 0.3692 (3) | 0.0798 (18) | |
| C8 | 0.4959 (9) | 0.0466 (4) | 0.2850 (2) | 0.103 (2) | |
| H7A | 0.6192 | 0.0327 | 0.2789 | 0.155* | |
| H7B | 0.4325 | 0.0570 | 0.2513 | 0.155* | |
| H7C | 0.4452 | −0.0114 | 0.3034 | 0.155* | |
| C7 | 0.4812 (9) | 0.1451 (4) | 0.3194 (2) | 0.0763 (18) | |
| C9 | 0.4947 (9) | 0.2463 (4) | 0.2867 (2) | 0.0884 (19) | |
| H9A | 0.5475 | 0.2368 | 0.2521 | 0.106* | |
| H9B | 0.5516 | 0.3033 | 0.3064 | 0.106* | |
| C10 | 0.2863 (9) | 0.1759 (4) | 0.3310 (2) | 0.0788 (18) | |
| H10A | 0.2016 | 0.1192 | 0.3261 | 0.095* | |
| H10B | 0.2737 | 0.2093 | 0.3656 | 0.095* | |
| C11 | 0.2862 (9) | 0.2530 (4) | 0.2835 (2) | 0.0811 (18) | |
| H11 | 0.2394 | 0.2187 | 0.2508 | 0.097* | |
| C12 | 0.1991 (8) | 0.3552 (4) | 0.2899 (2) | 0.0748 (18) | |
| C13 | 0.0734 (9) | 0.3984 (4) | 0.2568 (2) | 0.097 (2) | |
| H13 | 0.0241 | 0.3657 | 0.2264 | 0.117* | |
| C14 | 0.1626 (8) | 0.5056 (4) | 0.3315 (2) | 0.0730 (17) | |
| C15 | 0.1622 (8) | 0.6881 (4) | 0.3572 (2) | 0.082 (2) | |
| C16 | 0.1878 (8) | 0.7630 (4) | 0.4043 (2) | 0.0866 (19) | |
| H16A | 0.3040 | 0.7515 | 0.4217 | 0.104* | |
| H16B | 0.0976 | 0.7497 | 0.4303 | 0.104* | |
| C17 | 0.2259 (8) | 0.4801 (4) | 0.4471 (2) | 0.0809 (19) | |
| H17 | 0.2139 | 0.4198 | 0.4265 | 0.097* | |
| C18 | 0.2573 (8) | 0.4748 (4) | 0.5056 (2) | 0.0760 (17) | |
| C19 | 0.2719 (9) | 0.5651 (5) | 0.5343 (3) | 0.108 (3) | |
| H19 | 0.2600 | 0.6288 | 0.5167 | 0.129* | |
| C20 | 0.3046 (11) | 0.5631 (6) | 0.5900 (3) | 0.135 (3) | |
| H20 | 0.3144 | 0.6255 | 0.6089 | 0.162* | |
| C21 | 0.3222 (10) | 0.4714 (7) | 0.6168 (3) | 0.123 (3) | |
| H21 | 0.3454 | 0.4713 | 0.6537 | 0.148* | |
| C22 | 0.3057 (8) | 0.3774 (5) | 0.5892 (2) | 0.0807 (18) | |
| C23 | 0.2711 (8) | 0.3776 (4) | 0.5329 (2) | 0.0745 (17) | |
| C24 | 0.2565 (8) | 0.2805 (4) | 0.5059 (2) | 0.092 (2) | |
| H24 | 0.2392 | 0.2786 | 0.4687 | 0.111* | |
| C25 | 0.2679 (10) | 0.1900 (5) | 0.5343 (3) | 0.106 (2) | |
| H25 | 0.2522 | 0.1270 | 0.5163 | 0.127* | |
| C26 | 0.3021 (10) | 0.1887 (6) | 0.5893 (3) | 0.115 (3) | |
| H26 | 0.3126 | 0.1257 | 0.6076 | 0.138* | |
| C27 | 0.3200 (9) | 0.2792 (7) | 0.6158 (3) | 0.106 (2) | |
| H27 | 0.3424 | 0.2779 | 0.6527 | 0.127* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.187 (2) | 0.0619 (9) | 0.1246 (15) | −0.0055 (11) | −0.0343 (14) | 0.0081 (10) |
| S1 | 0.1371 (17) | 0.1007 (12) | 0.0888 (12) | 0.0283 (12) | −0.0443 (12) | −0.0148 (10) |
| N1 | 0.097 (4) | 0.059 (3) | 0.061 (3) | 0.004 (3) | −0.011 (3) | −0.002 (2) |
| N2 | 0.106 (4) | 0.058 (3) | 0.052 (3) | 0.011 (3) | −0.002 (3) | 0.003 (2) |
| N3 | 0.107 (4) | 0.069 (3) | 0.051 (3) | −0.006 (3) | −0.009 (3) | 0.004 (2) |
| O1 | 0.190 (5) | 0.081 (3) | 0.069 (3) | 0.020 (3) | −0.018 (3) | 0.014 (2) |
| C1 | 0.104 (7) | 0.186 (8) | 0.093 (6) | −0.001 (6) | 0.000 (6) | −0.024 (5) |
| C2 | 0.092 (7) | 0.232 (10) | 0.097 (6) | −0.001 (6) | 0.000 (6) | −0.026 (7) |
| C3 | 0.111 (8) | 0.120 (6) | 0.097 (6) | 0.024 (6) | −0.013 (6) | −0.029 (5) |
| C4 | 0.123 (7) | 0.086 (4) | 0.097 (6) | −0.002 (5) | −0.018 (5) | 0.005 (4) |
| C5 | 0.112 (6) | 0.079 (4) | 0.080 (5) | −0.007 (4) | −0.014 (5) | 0.003 (4) |
| C6 | 0.096 (6) | 0.067 (4) | 0.077 (5) | 0.006 (4) | 0.001 (4) | −0.017 (4) |
| C8 | 0.129 (6) | 0.091 (4) | 0.089 (5) | 0.017 (4) | −0.008 (4) | −0.029 (4) |
| C7 | 0.094 (6) | 0.068 (4) | 0.067 (4) | 0.004 (3) | 0.000 (4) | −0.011 (3) |
| C9 | 0.106 (6) | 0.083 (4) | 0.077 (4) | −0.005 (4) | 0.010 (4) | −0.016 (4) |
| C10 | 0.099 (6) | 0.071 (3) | 0.067 (4) | −0.004 (4) | 0.002 (4) | −0.003 (3) |
| C11 | 0.110 (6) | 0.064 (4) | 0.069 (4) | 0.000 (4) | −0.005 (4) | −0.007 (3) |
| C12 | 0.105 (5) | 0.059 (3) | 0.060 (4) | −0.006 (3) | 0.002 (4) | −0.006 (3) |
| C13 | 0.128 (6) | 0.096 (4) | 0.067 (4) | 0.004 (4) | −0.023 (4) | −0.013 (4) |
| C14 | 0.108 (5) | 0.063 (4) | 0.046 (3) | −0.003 (4) | −0.013 (3) | 0.003 (3) |
| C15 | 0.112 (6) | 0.064 (4) | 0.069 (4) | 0.007 (3) | −0.008 (4) | 0.008 (3) |
| C16 | 0.114 (6) | 0.061 (3) | 0.084 (4) | 0.006 (3) | −0.010 (4) | 0.008 (3) |
| C17 | 0.126 (6) | 0.067 (4) | 0.048 (4) | −0.013 (4) | −0.012 (3) | 0.011 (3) |
| C18 | 0.100 (5) | 0.067 (4) | 0.060 (4) | −0.009 (3) | −0.005 (3) | −0.003 (3) |
| C19 | 0.166 (8) | 0.077 (4) | 0.077 (5) | −0.001 (4) | −0.025 (5) | −0.007 (4) |
| C20 | 0.198 (9) | 0.118 (6) | 0.087 (6) | −0.016 (6) | −0.029 (6) | −0.023 (5) |
| C21 | 0.163 (8) | 0.145 (7) | 0.061 (5) | −0.014 (6) | −0.021 (5) | −0.007 (5) |
| C22 | 0.089 (5) | 0.107 (5) | 0.046 (4) | 0.000 (4) | −0.003 (3) | 0.012 (4) |
| C23 | 0.082 (5) | 0.079 (4) | 0.063 (4) | −0.006 (3) | −0.002 (3) | 0.006 (3) |
| C24 | 0.126 (6) | 0.078 (4) | 0.071 (4) | 0.008 (4) | −0.010 (4) | 0.012 (4) |
| C25 | 0.145 (7) | 0.082 (5) | 0.091 (5) | 0.006 (4) | −0.003 (5) | 0.021 (4) |
| C26 | 0.128 (7) | 0.108 (6) | 0.108 (7) | 0.008 (5) | 0.005 (5) | 0.037 (5) |
| C27 | 0.107 (6) | 0.138 (6) | 0.073 (5) | 0.010 (5) | −0.006 (4) | 0.032 (5) |
Geometric parameters (Å, °)
| Cl1—C16 | 1.766 (5) | C10—C11 | 1.541 (7) |
| S1—C13 | 1.686 (6) | C10—H10A | 0.9700 |
| S1—C14 | 1.720 (5) | C10—H10B | 0.9700 |
| N1—C14 | 1.301 (6) | C11—C12 | 1.476 (7) |
| N1—C12 | 1.392 (6) | C11—H11 | 0.9800 |
| N2—C15 | 1.379 (6) | C12—C13 | 1.349 (7) |
| N2—N3 | 1.393 (5) | C13—H13 | 0.9300 |
| N2—C14 | 1.412 (6) | C15—C16 | 1.523 (7) |
| N3—C17 | 1.257 (5) | C16—H16A | 0.9700 |
| O1—C15 | 1.190 (6) | C16—H16B | 0.9700 |
| C1—C6 | 1.380 (9) | C17—C18 | 1.470 (7) |
| C1—C2 | 1.425 (9) | C17—H17 | 0.9300 |
| C1—H1 | 0.9300 | C18—C19 | 1.364 (7) |
| C2—C3 | 1.321 (9) | C18—C23 | 1.422 (7) |
| C2—H2 | 0.9300 | C19—C20 | 1.403 (8) |
| C3—C4 | 1.353 (9) | C19—H19 | 0.9300 |
| C3—H3 | 0.9300 | C20—C21 | 1.356 (8) |
| C4—C5 | 1.401 (8) | C20—H20 | 0.9300 |
| C4—H4 | 0.9300 | C21—C22 | 1.391 (8) |
| C5—C6 | 1.356 (8) | C21—H21 | 0.9300 |
| C5—H5 | 0.9300 | C22—C23 | 1.417 (7) |
| C6—C7 | 1.532 (8) | C22—C27 | 1.425 (8) |
| C8—C7 | 1.532 (6) | C23—C24 | 1.418 (7) |
| C8—H7A | 0.9600 | C24—C25 | 1.361 (7) |
| C8—H7B | 0.9600 | C24—H24 | 0.9300 |
| C8—H7C | 0.9600 | C25—C26 | 1.384 (8) |
| C7—C9 | 1.539 (7) | C25—H25 | 0.9300 |
| C7—C10 | 1.552 (7) | C26—C27 | 1.340 (8) |
| C9—C11 | 1.565 (8) | C26—H26 | 0.9300 |
| C9—H9A | 0.9700 | C27—H27 | 0.9300 |
| C9—H9B | 0.9700 | ||
| C13—S1—C14 | 89.2 (3) | C9—C11—H11 | 110.0 |
| C14—N1—C12 | 110.4 (4) | C13—C12—N1 | 113.8 (5) |
| C15—N2—N3 | 113.2 (4) | C13—C12—C11 | 126.9 (5) |
| C15—N2—C14 | 120.6 (4) | N1—C12—C11 | 119.3 (5) |
| N3—N2—C14 | 126.0 (4) | C12—C13—S1 | 111.8 (4) |
| C17—N3—N2 | 123.5 (4) | C12—C13—H13 | 124.1 |
| C6—C1—C2 | 118.2 (8) | S1—C13—H13 | 124.1 |
| C6—C1—H1 | 120.9 | N1—C14—N2 | 123.4 (4) |
| C2—C1—H1 | 120.9 | N1—C14—S1 | 114.8 (4) |
| C3—C2—C1 | 121.9 (8) | N2—C14—S1 | 121.8 (4) |
| C3—C2—H2 | 119.1 | O1—C15—N2 | 122.4 (5) |
| C1—C2—H2 | 119.1 | O1—C15—C16 | 123.6 (5) |
| C2—C3—C4 | 120.8 (8) | N2—C15—C16 | 114.1 (5) |
| C2—C3—H3 | 119.6 | C15—C16—Cl1 | 110.6 (4) |
| C4—C3—H3 | 119.6 | C15—C16—H16A | 109.5 |
| C3—C4—C5 | 118.2 (8) | Cl1—C16—H16A | 109.5 |
| C3—C4—H4 | 120.9 | C15—C16—H16B | 109.5 |
| C5—C4—H4 | 120.9 | Cl1—C16—H16B | 109.5 |
| C6—C5—C4 | 122.9 (7) | H16A—C16—H16B | 108.1 |
| C6—C5—H5 | 118.5 | N3—C17—C18 | 117.9 (5) |
| C4—C5—H5 | 118.5 | N3—C17—H17 | 121.0 |
| C5—C6—C1 | 118.0 (6) | C18—C17—H17 | 121.0 |
| C5—C6—C7 | 122.0 (7) | C19—C18—C23 | 119.4 (5) |
| C1—C6—C7 | 119.9 (7) | C19—C18—C17 | 119.1 (5) |
| C7—C8—H7A | 109.5 | C23—C18—C17 | 121.5 (5) |
| C7—C8—H7B | 109.5 | C18—C19—C20 | 120.8 (6) |
| H7A—C8—H7B | 109.5 | C18—C19—H19 | 119.6 |
| C7—C8—H7C | 109.5 | C20—C19—H19 | 119.6 |
| H7A—C8—H7C | 109.5 | C21—C20—C19 | 120.9 (6) |
| H7B—C8—H7C | 109.5 | C21—C20—H20 | 119.6 |
| C6—C7—C8 | 109.1 (5) | C19—C20—H20 | 119.6 |
| C6—C7—C9 | 116.8 (5) | C20—C21—C22 | 120.2 (6) |
| C8—C7—C9 | 113.0 (5) | C20—C21—H21 | 119.9 |
| C6—C7—C10 | 115.1 (5) | C22—C21—H21 | 119.9 |
| C8—C7—C10 | 113.7 (5) | C21—C22—C23 | 119.9 (6) |
| C9—C7—C10 | 88.0 (4) | C21—C22—C27 | 122.1 (6) |
| C7—C9—C11 | 89.2 (5) | C23—C22—C27 | 118.0 (6) |
| C7—C9—H9A | 113.8 | C22—C23—C24 | 118.5 (5) |
| C11—C9—H9A | 113.8 | C22—C23—C18 | 118.9 (5) |
| C7—C9—H9B | 113.8 | C24—C23—C18 | 122.7 (5) |
| C11—C9—H9B | 113.8 | C25—C24—C23 | 119.9 (5) |
| H9A—C9—H9B | 111.0 | C25—C24—H24 | 120.0 |
| C11—C10—C7 | 89.7 (5) | C23—C24—H24 | 120.0 |
| C11—C10—H10A | 113.7 | C24—C25—C26 | 122.2 (7) |
| C7—C10—H10A | 113.7 | C24—C25—H25 | 118.9 |
| C11—C10—H10B | 113.7 | C26—C25—H25 | 118.9 |
| C7—C10—H10B | 113.7 | C27—C26—C25 | 119.2 (7) |
| H10A—C10—H10B | 110.9 | C27—C26—H26 | 120.4 |
| C12—C11—C10 | 118.3 (5) | C25—C26—H26 | 120.4 |
| C12—C11—C9 | 119.2 (5) | C26—C27—C22 | 122.2 (6) |
| C10—C11—C9 | 87.4 (4) | C26—C27—H27 | 118.9 |
| C12—C11—H11 | 110.0 | C22—C27—H27 | 118.9 |
| C10—C11—H11 | 110.0 | ||
| C15—N2—N3—C17 | 174.5 (6) | C15—N2—C14—N1 | 149.7 (6) |
| C14—N2—N3—C17 | −1.6 (9) | N3—N2—C14—N1 | −34.4 (9) |
| C6—C1—C2—C3 | 0.9 (12) | C15—N2—C14—S1 | −31.5 (8) |
| C1—C2—C3—C4 | −1.0 (13) | N3—N2—C14—S1 | 144.4 (5) |
| C2—C3—C4—C5 | 0.6 (12) | C13—S1—C14—N1 | 0.5 (5) |
| C3—C4—C5—C6 | −0.2 (10) | C13—S1—C14—N2 | −178.4 (5) |
| C4—C5—C6—C1 | 0.1 (10) | N3—N2—C15—O1 | −176.4 (6) |
| C4—C5—C6—C7 | −179.9 (5) | C14—N2—C15—O1 | 0.0 (10) |
| C2—C1—C6—C5 | −0.4 (10) | N3—N2—C15—C16 | 3.1 (7) |
| C2—C1—C6—C7 | 179.5 (6) | C14—N2—C15—C16 | 179.4 (5) |
| C5—C6—C7—C8 | −100.5 (7) | O1—C15—C16—Cl1 | −4.2 (9) |
| C1—C6—C7—C8 | 79.6 (7) | N2—C15—C16—Cl1 | 176.3 (4) |
| C5—C6—C7—C9 | 129.9 (6) | N2—N3—C17—C18 | −179.0 (5) |
| C1—C6—C7—C9 | −50.1 (8) | N3—C17—C18—C19 | 0.8 (9) |
| C5—C6—C7—C10 | 28.7 (8) | N3—C17—C18—C23 | −179.8 (6) |
| C1—C6—C7—C10 | −151.2 (6) | C23—C18—C19—C20 | 1.6 (10) |
| C6—C7—C9—C11 | −135.0 (6) | C17—C18—C19—C20 | −179.0 (7) |
| C8—C7—C9—C11 | 97.2 (5) | C18—C19—C20—C21 | −0.1 (13) |
| C10—C7—C9—C11 | −17.8 (4) | C19—C20—C21—C22 | −0.8 (13) |
| C6—C7—C10—C11 | 136.9 (5) | C20—C21—C22—C23 | 0.2 (11) |
| C8—C7—C10—C11 | −96.2 (5) | C20—C21—C22—C27 | −178.8 (8) |
| C9—C7—C10—C11 | 18.1 (4) | C21—C22—C23—C24 | 179.5 (6) |
| C7—C10—C11—C12 | −139.8 (5) | C27—C22—C23—C24 | −1.4 (9) |
| C7—C10—C11—C9 | −17.8 (4) | C21—C22—C23—C18 | 1.2 (9) |
| C7—C9—C11—C12 | 139.1 (5) | C27—C22—C23—C18 | −179.7 (6) |
| C7—C9—C11—C10 | 17.9 (4) | C19—C18—C23—C22 | −2.1 (9) |
| C14—N1—C12—C13 | −1.6 (7) | C17—C18—C23—C22 | 178.5 (6) |
| C14—N1—C12—C11 | 177.6 (5) | C19—C18—C23—C24 | 179.7 (6) |
| C10—C11—C12—C13 | −127.6 (7) | C17—C18—C23—C24 | 0.3 (9) |
| C9—C11—C12—C13 | 128.4 (6) | C22—C23—C24—C25 | 2.9 (9) |
| C10—C11—C12—N1 | 53.3 (8) | C18—C23—C24—C25 | −178.9 (6) |
| C9—C11—C12—N1 | −50.8 (7) | C23—C24—C25—C26 | −3.2 (11) |
| N1—C12—C13—S1 | 2.0 (7) | C24—C25—C26—C27 | 1.8 (12) |
| C11—C12—C13—S1 | −177.2 (5) | C25—C26—C27—C22 | −0.3 (12) |
| C14—S1—C13—C12 | −1.4 (5) | C21—C22—C27—C26 | 179.2 (8) |
| C12—N1—C14—N2 | 179.4 (5) | C23—C22—C27—C26 | 0.2 (10) |
| C12—N1—C14—S1 | 0.5 (6) |
Hydrogen-bond geometry (Å, °)
| Cg1 and Cg2 are the centroids of the C12/C13/S1/C14/N1 and C22–C27 rings, respectively. |
| D—H···A | D—H | H···A | D···A | D—H···A |
| C13—H13···O1i | 0.93 | 2.41 | 3.243 (7) | 148. |
| C8—H7A···Cg1ii | 0.96 | 2.94 | 3.748 (3) | 143 |
| C16—H16B···Cg2iii | 0.97 | 2.93 | 3.740 (8) | 142 |
Symmetry codes: (i) −x, y−1/2, −z+1/2; (ii) −x+1, y−1/2, −z+1/2; (iii) −x, −y+1, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: ZS2087).
References
- Demir, S., Dinçer, M., Çukurovalı, A. & Yılmaz, I. (2006). Acta Cryst. E62, o298–o299.
- Farrugia, L. J. (1997). J. Appl. Cryst. 30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst. 32, 837–838.
- Feng, D.-J., Wang, P., Li, X.-Q. & Li, Z.-T. (2006). Chin. J. Chem. 24, 1200–1208.
- Kamal, A., Khan, N. A., Reddy, K. S. & Rohini, K. (2007). Bioorg. Med. Chem. 15, 1004–1013. [DOI] [PubMed]
- Masunari, A. & Tavares, L. C. (2007). Bioorg. Med. Chem. 15, 4229–4236. [DOI] [PubMed]
- Rando, D. G., Sato, D. N., Siqueira, L., Malvezzi, A., Leite, C. Q. F., do Amaral, A. T., Ferreira, E. I. & Tavares, L. C. (2002). Bioorg. Med. Chem. 10, 557–560. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Stoe & Cie (2002). X-AREA and X-RED32 Stoe & Cie, Darmstadt, Germany.
- Yang, Y., Hu, H.-Y. & Chen, C.-F. (2007). Tetrahedron Lett. 48, 3505–3509.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536811000183/zs2087sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811000183/zs2087Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report