Abstract
In the title compound, C29H24N4O2S, intramolecular O—H⋯N hydrogen bonding influences the molecular conformation; the naphthol system and triazole ring form a dihedral angle of 3.88 (7)°. In the crystal, π–π interactions between the five- and six-membered rings of neighbouring molecules [centroid–centroid distances = 3.541 (3) and 3.711 (3) Å] consolidate the crystal packing.
Related literature
For details of the pharmacological properties of Mannich bases, see: Joshi et al. (2004 ▶); Ferlin et al. (2002 ▶); Holla et al. (2003 ▶). For their application in the polymer indusry, see: Negm et al. (2005 ▶). For standard bond lengths, see: Allen et al. (1987 ▶).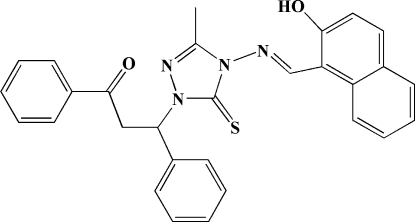
Experimental
Crystal data
C29H24N4O2S
M r = 492.58
Monoclinic,

a = 7.8192 (16) Å
b = 20.248 (4) Å
c = 15.360 (3) Å
β = 94.69 (3)°
V = 2423.7 (8) Å3
Z = 4
Mo Kα radiation
μ = 0.17 mm−1
T = 153 K
0.18 × 0.16 × 0.10 mm
Data collection
Rigaku Saturn CCD area-detector diffractometer
Absorption correction: multi-scan (CrystalClear; Rigaku/MSC, 2005 ▶) T min = 0.970, T max = 0.983
23596 measured reflections
5746 independent reflections
4491 reflections with I > 2σ(I)
R int = 0.048
Refinement
R[F 2 > 2σ(F 2)] = 0.054
wR(F 2) = 0.127
S = 0.96
5746 reflections
331 parameters
3 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.31 e Å−3
Δρmin = −0.30 e Å−3
Data collection: CrystalClear (Rigaku/MSC, 2005 ▶); cell refinement: CrystalClear; data reduction: CrystalClear; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810052979/cv5020sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810052979/cv5020Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯N1 | 0.89 (3) | 1.83 (3) | 2.610 (2) | 145 (2) |
Acknowledgments
We gratefully acknowledge the financial support of the Science Fund for Young Scholars of Heilongjiang Province of China under grant No. QC2009C61.
supplementary crystallographic information
Comment
Mannich bases have been reported as potential biological agents and received considerable attention due to their pharmacological properties - antitubercular (Joshi et al., 2004), vasorelaxing (Ferlin et al., 2002), anticancer (Holla et al., 2003), and due to their applications in the polymer industry as paints and surface active agents (Negm et al., 2005). Herein we report the synthesis and crystal structure of the title compound, (I).
In (I) (Fig. 1), the bond lengths and angles are found to have normal values (Allen et al., 1987). An intramolecular O—H···N hydrogen bond results in the formation of a planar (r.m.s. deviation = 0.0262 (2) Å) six-membered ring (Table 2) and influences the molecular conformation - the naphthol system and triazole ring form a dihedral angle of 3.88 (7)°. Two phenyl rings are located on the two sides of the triazole ring. They form a dihedral angle of 34.2 (3)°.
In the crystal structure, π-π interactions (Table 1) between the five- and six-mebered rings from the neighbouring molecules consolidate the crystal packing.
Experimental
The title compound was synthesized by the reaction of the chalcone (2.0 mmol) with its corresponding Schiff base, which was in turn obtained by refluxing 4-amino-1-methyl-4H-1,2,4-triazole-5-thiol (2.0 mmol), 2-hydroxynaphthalene-1 -carbaldehyde (2.0 mmol) in ethanol. A mixture of Schiff base and chalcone in ethanol was stirring for 24 h.The reaction progress was monitored via TLC. The resulting precipitate was filtered off, washed with cold ethanol, dried and purified to give the target product as colorless solid in 84% yield. Crystals of (I) suitable for single-crystal X-ray analysis were grown by slow evaporation of a solution in chloroform-ethanol (1:1).
Refinement
The H atom attached to O atom was located on a difference map and isotropically refined. Other H atoms were positioned geometrically (C—H = 0.95–0.99 Å) and refined as riding, with Uiso(H) = 1.2-1.5 Ueq of the parent atom.
Figures
Fig. 1.
View of the molecule of (I) showing the atom-labelling scheme. Displacement ellipsoids are drawn at the 60% probability level. Dashed line denotes the hydrogen bond.
Crystal data
| C29H24N4O2S | F(000) = 1032 |
| Mr = 492.58 | Dx = 1.350 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 6032 reflections |
| a = 7.8192 (16) Å | θ = 1.7–27.9° |
| b = 20.248 (4) Å | µ = 0.17 mm−1 |
| c = 15.360 (3) Å | T = 153 K |
| β = 94.69 (3)° | Prism, colourless |
| V = 2423.7 (8) Å3 | 0.18 × 0.16 × 0.10 mm |
| Z = 4 |
Data collection
| Rigaku Saturn CCD area-detector diffractometer | 5746 independent reflections |
| Radiation source: rotating anode | 4491 reflections with I > 2σ(I) |
| multilayer | Rint = 0.048 |
| Detector resolution: 7.31 pixels mm-1 | θmax = 27.9°, θmin = 1.7° |
| φ and ω scans | h = −10→10 |
| Absorption correction: multi-scan (CrystalClear; Rigaku/MSC, 2005) | k = −26→26 |
| Tmin = 0.970, Tmax = 0.983 | l = −17→20 |
| 23596 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.054 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.127 | w = 1/[σ2(Fo2) + (0.06P)2 + 1.1394P] where P = (Fo2 + 2Fc2)/3 |
| S = 0.96 | (Δ/σ)max = 0.001 |
| 5746 reflections | Δρmax = 0.31 e Å−3 |
| 331 parameters | Δρmin = −0.30 e Å−3 |
| 3 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0261 (17) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.28857 (5) | 0.20831 (2) | −0.04504 (3) | 0.02895 (15) | |
| O1 | 0.77192 (16) | −0.01503 (7) | −0.07589 (10) | 0.0358 (3) | |
| H1 | 0.760 (3) | 0.0246 (14) | −0.0519 (16) | 0.071 (9)* | |
| O2 | 0.64680 (17) | 0.36195 (7) | −0.03561 (8) | 0.0396 (4) | |
| N1 | 0.61510 (18) | 0.09480 (7) | −0.04258 (9) | 0.0249 (3) | |
| N2 | 0.60991 (17) | 0.15359 (7) | 0.00352 (9) | 0.0210 (3) | |
| N3 | 0.55703 (17) | 0.24391 (7) | 0.06788 (9) | 0.0227 (3) | |
| N4 | 0.72369 (17) | 0.22717 (7) | 0.09686 (9) | 0.0249 (3) | |
| C1 | 0.4776 (2) | 0.07206 (9) | −0.08296 (11) | 0.0246 (4) | |
| H1A | 0.3737 | 0.0963 | −0.0827 | 0.030* | |
| C2 | 0.4795 (2) | 0.00970 (8) | −0.12901 (11) | 0.0237 (4) | |
| C3 | 0.3266 (2) | −0.01202 (9) | −0.17983 (11) | 0.0252 (4) | |
| C4 | 0.1714 (2) | 0.02452 (9) | −0.18680 (12) | 0.0288 (4) | |
| H4 | 0.1656 | 0.0655 | −0.1570 | 0.035* | |
| C5 | 0.0298 (3) | 0.00162 (10) | −0.23590 (12) | 0.0351 (5) | |
| H5 | −0.0730 | 0.0269 | −0.2393 | 0.042* | |
| C6 | 0.0336 (3) | −0.05847 (11) | −0.28135 (12) | 0.0401 (5) | |
| H6 | −0.0655 | −0.0736 | −0.3155 | 0.048* | |
| C7 | 0.1806 (3) | −0.09494 (10) | −0.27597 (12) | 0.0374 (5) | |
| H7 | 0.1831 | −0.1357 | −0.3065 | 0.045* | |
| C8 | 0.3299 (2) | −0.07301 (9) | −0.22563 (11) | 0.0310 (4) | |
| C9 | 0.4830 (3) | −0.11108 (9) | −0.21919 (12) | 0.0350 (5) | |
| H9 | 0.4860 | −0.1515 | −0.2505 | 0.042* | |
| C10 | 0.6248 (3) | −0.09121 (9) | −0.16955 (13) | 0.0340 (5) | |
| H10 | 0.7247 | −0.1181 | −0.1654 | 0.041* | |
| C11 | 0.6248 (2) | −0.03069 (9) | −0.12387 (11) | 0.0280 (4) | |
| C12 | 0.4817 (2) | 0.20104 (8) | 0.00885 (11) | 0.0218 (4) | |
| C13 | 0.7525 (2) | 0.17231 (9) | 0.05596 (11) | 0.0228 (4) | |
| C14 | 0.9162 (2) | 0.13542 (9) | 0.06260 (12) | 0.0287 (4) | |
| H14A | 0.9996 | 0.1582 | 0.1032 | 0.043* | |
| H14B | 0.9607 | 0.1329 | 0.0049 | 0.043* | |
| H14C | 0.8968 | 0.0907 | 0.0841 | 0.043* | |
| C15 | 0.4795 (2) | 0.30482 (8) | 0.09933 (11) | 0.0250 (4) | |
| H15 | 0.3971 | 0.3220 | 0.0515 | 0.030* | |
| C16 | 0.3790 (2) | 0.29017 (8) | 0.17815 (11) | 0.0246 (4) | |
| C17 | 0.2021 (2) | 0.28203 (9) | 0.16602 (12) | 0.0304 (4) | |
| H17 | 0.1450 | 0.2880 | 0.1096 | 0.037* | |
| C18 | 0.1083 (2) | 0.26531 (10) | 0.23552 (13) | 0.0343 (5) | |
| H18 | −0.0125 | 0.2598 | 0.2264 | 0.041* | |
| C19 | 0.1902 (2) | 0.25662 (10) | 0.31833 (13) | 0.0350 (5) | |
| H19 | 0.1264 | 0.2444 | 0.3658 | 0.042* | |
| C20 | 0.3654 (2) | 0.26592 (10) | 0.33109 (12) | 0.0329 (4) | |
| H20 | 0.4217 | 0.2607 | 0.3878 | 0.039* | |
| C21 | 0.4598 (2) | 0.28280 (9) | 0.26172 (12) | 0.0297 (4) | |
| H21 | 0.5802 | 0.2894 | 0.2713 | 0.036* | |
| C22 | 0.6197 (2) | 0.35606 (9) | 0.11750 (11) | 0.0282 (4) | |
| H22A | 0.5679 | 0.3971 | 0.1386 | 0.034* | |
| H22B | 0.7029 | 0.3396 | 0.1645 | 0.034* | |
| C23 | 0.7142 (2) | 0.37200 (9) | 0.03791 (12) | 0.0292 (4) | |
| C24 | 0.8901 (2) | 0.39992 (8) | 0.05155 (11) | 0.0267 (4) | |
| C25 | 0.9478 (2) | 0.42988 (9) | 0.13012 (12) | 0.0313 (4) | |
| H25 | 0.8764 | 0.4305 | 0.1773 | 0.038* | |
| C26 | 1.1086 (2) | 0.45870 (10) | 0.13988 (12) | 0.0343 (5) | |
| H26 | 1.1462 | 0.4801 | 0.1931 | 0.041* | |
| C27 | 1.2149 (2) | 0.45643 (10) | 0.07221 (13) | 0.0339 (4) | |
| H27 | 1.3257 | 0.4760 | 0.0790 | 0.041* | |
| C28 | 1.1596 (2) | 0.42573 (10) | −0.00498 (13) | 0.0356 (5) | |
| H28 | 1.2336 | 0.4235 | −0.0509 | 0.043* | |
| C29 | 0.9982 (2) | 0.39815 (9) | −0.01643 (12) | 0.0332 (4) | |
| H29 | 0.9606 | 0.3780 | −0.0705 | 0.040* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0224 (3) | 0.0312 (3) | 0.0323 (3) | 0.00355 (18) | −0.00375 (19) | −0.00608 (19) |
| O1 | 0.0287 (8) | 0.0362 (8) | 0.0423 (8) | 0.0057 (6) | 0.0020 (6) | −0.0040 (7) |
| O2 | 0.0469 (8) | 0.0435 (9) | 0.0273 (8) | −0.0095 (7) | −0.0036 (6) | 0.0027 (6) |
| N1 | 0.0272 (8) | 0.0238 (8) | 0.0241 (8) | 0.0011 (6) | 0.0048 (6) | −0.0021 (6) |
| N2 | 0.0205 (7) | 0.0225 (7) | 0.0203 (7) | 0.0015 (6) | 0.0026 (6) | −0.0005 (5) |
| N3 | 0.0197 (7) | 0.0238 (7) | 0.0241 (8) | 0.0012 (6) | −0.0013 (6) | −0.0012 (6) |
| N4 | 0.0200 (7) | 0.0269 (8) | 0.0273 (8) | 0.0017 (6) | −0.0018 (6) | 0.0015 (6) |
| C1 | 0.0253 (9) | 0.0251 (9) | 0.0238 (9) | 0.0022 (7) | 0.0037 (7) | 0.0000 (7) |
| C2 | 0.0283 (9) | 0.0220 (8) | 0.0216 (9) | 0.0004 (7) | 0.0065 (7) | 0.0022 (7) |
| C3 | 0.0316 (10) | 0.0252 (9) | 0.0194 (9) | −0.0038 (7) | 0.0069 (7) | 0.0012 (7) |
| C4 | 0.0320 (10) | 0.0292 (9) | 0.0253 (9) | −0.0031 (8) | 0.0025 (8) | 0.0015 (7) |
| C5 | 0.0347 (11) | 0.0413 (11) | 0.0284 (10) | −0.0054 (9) | −0.0017 (8) | 0.0047 (8) |
| C6 | 0.0463 (13) | 0.0472 (13) | 0.0260 (10) | −0.0173 (10) | −0.0031 (9) | 0.0020 (9) |
| C7 | 0.0547 (13) | 0.0327 (11) | 0.0252 (10) | −0.0127 (10) | 0.0052 (9) | −0.0018 (8) |
| C8 | 0.0441 (12) | 0.0273 (9) | 0.0228 (9) | −0.0063 (8) | 0.0094 (8) | 0.0015 (7) |
| C9 | 0.0551 (13) | 0.0219 (9) | 0.0298 (10) | 0.0003 (9) | 0.0151 (9) | −0.0016 (7) |
| C10 | 0.0409 (11) | 0.0252 (9) | 0.0374 (11) | 0.0073 (8) | 0.0121 (9) | 0.0006 (8) |
| C11 | 0.0316 (10) | 0.0273 (9) | 0.0260 (9) | 0.0021 (8) | 0.0088 (8) | 0.0034 (7) |
| C12 | 0.0206 (9) | 0.0230 (8) | 0.0222 (9) | −0.0010 (7) | 0.0037 (7) | −0.0003 (7) |
| C13 | 0.0207 (8) | 0.0256 (9) | 0.0223 (9) | −0.0023 (7) | 0.0019 (7) | 0.0033 (7) |
| C14 | 0.0220 (9) | 0.0307 (10) | 0.0332 (10) | 0.0021 (8) | 0.0010 (7) | 0.0031 (8) |
| C15 | 0.0257 (9) | 0.0224 (9) | 0.0265 (9) | 0.0034 (7) | 0.0005 (7) | −0.0020 (7) |
| C16 | 0.0276 (9) | 0.0201 (8) | 0.0260 (9) | 0.0029 (7) | 0.0012 (7) | −0.0030 (7) |
| C17 | 0.0287 (10) | 0.0328 (10) | 0.0291 (10) | 0.0024 (8) | −0.0015 (8) | −0.0029 (8) |
| C18 | 0.0266 (10) | 0.0385 (11) | 0.0383 (11) | 0.0007 (8) | 0.0059 (8) | −0.0059 (9) |
| C19 | 0.0383 (11) | 0.0366 (11) | 0.0312 (11) | −0.0004 (9) | 0.0103 (9) | −0.0065 (8) |
| C20 | 0.0379 (11) | 0.0353 (11) | 0.0252 (10) | 0.0014 (9) | 0.0016 (8) | −0.0040 (8) |
| C21 | 0.0287 (10) | 0.0304 (10) | 0.0297 (10) | 0.0011 (8) | 0.0003 (8) | −0.0050 (8) |
| C22 | 0.0321 (10) | 0.0252 (9) | 0.0270 (10) | −0.0006 (8) | 0.0005 (8) | −0.0038 (7) |
| C23 | 0.0381 (11) | 0.0226 (9) | 0.0263 (10) | 0.0009 (8) | −0.0002 (8) | 0.0015 (7) |
| C24 | 0.0332 (10) | 0.0199 (8) | 0.0271 (9) | 0.0015 (7) | 0.0038 (8) | 0.0015 (7) |
| C25 | 0.0342 (10) | 0.0316 (10) | 0.0284 (10) | 0.0008 (8) | 0.0055 (8) | −0.0037 (8) |
| C26 | 0.0354 (11) | 0.0331 (10) | 0.0342 (11) | −0.0013 (9) | 0.0008 (8) | −0.0050 (8) |
| C27 | 0.0309 (10) | 0.0299 (10) | 0.0417 (11) | 0.0006 (8) | 0.0064 (8) | 0.0041 (8) |
| C28 | 0.0388 (11) | 0.0369 (11) | 0.0328 (11) | 0.0050 (9) | 0.0129 (9) | 0.0045 (9) |
| C29 | 0.0430 (11) | 0.0313 (10) | 0.0252 (10) | 0.0011 (9) | 0.0032 (8) | 0.0006 (8) |
Geometric parameters (Å, °)
| S1—C12 | 1.6679 (18) | C14—H14A | 0.9800 |
| O1—C11 | 1.352 (2) | C14—H14B | 0.9800 |
| O1—H1 | 0.89 (3) | C14—H14C | 0.9800 |
| O2—C23 | 1.223 (2) | C15—C22 | 1.518 (2) |
| N1—C1 | 1.282 (2) | C15—C16 | 1.526 (2) |
| N1—N2 | 1.3873 (19) | C15—H15 | 1.0000 |
| N2—C13 | 1.374 (2) | C16—C17 | 1.390 (2) |
| N2—C12 | 1.396 (2) | C16—C21 | 1.392 (3) |
| N3—C12 | 1.355 (2) | C17—C18 | 1.386 (3) |
| N3—N4 | 1.3842 (19) | C17—H17 | 0.9500 |
| N3—C15 | 1.473 (2) | C18—C19 | 1.388 (3) |
| N4—C13 | 1.305 (2) | C18—H18 | 0.9500 |
| C1—C2 | 1.448 (2) | C19—C20 | 1.380 (3) |
| C1—H1A | 0.9500 | C19—H19 | 0.9500 |
| C2—C11 | 1.397 (2) | C20—C21 | 1.388 (2) |
| C2—C3 | 1.442 (2) | C20—H20 | 0.9500 |
| C3—C4 | 1.418 (2) | C21—H21 | 0.9500 |
| C3—C8 | 1.423 (2) | C22—C23 | 1.514 (2) |
| C4—C5 | 1.369 (3) | C22—H22A | 0.9900 |
| C4—H4 | 0.9500 | C22—H22B | 0.9900 |
| C5—C6 | 1.404 (3) | C23—C24 | 1.486 (2) |
| C5—H5 | 0.9500 | C24—C25 | 1.393 (2) |
| C6—C7 | 1.363 (3) | C24—C29 | 1.397 (2) |
| C6—H6 | 0.9500 | C25—C26 | 1.383 (3) |
| C7—C8 | 1.417 (3) | C25—H25 | 0.9500 |
| C7—H7 | 0.9500 | C26—C27 | 1.384 (3) |
| C8—C9 | 1.421 (3) | C26—H26 | 0.9500 |
| C9—C10 | 1.354 (3) | C27—C28 | 1.377 (3) |
| C9—H9 | 0.9500 | C27—H27 | 0.9500 |
| C10—C11 | 1.412 (2) | C28—C29 | 1.378 (3) |
| C10—H10 | 0.9500 | C28—H28 | 0.9500 |
| C13—C14 | 1.478 (2) | C29—H29 | 0.9500 |
| Cg1···Cg1i | 3.541 (3) | Cg2···Cg3ii | 3.711 (3) |
| C11—O1—H1 | 109.0 (16) | H14B—C14—H14C | 109.5 |
| C1—N1—N2 | 119.78 (14) | N3—C15—C22 | 108.79 (13) |
| C13—N2—N1 | 118.65 (13) | N3—C15—C16 | 110.50 (14) |
| C13—N2—C12 | 109.15 (14) | C22—C15—C16 | 113.46 (14) |
| N1—N2—C12 | 132.19 (14) | N3—C15—H15 | 108.0 |
| C12—N3—N4 | 113.88 (14) | C22—C15—H15 | 108.0 |
| C12—N3—C15 | 125.99 (14) | C16—C15—H15 | 108.0 |
| N4—N3—C15 | 120.09 (13) | C17—C16—C21 | 118.86 (16) |
| C13—N4—N3 | 104.41 (13) | C17—C16—C15 | 119.23 (15) |
| N1—C1—C2 | 120.53 (16) | C21—C16—C15 | 121.88 (16) |
| N1—C1—H1A | 119.7 | C18—C17—C16 | 120.61 (17) |
| C2—C1—H1A | 119.7 | C18—C17—H17 | 119.7 |
| C11—C2—C3 | 119.12 (16) | C16—C17—H17 | 119.7 |
| C11—C2—C1 | 121.57 (16) | C17—C18—C19 | 120.22 (18) |
| C3—C2—C1 | 119.30 (15) | C17—C18—H18 | 119.9 |
| C4—C3—C8 | 117.74 (16) | C19—C18—H18 | 119.9 |
| C4—C3—C2 | 123.25 (16) | C20—C19—C18 | 119.41 (18) |
| C8—C3—C2 | 119.01 (16) | C20—C19—H19 | 120.3 |
| C5—C4—C3 | 120.92 (18) | C18—C19—H19 | 120.3 |
| C5—C4—H4 | 119.5 | C19—C20—C21 | 120.57 (18) |
| C3—C4—H4 | 119.5 | C19—C20—H20 | 119.7 |
| C4—C5—C6 | 121.22 (19) | C21—C20—H20 | 119.7 |
| C4—C5—H5 | 119.4 | C20—C21—C16 | 120.30 (17) |
| C6—C5—H5 | 119.4 | C20—C21—H21 | 119.8 |
| C7—C6—C5 | 119.41 (19) | C16—C21—H21 | 119.8 |
| C7—C6—H6 | 120.3 | C23—C22—C15 | 112.96 (14) |
| C5—C6—H6 | 120.3 | C23—C22—H22A | 109.0 |
| C6—C7—C8 | 121.07 (19) | C15—C22—H22A | 109.0 |
| C6—C7—H7 | 119.5 | C23—C22—H22B | 109.0 |
| C8—C7—H7 | 119.5 | C15—C22—H22B | 109.0 |
| C7—C8—C9 | 121.31 (18) | H22A—C22—H22B | 107.8 |
| C7—C8—C3 | 119.64 (18) | O2—C23—C24 | 121.14 (16) |
| C9—C8—C3 | 119.05 (17) | O2—C23—C22 | 120.53 (16) |
| C10—C9—C8 | 121.64 (18) | C24—C23—C22 | 118.33 (15) |
| C10—C9—H9 | 119.2 | C25—C24—C29 | 119.08 (17) |
| C8—C9—H9 | 119.2 | C25—C24—C23 | 121.24 (16) |
| C9—C10—C11 | 120.27 (18) | C29—C24—C23 | 119.65 (16) |
| C9—C10—H10 | 119.9 | C26—C25—C24 | 120.33 (17) |
| C11—C10—H10 | 119.9 | C26—C25—H25 | 119.8 |
| O1—C11—C2 | 123.05 (17) | C24—C25—H25 | 119.8 |
| O1—C11—C10 | 116.08 (16) | C25—C26—C27 | 120.08 (18) |
| C2—C11—C10 | 120.87 (17) | C25—C26—H26 | 120.0 |
| N3—C12—N2 | 101.83 (14) | C27—C26—H26 | 120.0 |
| N3—C12—S1 | 127.02 (13) | C28—C27—C26 | 119.75 (18) |
| N2—C12—S1 | 131.07 (13) | C28—C27—H27 | 120.1 |
| N4—C13—N2 | 110.67 (15) | C26—C27—H27 | 120.1 |
| N4—C13—C14 | 125.43 (16) | C27—C28—C29 | 120.85 (17) |
| N2—C13—C14 | 123.88 (16) | C27—C28—H28 | 119.6 |
| C13—C14—H14A | 109.5 | C29—C28—H28 | 119.6 |
| C13—C14—H14B | 109.5 | C28—C29—C24 | 119.88 (17) |
| H14A—C14—H14B | 109.5 | C28—C29—H29 | 120.1 |
| C13—C14—H14C | 109.5 | C24—C29—H29 | 120.1 |
| H14A—C14—H14C | 109.5 | ||
| C1—N1—N2—C13 | 171.14 (15) | N3—N4—C13—N2 | −0.84 (18) |
| C1—N1—N2—C12 | −8.5 (3) | N3—N4—C13—C14 | 177.86 (15) |
| C12—N3—N4—C13 | −0.75 (19) | N1—N2—C13—N4 | −177.60 (14) |
| C15—N3—N4—C13 | −178.93 (14) | C12—N2—C13—N4 | 2.09 (19) |
| N2—N1—C1—C2 | −177.96 (14) | N1—N2—C13—C14 | 3.7 (2) |
| N1—C1—C2—C11 | 6.5 (3) | C12—N2—C13—C14 | −176.63 (15) |
| N1—C1—C2—C3 | −174.89 (15) | C12—N3—C15—C22 | −146.60 (16) |
| C11—C2—C3—C4 | 178.02 (15) | N4—N3—C15—C22 | 31.3 (2) |
| C1—C2—C3—C4 | −0.6 (2) | C12—N3—C15—C16 | 88.2 (2) |
| C11—C2—C3—C8 | −2.0 (2) | N4—N3—C15—C16 | −93.84 (17) |
| C1—C2—C3—C8 | 179.36 (15) | N3—C15—C16—C17 | −98.27 (18) |
| C8—C3—C4—C5 | 0.1 (2) | C22—C15—C16—C17 | 139.25 (17) |
| C2—C3—C4—C5 | −179.94 (16) | N3—C15—C16—C21 | 79.84 (19) |
| C3—C4—C5—C6 | −0.4 (3) | C22—C15—C16—C21 | −42.6 (2) |
| C4—C5—C6—C7 | 0.5 (3) | C21—C16—C17—C18 | −1.5 (3) |
| C5—C6—C7—C8 | −0.3 (3) | C15—C16—C17—C18 | 176.63 (17) |
| C6—C7—C8—C9 | 179.46 (17) | C16—C17—C18—C19 | 0.1 (3) |
| C6—C7—C8—C3 | 0.0 (3) | C17—C18—C19—C20 | 1.1 (3) |
| C4—C3—C8—C7 | 0.1 (2) | C18—C19—C20—C21 | −1.0 (3) |
| C2—C3—C8—C7 | −179.90 (15) | C19—C20—C21—C16 | −0.4 (3) |
| C4—C3—C8—C9 | −179.36 (15) | C17—C16—C21—C20 | 1.7 (3) |
| C2—C3—C8—C9 | 0.7 (2) | C15—C16—C21—C20 | −176.46 (16) |
| C7—C8—C9—C10 | −178.40 (17) | N3—C15—C22—C23 | 58.68 (19) |
| C3—C8—C9—C10 | 1.0 (3) | C16—C15—C22—C23 | −177.89 (14) |
| C8—C9—C10—C11 | −1.3 (3) | C15—C22—C23—O2 | 22.8 (2) |
| C3—C2—C11—O1 | −178.19 (15) | C15—C22—C23—C24 | −157.10 (15) |
| C1—C2—C11—O1 | 0.4 (3) | O2—C23—C24—C25 | 160.25 (18) |
| C3—C2—C11—C10 | 1.7 (2) | C22—C23—C24—C25 | −19.9 (2) |
| C1—C2—C11—C10 | −179.67 (16) | O2—C23—C24—C29 | −17.6 (3) |
| C9—C10—C11—O1 | 179.87 (16) | C22—C23—C24—C29 | 162.27 (17) |
| C9—C10—C11—C2 | −0.1 (3) | C29—C24—C25—C26 | 1.4 (3) |
| N4—N3—C12—N2 | 1.92 (18) | C23—C24—C25—C26 | −176.53 (17) |
| C15—N3—C12—N2 | 179.96 (14) | C24—C25—C26—C27 | −1.6 (3) |
| N4—N3—C12—S1 | −175.26 (12) | C25—C26—C27—C28 | 0.4 (3) |
| C15—N3—C12—S1 | 2.8 (2) | C26—C27—C28—C29 | 1.2 (3) |
| C13—N2—C12—N3 | −2.33 (17) | C27—C28—C29—C24 | −1.4 (3) |
| N1—N2—C12—N3 | 177.30 (15) | C25—C24—C29—C28 | 0.1 (3) |
| C13—N2—C12—S1 | 174.67 (13) | C23—C24—C29—C28 | 178.06 (17) |
| N1—N2—C12—S1 | −5.7 (3) |
Symmetry codes: (i) −x+3/2, y+3/2, −z−1/2; (ii) −x+1/2, y+1/2, −z−1/2.
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···N1 | 0.89 (3) | 1.83 (3) | 2.610 (2) | 145 (2) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: CV5020).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Ferlin, M. G., Chiarelotto, G., Antonucci, F., Caparrotta, L. & Froldi, G. (2002). Eur. J. Med. Chem. 37, 427–434. [DOI] [PubMed]
- Holla, B. S., Veerendra, B., Shivananda, M. K. & Poojary, B. (2003). Eur. J. Med. Chem. 38, 759–767. [DOI] [PubMed]
- Joshi, S., Khosla, N. & Tiwari, P. (2004). Bioorg. Med. Chem. 12, 571–576. [DOI] [PubMed]
- Negm, N. A., Morsy, S. M. I. & Said, M. M. (2005). Bioorg. Med. Chem. Lett. 13, 5921–5926. [DOI] [PubMed]
- Rigaku/MSC (2005). CrystalClear Rigaku/MSC, The Woodlands, Texas, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810052979/cv5020sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810052979/cv5020Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



