Abstract
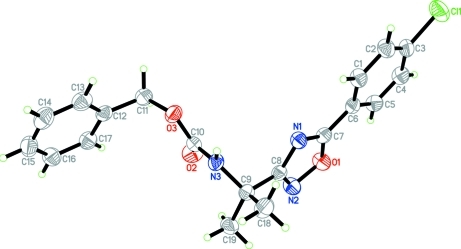
In the title 1,2,4-oxadiazole derivative, C19H18ClN3O3, the 1,2,4-oxadiazole ring makes dihedral angles of 12.83 (8) and 4.89 (8)°, respectively, with the benzyl and 4-chlorophenyl rings, while the dihedral angle between the benzyl and 4-chlorophenyl rings is 11.53 (7)°. In the crystal, molecules are linked by N—H⋯N hydrogen bonds into helical chains along the b axis. A weak C—H⋯π interaction is also present.
Related literature
For bond-length data, see: Allen et al. (1987 ▶). For background to and applications of 1,2,4-oxadiazole derivatives, see: Chen et al. (1994 ▶); Chimirri et al. (1996 ▶); Clitherow et al. (1996 ▶); Nicolaides et al. (1998 ▶); Saunders et al. (1990 ▶); Showell et al. (1991 ▶); Swain et al. (1991 ▶); Tully et al. (1991 ▶); Watjen et al. (1989 ▶). For a related structure, see: Fun et al. (2011 ▶).
Experimental
Crystal data
C19H18ClN3O3
M r = 371.81
Orthorhombic,
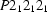
a = 7.7501 (1) Å
b = 11.0052 (2) Å
c = 20.9834 (3) Å
V = 1789.70 (5) Å3
Z = 4
Mo Kα radiation
μ = 0.24 mm−1
T = 297 K
0.41 × 0.35 × 0.21 mm
Data collection
Bruker APEXII CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2005 ▶) T min = 0.908, T max = 0.951
15995 measured reflections
4556 independent reflections
3620 reflections with I > 2σ(I)
R int = 0.028
Refinement
R[F 2 > 2σ(F 2)] = 0.036
wR(F 2) = 0.087
S = 1.02
4556 reflections
241 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.14 e Å−3
Δρmin = −0.21 e Å−3
Absolute structure: Flack (1983 ▶), 1950 Friedel pairs
Flack parameter: 0.01 (6)
Data collection: APEX2 (Bruker, 2005 ▶); cell refinement: SAINT (Bruker, 2005 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536811001504/is2659sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811001504/is2659Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg1 is the centroid of the C12–C17 ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N3—H1N3⋯N2i | 0.840 (16) | 2.428 (15) | 3.2492 (18) | 165.7 (14) |
| C16—H16A⋯Cg1ii | 0.93 | 2.91 | 3.6700 (18) | 140 |
Symmetry codes: (i) 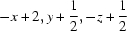 ; (ii)
; (ii) 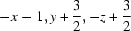 .
.
Acknowledgments
VS, GKN and BP thank Mangalore University for a research grant. SC thanks the Prince of Songkla University for generous support. The authors thank Universiti Sains Malaysia for the Research University Grant No. 1001/PFIZIK/811160.
supplementary crystallographic information
Comment
The 1,2,4-oxadiazole ring occurs frequently in biologically active synthetic compounds and 1,2,4-oxadiazole derivatives are suggested as potential agonists for cortical muscarinic (Saunders et al., 1990; Showell et al., 1991), benzodiazepine (Watjen et al., 1989; Tully et al., 1991) and 5-HT1D (5-hydroxytryptamine) as receptors (Chen et al., 1994) as well as antagonists for 5-HT3 (Swain et al., 1991) or histamine H3 receptors (Clitherow et al., 1996). They also demonstrate anti-inflammatory (Nicolaides et al., 1998) and antitumor activities (Chimirri et al., 1996). These interesting and important properties of 1,2,4-oxadiazole derivatives lead us to synthesize the title compound (I) and its crystal structure was reported.
In the molecule of (I), (Fig. 1), the 1,2,4-oxadiazole ring is nearly co-planar with the 4-chlorophenyl with a dihedral angle between the two rings being 4.89 (8)°, whereas it is inclined to the benzyl (C11–C17) unit with a dihedral angle of 12.48 (9)°. The dihedral angle between the benzyl group and the phenyl (C1–C6) ring is 11.53 (7)°. The oxycarbonylamino unit (atoms C10, N3, O2 and O3) is planar with an r.m.s. 0.0009 (1) Å. This unit makes dihedral angles of 79.99 (9), 89.15 (9) and 89.64 (9)°, respectively, with the benzyl, 1,2,4-oxadiazole and 4-chlorophenyl rings. The bond distances are of normal values (Allen et al., 1987) and are comparable to the related structure (Fun et al., 2011).
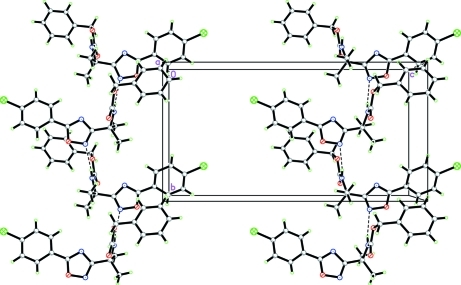
In the crystal packing (Fig. 2), the molecules are linked by by N—H···N hydrogen bonds (Table 1) into helical chains along the b axis. The crystal is stabilized by N—H···O hydrogen bonds and weak C—H···π interactions (Table 1).
Experimental
The title compound was synthesized by taking benzyl [(2E)-2-amino-2-(hydroxyimino)-1,1-dimethylethyl]carbamate (0.1 mole) in 5 mol volume of pyridine and treated with 0.1 mole of 4-chlorobenzoylchloride. The reaction mixture is heated to reflux for 3–4 hrs. After the reaction is completed, the excess pyridine is distilled out under low pressure and the resultant quenched onto ice water. The solid thus formed is filtered and washed with water. The sample is dried and recrystallized in ethanol yielding 95% product. Colorless plate-shaped single crystals of the title compound suitable for x-ray structure determination were recrystalized from ethanol by the slow evaporation of the solvent at room temperature after several days (m.p. 405–409 K).
Refinement
NH hydrogen atom is located in a difference map and refined isotropically. The remaining H atoms were positioned geometrically and allowed to ride on their parent atoms, with d(C—H) = 0.93 Å for aromatic and 0.96 Å for CH3 atoms. The Uiso values were constrained to be 1.5Ueq of the carrier atom for methyl H atoms and 1.2Ueq for the remaining H atoms. A rotating group model was used for the methyl groups. A total of 1950 Friedel pairs were used to determined the absolute configuration
Figures
Fig. 1.
The molecular structure of the title compound, showing 40% probability displacement ellipsoids and the atom-numbering scheme.
Fig. 2.
The crystal packing of the title compound viewed along the a axis, showing helical chains along the b axis. N—H···N hydrogen bonds are shown as dashed lines.
Crystal data
| C19H18ClN3O3 | Dx = 1.380 Mg m−3 |
| Mr = 371.81 | Melting point = 405–409 K |
| Orthorhombic, P212121 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 4556 reflections |
| a = 7.7501 (1) Å | θ = 1.9–28.5° |
| b = 11.0052 (2) Å | µ = 0.24 mm−1 |
| c = 20.9834 (3) Å | T = 297 K |
| V = 1789.70 (5) Å3 | Plate, colorless |
| Z = 4 | 0.41 × 0.35 × 0.21 mm |
| F(000) = 776 |
Data collection
| Bruker APEXII CCD area-detector diffractometer | 4556 independent reflections |
| Radiation source: sealed tube | 3620 reflections with I > 2σ(I) |
| graphite | Rint = 0.028 |
| φ and ω scans | θmax = 28.5°, θmin = 1.9° |
| Absorption correction: multi-scan (SADABS; Bruker, 2005) | h = −10→8 |
| Tmin = 0.908, Tmax = 0.951 | k = −14→14 |
| 15995 measured reflections | l = −23→28 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.036 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.087 | w = 1/[σ2(Fo2) + (0.0458P)2 + 0.0092P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.02 | (Δ/σ)max = 0.001 |
| 4556 reflections | Δρmax = 0.14 e Å−3 |
| 241 parameters | Δρmin = −0.21 e Å−3 |
| 0 restraints | Absolute structure: Flack (1983), 1950 Friedel pairs |
| Primary atom site location: structure-invariant direct methods | Flack parameter: 0.01 (6) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 1.06337 (9) | 0.74415 (5) | −0.14993 (2) | 0.08010 (19) | |
| O1 | 0.90644 (16) | 0.42996 (9) | 0.11883 (5) | 0.0490 (3) | |
| O2 | 0.62272 (16) | 0.60054 (11) | 0.29142 (7) | 0.0631 (4) | |
| O3 | 0.71211 (14) | 0.79437 (9) | 0.30763 (6) | 0.0501 (3) | |
| N1 | 1.00418 (18) | 0.59694 (10) | 0.16396 (6) | 0.0422 (3) | |
| N2 | 0.89957 (19) | 0.41084 (11) | 0.18577 (6) | 0.0487 (3) | |
| N3 | 0.90781 (17) | 0.65050 (11) | 0.29774 (6) | 0.0401 (3) | |
| C1 | 1.0718 (2) | 0.70094 (14) | 0.03799 (7) | 0.0470 (4) | |
| H1A | 1.1094 | 0.7446 | 0.0733 | 0.056* | |
| C2 | 1.0945 (2) | 0.74801 (15) | −0.02232 (8) | 0.0522 (4) | |
| H2A | 1.1470 | 0.8233 | −0.0279 | 0.063* | |
| C3 | 1.0385 (2) | 0.68209 (16) | −0.07420 (7) | 0.0499 (4) | |
| C4 | 0.9627 (2) | 0.56952 (14) | −0.06747 (8) | 0.0517 (4) | |
| H4A | 0.9283 | 0.5255 | −0.1031 | 0.062* | |
| C5 | 0.9386 (2) | 0.52308 (13) | −0.00694 (7) | 0.0469 (4) | |
| H5A | 0.8859 | 0.4478 | −0.0017 | 0.056* | |
| C6 | 0.9930 (2) | 0.58868 (12) | 0.04632 (7) | 0.0393 (3) | |
| C7 | 0.96954 (19) | 0.54258 (12) | 0.11114 (7) | 0.0386 (3) | |
| C8 | 0.9582 (2) | 0.51195 (12) | 0.20882 (7) | 0.0371 (3) | |
| C9 | 0.9872 (2) | 0.53376 (12) | 0.27932 (7) | 0.0417 (4) | |
| C10 | 0.7365 (2) | 0.67374 (13) | 0.29808 (7) | 0.0414 (3) | |
| C11 | 0.5372 (2) | 0.83404 (16) | 0.32187 (8) | 0.0528 (4) | |
| H11A | 0.4552 | 0.7744 | 0.3064 | 0.063* | |
| H11B | 0.5141 | 0.9106 | 0.3007 | 0.063* | |
| C12 | 0.51699 (19) | 0.84918 (13) | 0.39282 (7) | 0.0423 (3) | |
| C13 | 0.5898 (2) | 0.94799 (14) | 0.42406 (9) | 0.0545 (4) | |
| H13A | 0.6480 | 1.0070 | 0.4008 | 0.065* | |
| C14 | 0.5766 (3) | 0.95920 (16) | 0.48927 (9) | 0.0621 (5) | |
| H14A | 0.6270 | 1.0253 | 0.5096 | 0.074* | |
| C15 | 0.4903 (3) | 0.87436 (17) | 0.52416 (9) | 0.0591 (5) | |
| H15A | 0.4826 | 0.8825 | 0.5682 | 0.071* | |
| C16 | 0.4146 (2) | 0.77656 (16) | 0.49429 (8) | 0.0572 (4) | |
| H16A | 0.3541 | 0.7191 | 0.5179 | 0.069* | |
| C17 | 0.4292 (2) | 0.76457 (13) | 0.42877 (8) | 0.0496 (4) | |
| H17A | 0.3788 | 0.6981 | 0.4087 | 0.060* | |
| C18 | 1.1805 (2) | 0.54871 (16) | 0.28978 (9) | 0.0562 (5) | |
| H18A | 1.2220 | 0.6155 | 0.2647 | 0.084* | |
| H18B | 1.2389 | 0.4756 | 0.2773 | 0.084* | |
| H18C | 1.2024 | 0.5645 | 0.3340 | 0.084* | |
| C19 | 0.9170 (3) | 0.42786 (14) | 0.31875 (8) | 0.0592 (5) | |
| H19A | 0.7970 | 0.4162 | 0.3092 | 0.089* | |
| H19B | 0.9302 | 0.4456 | 0.3633 | 0.089* | |
| H19C | 0.9799 | 0.3552 | 0.3085 | 0.089* | |
| H1N3 | 0.973 (2) | 0.7115 (14) | 0.2985 (7) | 0.036 (4)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.1106 (5) | 0.0857 (4) | 0.0440 (2) | 0.0012 (3) | 0.0132 (3) | 0.0075 (2) |
| O1 | 0.0635 (7) | 0.0416 (5) | 0.0418 (6) | −0.0124 (5) | −0.0050 (6) | −0.0044 (4) |
| O2 | 0.0508 (7) | 0.0552 (7) | 0.0833 (10) | −0.0086 (6) | −0.0013 (7) | 0.0010 (6) |
| O3 | 0.0486 (7) | 0.0433 (5) | 0.0584 (7) | 0.0076 (5) | 0.0118 (6) | 0.0012 (5) |
| N1 | 0.0508 (8) | 0.0373 (6) | 0.0384 (7) | −0.0039 (5) | 0.0004 (6) | −0.0046 (5) |
| N2 | 0.0609 (9) | 0.0431 (7) | 0.0422 (7) | −0.0097 (6) | −0.0013 (7) | −0.0010 (5) |
| N3 | 0.0438 (8) | 0.0338 (6) | 0.0425 (7) | −0.0013 (6) | 0.0046 (6) | −0.0040 (5) |
| C1 | 0.0536 (10) | 0.0451 (8) | 0.0422 (8) | −0.0064 (7) | −0.0001 (8) | −0.0097 (6) |
| C2 | 0.0575 (10) | 0.0478 (8) | 0.0514 (9) | −0.0079 (8) | 0.0081 (8) | −0.0012 (7) |
| C3 | 0.0530 (10) | 0.0577 (9) | 0.0389 (8) | 0.0070 (8) | 0.0062 (8) | −0.0006 (7) |
| C4 | 0.0598 (11) | 0.0534 (9) | 0.0420 (8) | 0.0038 (8) | −0.0090 (8) | −0.0110 (7) |
| C5 | 0.0536 (11) | 0.0415 (8) | 0.0457 (8) | −0.0013 (7) | −0.0070 (8) | −0.0065 (6) |
| C6 | 0.0399 (8) | 0.0388 (7) | 0.0390 (8) | 0.0034 (6) | 0.0000 (7) | −0.0060 (5) |
| C7 | 0.0376 (8) | 0.0352 (7) | 0.0431 (8) | −0.0002 (6) | −0.0026 (7) | −0.0057 (6) |
| C8 | 0.0374 (8) | 0.0328 (6) | 0.0410 (7) | 0.0000 (6) | 0.0007 (6) | −0.0021 (5) |
| C9 | 0.0507 (10) | 0.0357 (7) | 0.0389 (8) | 0.0043 (6) | 0.0017 (7) | −0.0032 (6) |
| C10 | 0.0496 (9) | 0.0419 (7) | 0.0327 (7) | 0.0033 (7) | 0.0047 (7) | 0.0044 (6) |
| C11 | 0.0474 (10) | 0.0591 (10) | 0.0518 (10) | 0.0146 (8) | 0.0031 (8) | 0.0053 (7) |
| C12 | 0.0345 (8) | 0.0425 (7) | 0.0499 (9) | 0.0088 (6) | 0.0019 (7) | 0.0026 (6) |
| C13 | 0.0503 (10) | 0.0438 (8) | 0.0692 (12) | −0.0025 (8) | 0.0048 (9) | 0.0023 (8) |
| C14 | 0.0618 (12) | 0.0539 (10) | 0.0705 (12) | 0.0008 (9) | −0.0075 (11) | −0.0185 (8) |
| C15 | 0.0585 (12) | 0.0680 (11) | 0.0507 (10) | 0.0107 (9) | 0.0034 (9) | −0.0074 (8) |
| C16 | 0.0589 (11) | 0.0567 (10) | 0.0561 (10) | −0.0002 (9) | 0.0101 (9) | 0.0080 (7) |
| C17 | 0.0506 (10) | 0.0423 (8) | 0.0560 (9) | −0.0043 (8) | 0.0001 (8) | −0.0035 (7) |
| C18 | 0.0540 (11) | 0.0618 (10) | 0.0528 (10) | 0.0117 (8) | −0.0128 (9) | −0.0138 (8) |
| C19 | 0.0924 (15) | 0.0401 (8) | 0.0452 (9) | 0.0039 (9) | 0.0064 (10) | 0.0028 (6) |
Geometric parameters (Å, °)
| Cl1—C3 | 1.7404 (16) | C8—C9 | 1.515 (2) |
| O1—C7 | 1.3422 (16) | C9—C18 | 1.523 (2) |
| O1—N2 | 1.4213 (17) | C9—C19 | 1.529 (2) |
| O2—C10 | 1.2023 (19) | C11—C12 | 1.506 (2) |
| O3—C10 | 1.3558 (18) | C11—H11A | 0.9700 |
| O3—C11 | 1.455 (2) | C11—H11B | 0.9700 |
| N1—C7 | 1.2877 (18) | C12—C17 | 1.378 (2) |
| N1—C8 | 1.3740 (19) | C12—C13 | 1.390 (2) |
| N2—C8 | 1.2955 (18) | C13—C14 | 1.378 (3) |
| N3—C10 | 1.352 (2) | C13—H13A | 0.9300 |
| N3—C9 | 1.4758 (18) | C14—C15 | 1.362 (3) |
| N3—H1N3 | 0.841 (16) | C14—H14A | 0.9300 |
| C1—C2 | 1.379 (2) | C15—C16 | 1.377 (3) |
| C1—C6 | 1.389 (2) | C15—H15A | 0.9300 |
| C1—H1A | 0.9300 | C16—C17 | 1.386 (2) |
| C2—C3 | 1.378 (2) | C16—H16A | 0.9300 |
| C2—H2A | 0.9300 | C17—H17A | 0.9300 |
| C3—C4 | 1.378 (2) | C18—H18A | 0.9600 |
| C4—C5 | 1.382 (2) | C18—H18B | 0.9600 |
| C4—H4A | 0.9300 | C18—H18C | 0.9600 |
| C5—C6 | 1.3957 (19) | C19—H19A | 0.9600 |
| C5—H5A | 0.9300 | C19—H19B | 0.9600 |
| C6—C7 | 1.463 (2) | C19—H19C | 0.9600 |
| C7—O1—N2 | 105.61 (10) | O2—C10—O3 | 124.84 (16) |
| C10—O3—C11 | 116.99 (13) | N3—C10—O3 | 108.82 (14) |
| C7—N1—C8 | 102.66 (12) | O3—C11—C12 | 109.46 (13) |
| C8—N2—O1 | 103.21 (11) | O3—C11—H11A | 109.8 |
| C10—N3—C9 | 125.11 (13) | C12—C11—H11A | 109.8 |
| C10—N3—H1N3 | 116.2 (10) | O3—C11—H11B | 109.8 |
| C9—N3—H1N3 | 116.6 (10) | C12—C11—H11B | 109.8 |
| C2—C1—C6 | 120.39 (14) | H11A—C11—H11B | 108.2 |
| C2—C1—H1A | 119.8 | C17—C12—C13 | 118.11 (15) |
| C6—C1—H1A | 119.8 | C17—C12—C11 | 121.18 (14) |
| C3—C2—C1 | 119.14 (15) | C13—C12—C11 | 120.70 (15) |
| C3—C2—H2A | 120.4 | C14—C13—C12 | 120.56 (16) |
| C1—C2—H2A | 120.4 | C14—C13—H13A | 119.7 |
| C2—C3—C4 | 121.76 (15) | C12—C13—H13A | 119.7 |
| C2—C3—Cl1 | 118.67 (13) | C15—C14—C13 | 120.61 (17) |
| C4—C3—Cl1 | 119.56 (13) | C15—C14—H14A | 119.7 |
| C3—C4—C5 | 118.97 (14) | C13—C14—H14A | 119.7 |
| C3—C4—H4A | 120.5 | C14—C15—C16 | 120.02 (18) |
| C5—C4—H4A | 120.5 | C14—C15—H15A | 120.0 |
| C4—C5—C6 | 120.26 (15) | C16—C15—H15A | 120.0 |
| C4—C5—H5A | 119.9 | C15—C16—C17 | 119.42 (16) |
| C6—C5—H5A | 119.9 | C15—C16—H16A | 120.3 |
| C1—C6—C5 | 119.47 (14) | C17—C16—H16A | 120.3 |
| C1—C6—C7 | 118.68 (13) | C12—C17—C16 | 121.27 (15) |
| C5—C6—C7 | 121.84 (14) | C12—C17—H17A | 119.4 |
| N1—C7—O1 | 113.68 (13) | C16—C17—H17A | 119.4 |
| N1—C7—C6 | 127.82 (13) | C9—C18—H18A | 109.5 |
| O1—C7—C6 | 118.50 (12) | C9—C18—H18B | 109.5 |
| N2—C8—N1 | 114.83 (13) | H18A—C18—H18B | 109.5 |
| N2—C8—C9 | 123.53 (13) | C9—C18—H18C | 109.5 |
| N1—C8—C9 | 121.50 (12) | H18A—C18—H18C | 109.5 |
| N3—C9—C8 | 109.39 (12) | H18B—C18—H18C | 109.5 |
| N3—C9—C18 | 106.14 (13) | C9—C19—H19A | 109.5 |
| C8—C9—C18 | 107.66 (13) | C9—C19—H19B | 109.5 |
| N3—C9—C19 | 111.94 (13) | H19A—C19—H19B | 109.5 |
| C8—C9—C19 | 110.79 (12) | C9—C19—H19C | 109.5 |
| C18—C9—C19 | 110.72 (15) | H19A—C19—H19C | 109.5 |
| O2—C10—N3 | 126.34 (14) | H19B—C19—H19C | 109.5 |
| C7—O1—N2—C8 | −0.08 (16) | C10—N3—C9—C18 | −177.07 (15) |
| C6—C1—C2—C3 | −0.2 (3) | C10—N3—C9—C19 | −56.2 (2) |
| C1—C2—C3—C4 | −1.0 (3) | N2—C8—C9—N3 | −131.00 (16) |
| C1—C2—C3—Cl1 | 178.49 (14) | N1—C8—C9—N3 | 53.48 (19) |
| C2—C3—C4—C5 | 1.6 (3) | N2—C8—C9—C18 | 114.08 (17) |
| Cl1—C3—C4—C5 | −177.88 (13) | N1—C8—C9—C18 | −61.44 (18) |
| C3—C4—C5—C6 | −1.0 (3) | N2—C8—C9—C19 | −7.1 (2) |
| C2—C1—C6—C5 | 0.7 (2) | N1—C8—C9—C19 | 177.35 (15) |
| C2—C1—C6—C7 | −179.32 (15) | C9—N3—C10—O2 | 9.9 (3) |
| C4—C5—C6—C1 | −0.1 (2) | C9—N3—C10—O3 | −170.43 (13) |
| C4—C5—C6—C7 | 179.93 (15) | C11—O3—C10—O2 | 11.3 (2) |
| C8—N1—C7—O1 | −0.38 (18) | C11—O3—C10—N3 | −168.37 (12) |
| C8—N1—C7—C6 | −179.60 (15) | C10—O3—C11—C12 | 98.05 (16) |
| N2—O1—C7—N1 | 0.30 (18) | O3—C11—C12—C17 | −104.15 (17) |
| N2—O1—C7—C6 | 179.60 (13) | O3—C11—C12—C13 | 74.49 (18) |
| C1—C6—C7—N1 | 4.6 (2) | C17—C12—C13—C14 | 1.1 (2) |
| C5—C6—C7—N1 | −175.44 (16) | C11—C12—C13—C14 | −177.57 (16) |
| C1—C6—C7—O1 | −174.60 (14) | C12—C13—C14—C15 | −0.7 (3) |
| C5—C6—C7—O1 | 5.4 (2) | C13—C14—C15—C16 | −0.3 (3) |
| O1—N2—C8—N1 | −0.16 (18) | C14—C15—C16—C17 | 0.9 (3) |
| O1—N2—C8—C9 | −175.95 (14) | C13—C12—C17—C16 | −0.5 (2) |
| C7—N1—C8—N2 | 0.33 (19) | C11—C12—C17—C16 | 178.18 (15) |
| C7—N1—C8—C9 | 176.22 (14) | C15—C16—C17—C12 | −0.5 (3) |
| C10—N3—C9—C8 | 67.03 (18) |
Hydrogen-bond geometry (Å, °)
| Cg1 is the centroid of the C12–C17 ring. |
| D—H···A | D—H | H···A | D···A | D—H···A |
| N3—H1N3···N2i | 0.840 (16) | 2.428 (15) | 3.2492 (18) | 165.7 (14) |
| C16—H16A···Cg1ii | 0.93 | 2.91 | 3.6700 (18) | 140 |
Symmetry codes: (i) −x+2, y+1/2, −z+1/2; (ii) −x−1, y+3/2, −z+3/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: IS2659).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Bruker (2005). APEX2, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Chen, C.-Y., Senanayake, C. H., Bill, T. J., Larsen, R. D., Verhoeven, T. R. & Reider, P. J. (1994). J. Org. Chem. 59, 3738–3741.
- Chimirri, A., Grasso, S., Montforte, A.-M., Rao, A. & Zappala, M. (1996). Farmaco, 51, 125–129. [PubMed]
- Clitherow, J. W., Beswick, P., Irving, W. J., Scopes, D. I. C., Barnes, J. C., Clapham, J., Brown, J. D., Evans, D. J. & Hayes, A. G. (1996). Bioorg. Med. Chem. Lett. 6, 833–838.
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Fun, H.-K., Sumangala, V., Prasad, D. J., Poojary, B. & Chantrapromma, S. (2011). Acta Cryst. E67, o274. [DOI] [PMC free article] [PubMed]
- Nicolaides, D. N., Fylaktakidou, K. C., Litinas, K. E. & Hadjipavlou-Litina, D. (1998). Eur. J. Med. Chem. 33, 715–724.
- Saunders, J., Cassidy, M., Freedman, S. B., Harley, E. A., Iversen, L. L., Kneen, C., MacLeod, A. M., Merchant, K. J., Snow, R. J. & Baker, R. (1990). J. Med. Chem. 33, 1128–1138. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Showell, G. A., Gibbons, T. L., Kneen, C. O., MacLeod, A. M., Merchant, K., Saunders, J., Freedman, S. B., Patel, S. & Baker, R. (1991). J. Med. Chem. 34, 1086–1094. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Swain, C. J., Baker, R., Kneen, C., Moseley, J., Saunders, J., Seward, E. M., Stevenson, G., Beer, M., Stanton, J. & Watling, K. (1991). J. Med. Chem. 34, 140–151. [DOI] [PubMed]
- Tully, W. R., Gardner, C. R., Gillespie, R. J. & Westwood, R. (1991). J. Med. Chem. 34, 2060–2062. [DOI] [PubMed]
- Watjen, F., Baker, R., Engelstoff, M., Herbert, R., MacLeod, A., Knight, A., Merchant, K., Moseley, J., Saunders, J., Swain, C. J., Wang, E. & Springer, J. P. (1989). J. Med. Chem. 32, 2282–2291. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536811001504/is2659sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811001504/is2659Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report