Abstract
In the monomeric title complex, [Cu(C6H3BrNO2)(NO3)(H2O)2], the CuII ion is coordinated by a bidentate 6-bromopicolinate ion, one nitrate ion and two water molecules in a geometry intermediate between five- and six-coordinate. Conventional O—H⋯O hydrogen bonds link the complex molecules, forming layers parallel to the ab plane.
Related literature
For general background to copper complexes with low-dimensionality synthesized by our group, see: Martins, Ramos Silva et al. (2008 ▶), Martins, Silva et al. (2008 ▶); Ramos Silva et al. (2001a
▶,b
▶,c
▶, 2005a
▶,b
▶). For a magnetic low-dimensional system with picolinic acid, see: Eppley et al. (1997 ▶). For a similar compound with magnetic properties, see: Kukovec et al. (2008 ▶).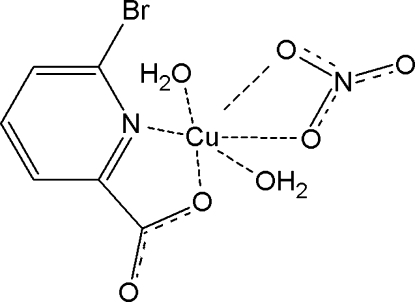
Experimental
Crystal data
[Cu(C6H3BrNO2)(NO3)(H2O)2]
M r = 362.59
Orthorhombic,

a = 9.0791 (14) Å
b = 14.035 (2) Å
c = 17.165 (2) Å
V = 2187.2 (6) Å3
Z = 8
Mo Kα radiation
μ = 5.68 mm−1
T = 293 K
0.40 × 0.10 × 0.08 mm
Data collection
Bruker APEX CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 2000 ▶) T min = 0.619, T max = 0.999
35919 measured reflections
3263 independent reflections
1849 reflections with I > 2σ(I)
R int = 0.069
Refinement
R[F 2 > 2σ(F 2)] = 0.038
wR(F 2) = 0.137
S = 1.03
3263 reflections
167 parameters
7 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 1.06 e Å−3
Δρmin = −1.19 e Å−3
Data collection: SMART (Bruker, 2003 ▶); cell refinement: SAINT (Bruker, 2003 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEPII (Johnson, 1976 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S160053681100064X/bt5450sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053681100064X/bt5450Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O6—H6A⋯O3i | 0.85 (1) | 2.03 (2) | 2.825 (4) | 156 (4) |
| O6—H6B⋯O2ii | 0.85 (2) | 1.86 (1) | 2.700 (4) | 166 (2) |
| O7—H7A⋯O4iii | 0.85 (1) | 2.06 (2) | 2.874 (5) | 163 (5) |
| O7—H7B⋯O1ii | 0.85 (3) | 2.08 (3) | 2.833 (4) | 148 (5) |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Acknowledgments
This work was supported by the Fundo Europeu de Desenvolvimento Regional-QREN-COMPETE through project PTDC/FIS/102284/2008-Fundação para a Ciência e a Tecnologia (FCT).
supplementary crystallographic information
Comment
The title compound was obtained within a project of synthesizing new molecular magnets (Martins, Silva et al., 2008; Martins, Ramos Silva et al., 2008; Ramos Silva et al., 2001a, 2001b, 2001c, 2005a, 2005b). Molecular based magnets can capitalize on the flexibility inherent in carbon chemistry. Such flexibility allows a rational choice of ligands to control the dimensionality of the system, so that quantum effects can be enhanced. Picolinic and hydroxypicolinic acid have been widely used as ligands in low- dimensional metallic systems (Eppley et al., 1997) but 6-bromopicolinic acid has been scarcely used. A different substituent in the pyridine ring may lead to significant electronic and steric effects enlarging the structural diversity. In fact, Kukovec et al. (2008) synthesized a copper (II) complex with 6-bromopicolinic acid as a bidentate ligand in which the magnetic exchange pathway is connected to the Br···π interaction.
In the title compound, the CuII ion is coordinated by a bromopicolinate ligand, two water molecules and a nitrate ion (Fig. 1). One of the Cu—O bonds is rather long [Cu1—O4 2.682 (3) °] so that the coordination about the copper ion is intermediate between five and six-coordination. If the latter bond is to be ignored, the remaining coordination stereochemistry is near a square pyramid. In that case, the copper ion is 0.3149 (5) Å above the least-squares plane of the basal coordinating atoms. The H-bond network is confined to layers parallel to the ab plane (Fig. 2, Table 1). The bromine also forms a short contact [3.066 (2) Å] with O2i [symmetry code: (i) -1 + x,y,z].
The magnetic susceptibility was measured using a SQUID magnetometer in function of temperature with an applied magnetic field of 2 T. The inverse susceptibility showed a linear dependence with temperature, excluding any interaction between magnetic centers.
Experimental
0.14 mmol of 6-bromo-2-pyridinecarboxaldehyde in dichloromethane (10 ml) was added to 0.12 mmol of Cu(NO3)2.3H2O in water (10 ml). After a few weeks, blue single crystals of the title compound were obtained.
Refinement
H atoms bound to C atoms were placed at calculated positions and were treated as riding on the parent atoms with C—H = 0.93 Å (aromatic) and with Uiso(H) = 1.2 Ueq(C). H atoms of water molecules O6 an O7 could not be correctly located in a difference Fourier map. They were placed at positions calculated to optimize H-bonds and refined using restraints [O—H = 0.85 (1) Å, H—H = 1.34 (1) Å] and Uiso(H) = 1.5Ueq(O). To avoid that water (O6) H atoms slip into density peaks around the heavy metal atom, a DFIX command was used to garantee a Cu···H distance of at least 2.50 (1) Å. There are maximum and minimum density peaks slightly above 1 e/Å3.
Figures
Fig. 1.
ORTEPII (Johnson, 1976) plot of the title compound. Displacement ellipsoids are drawn at the 50% level.
Fig. 2.
Packing of the molecules in the unit cell showing the H-bonds as dashed lines.
Crystal data
| [Cu(C6H3BrNO2)(NO3)(H2O)2] | F(000) = 1416 |
| Mr = 362.59 | Dx = 2.202 Mg m−3 |
| Orthorhombic, Pbca | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ac 2ab | Cell parameters from 6959 reflections |
| a = 9.0791 (14) Å | θ = 2.9–26.3° |
| b = 14.035 (2) Å | µ = 5.68 mm−1 |
| c = 17.165 (2) Å | T = 293 K |
| V = 2187.2 (6) Å3 | Needle, blue |
| Z = 8 | 0.40 × 0.10 × 0.08 mm |
Data collection
| Bruker APEX CCD area-detector diffractometer | 3263 independent reflections |
| Radiation source: fine-focus sealed tube | 1849 reflections with I > 2σ(I) |
| graphite | Rint = 0.069 |
| φ and ω scans | θmax = 31.4°, θmin = 2.4° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2000) | h = −12→11 |
| Tmin = 0.619, Tmax = 0.999 | k = −19→20 |
| 35919 measured reflections | l = −24→24 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.038 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.137 | w = 1/[σ2(Fo2) + (0.0722P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 1.03 | (Δ/σ)max = 0.001 |
| 3263 reflections | Δρmax = 1.06 e Å−3 |
| 167 parameters | Δρmin = −1.19 e Å−3 |
| 7 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0019 (3) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cu1 | 0.27639 (5) | 0.10769 (4) | 0.67505 (3) | 0.03377 (16) | |
| Br1 | −0.01236 (4) | 0.12831 (4) | 0.52718 (3) | 0.04471 (16) | |
| C1 | 0.5394 (5) | 0.1362 (3) | 0.6017 (2) | 0.0356 (10) | |
| C2 | 0.4323 (4) | 0.1249 (2) | 0.5356 (2) | 0.0291 (8) | |
| C3 | 0.4765 (5) | 0.1207 (4) | 0.4602 (3) | 0.0463 (12) | |
| H3 | 0.5763 | 0.1214 | 0.4479 | 0.056* | |
| C4 | 0.3723 (6) | 0.1155 (4) | 0.4020 (3) | 0.0542 (14) | |
| H4 | 0.4004 | 0.1107 | 0.3500 | 0.065* | |
| C5 | 0.2261 (5) | 0.1175 (4) | 0.4223 (3) | 0.0500 (13) | |
| H5 | 0.1533 | 0.1164 | 0.3843 | 0.060* | |
| C6 | 0.1890 (5) | 0.1212 (3) | 0.5006 (2) | 0.0367 (10) | |
| O1 | 0.4821 (3) | 0.1405 (2) | 0.66941 (16) | 0.0406 (8) | |
| O2 | 0.6707 (3) | 0.1410 (3) | 0.58800 (18) | 0.0529 (9) | |
| O3 | 0.1019 (3) | 0.0189 (2) | 0.67729 (14) | 0.0378 (7) | |
| O4 | 0.2795 (3) | −0.0834 (3) | 0.66960 (18) | 0.0516 (9) | |
| O5 | 0.0612 (4) | −0.1309 (2) | 0.7005 (2) | 0.0572 (9) | |
| O6 | 0.3112 (3) | 0.0737 (3) | 0.78475 (15) | 0.0505 (9) | |
| H6A | 0.3992 (12) | 0.074 (3) | 0.8019 (8) | 0.076* | |
| H6B | 0.264 (3) | 0.103 (2) | 0.8203 (5) | 0.076* | |
| O7 | 0.1350 (4) | 0.2239 (2) | 0.70395 (19) | 0.0494 (8) | |
| H7A | 0.156 (5) | 0.2826 (10) | 0.703 (3) | 0.074* | |
| H7B | 0.086 (4) | 0.222 (4) | 0.7459 (14) | 0.074* | |
| N1 | 0.2882 (4) | 0.1232 (2) | 0.55771 (19) | 0.0313 (8) | |
| N2 | 0.1477 (4) | −0.0683 (3) | 0.68317 (17) | 0.0356 (8) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cu1 | 0.0240 (3) | 0.0461 (4) | 0.0312 (2) | 0.0008 (2) | 0.00166 (17) | 0.0011 (2) |
| Br1 | 0.0287 (2) | 0.0573 (3) | 0.0481 (3) | 0.00161 (19) | −0.00589 (16) | 0.0033 (2) |
| C1 | 0.030 (2) | 0.040 (3) | 0.037 (2) | −0.0019 (18) | −0.0022 (16) | 0.0035 (17) |
| C2 | 0.028 (2) | 0.022 (2) | 0.0374 (19) | 0.0017 (16) | 0.0021 (15) | 0.0012 (15) |
| C3 | 0.035 (3) | 0.063 (3) | 0.041 (2) | −0.002 (2) | 0.0043 (17) | −0.001 (2) |
| C4 | 0.049 (3) | 0.079 (4) | 0.035 (2) | −0.004 (2) | 0.0032 (19) | −0.007 (2) |
| C5 | 0.043 (3) | 0.070 (4) | 0.037 (2) | −0.004 (2) | −0.0053 (18) | 0.002 (2) |
| C6 | 0.032 (2) | 0.045 (3) | 0.0328 (19) | 0.0002 (17) | −0.0041 (16) | 0.0018 (17) |
| O1 | 0.0272 (15) | 0.064 (2) | 0.0312 (14) | −0.0077 (14) | 0.0003 (10) | −0.0009 (13) |
| O2 | 0.0238 (16) | 0.091 (3) | 0.0443 (17) | −0.0025 (15) | 0.0015 (12) | 0.0099 (16) |
| O3 | 0.0269 (14) | 0.0397 (19) | 0.0468 (15) | 0.0008 (12) | −0.0003 (11) | 0.0047 (12) |
| O4 | 0.0397 (19) | 0.049 (2) | 0.066 (2) | 0.0137 (15) | 0.0097 (14) | 0.0014 (16) |
| O5 | 0.055 (2) | 0.052 (2) | 0.065 (2) | −0.0217 (17) | −0.0069 (17) | 0.0160 (16) |
| O6 | 0.0313 (16) | 0.087 (3) | 0.0326 (14) | 0.0088 (16) | 0.0015 (11) | 0.0027 (16) |
| O7 | 0.0525 (19) | 0.0418 (19) | 0.0541 (17) | 0.0043 (16) | 0.0184 (14) | −0.0001 (16) |
| N1 | 0.0277 (17) | 0.032 (2) | 0.0340 (16) | −0.0022 (14) | 0.0002 (13) | 0.0037 (13) |
| N2 | 0.037 (2) | 0.036 (2) | 0.0337 (16) | 0.0017 (17) | −0.0030 (14) | 0.0025 (14) |
Geometric parameters (Å, °)
| Cu1—O1 | 1.926 (3) | C4—C5 | 1.373 (7) |
| Cu1—O6 | 1.968 (3) | C4—H4 | 0.9300 |
| Cu1—O3 | 2.016 (3) | C5—C6 | 1.386 (6) |
| Cu1—N1 | 2.029 (3) | C5—H5 | 0.9300 |
| Cu1—O7 | 2.134 (3) | C6—N1 | 1.332 (5) |
| Br1—C6 | 1.886 (4) | O3—N2 | 1.296 (4) |
| C1—O2 | 1.218 (5) | O4—N2 | 1.238 (4) |
| C1—O1 | 1.274 (5) | O5—N2 | 1.215 (5) |
| C1—C2 | 1.503 (6) | O6—H6A | 0.851 (9) |
| C2—C3 | 1.356 (6) | O6—H6B | 0.85 (2) |
| C2—N1 | 1.363 (5) | O7—H7A | 0.845 (10) |
| C3—C4 | 1.378 (6) | O7—H7B | 0.85 (3) |
| C3—H3 | 0.9300 | ||
| O1—Cu1—O6 | 87.13 (12) | C3—C4—H4 | 120.7 |
| O1—Cu1—O3 | 155.59 (13) | C4—C5—C6 | 118.9 (4) |
| O6—Cu1—O3 | 87.61 (12) | C4—C5—H5 | 120.6 |
| O1—Cu1—N1 | 82.72 (12) | C6—C5—H5 | 120.6 |
| O6—Cu1—N1 | 165.35 (13) | N1—C6—C5 | 123.3 (4) |
| O3—Cu1—N1 | 97.29 (11) | N1—C6—Br1 | 118.4 (3) |
| O1—Cu1—O7 | 114.35 (14) | C5—C6—Br1 | 118.2 (3) |
| O6—Cu1—O7 | 93.40 (13) | C1—O1—Cu1 | 115.5 (3) |
| O3—Cu1—O7 | 89.74 (13) | N2—O3—Cu1 | 109.4 (2) |
| N1—Cu1—O7 | 100.38 (13) | Cu1—O6—H6A | 118.7 (11) |
| O2—C1—O1 | 125.0 (4) | Cu1—O6—H6B | 118.9 (11) |
| O2—C1—C2 | 119.6 (4) | H6A—O6—H6B | 103.2 (14) |
| O1—C1—C2 | 115.5 (4) | Cu1—O7—H7A | 127 (3) |
| C3—C2—N1 | 123.3 (4) | Cu1—O7—H7B | 119 (3) |
| C3—C2—C1 | 122.3 (4) | H7A—O7—H7B | 99 (4) |
| N1—C2—C1 | 114.3 (3) | C6—N1—C2 | 116.4 (3) |
| C2—C3—C4 | 119.4 (4) | C6—N1—Cu1 | 133.9 (3) |
| C2—C3—H3 | 120.3 | C2—N1—Cu1 | 109.2 (2) |
| C4—C3—H3 | 120.3 | O5—N2—O4 | 123.1 (4) |
| C5—C4—C3 | 118.6 (4) | O5—N2—O3 | 119.7 (4) |
| C5—C4—H4 | 120.7 | O4—N2—O3 | 117.2 (3) |
| O2—C1—C2—C3 | −0.6 (6) | O7—Cu1—O3—N2 | −162.2 (2) |
| O1—C1—C2—C3 | 179.0 (4) | C5—C6—N1—C2 | 2.3 (6) |
| O2—C1—C2—N1 | −178.1 (4) | Br1—C6—N1—C2 | −175.4 (3) |
| O1—C1—C2—N1 | 1.5 (5) | C5—C6—N1—Cu1 | −168.5 (3) |
| N1—C2—C3—C4 | 0.7 (7) | Br1—C6—N1—Cu1 | 13.8 (5) |
| C1—C2—C3—C4 | −176.6 (4) | C3—C2—N1—C6 | −2.8 (6) |
| C2—C3—C4—C5 | 1.9 (7) | C1—C2—N1—C6 | 174.7 (3) |
| C3—C4—C5—C6 | −2.3 (7) | C3—C2—N1—Cu1 | 170.2 (3) |
| C4—C5—C6—N1 | 0.2 (7) | C1—C2—N1—Cu1 | −12.3 (4) |
| C4—C5—C6—Br1 | 177.9 (4) | O1—Cu1—N1—C6 | −174.5 (4) |
| O2—C1—O1—Cu1 | −169.1 (4) | O6—Cu1—N1—C6 | 139.0 (5) |
| C2—C1—O1—Cu1 | 11.3 (5) | O3—Cu1—N1—C6 | 30.1 (4) |
| O6—Cu1—O1—C1 | 155.0 (3) | O7—Cu1—N1—C6 | −60.9 (4) |
| O3—Cu1—O1—C1 | 77.1 (4) | O1—Cu1—N1—C2 | 14.3 (2) |
| N1—Cu1—O1—C1 | −14.5 (3) | O6—Cu1—N1—C2 | −32.2 (6) |
| O7—Cu1—O1—C1 | −112.6 (3) | O3—Cu1—N1—C2 | −141.1 (2) |
| O1—Cu1—O3—N2 | 8.9 (4) | O7—Cu1—N1—C2 | 127.8 (2) |
| O6—Cu1—O3—N2 | −68.8 (2) | Cu1—O3—N2—O5 | 165.8 (3) |
| N1—Cu1—O3—N2 | 97.3 (2) | Cu1—O3—N2—O4 | −15.0 (4) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O6—H6A···O3i | 0.85 (1) | 2.03 (2) | 2.825 (4) | 156 (4) |
| O6—H6B···O2ii | 0.85 (2) | 1.86 (1) | 2.700 (4) | 166 (2) |
| O7—H7A···O4iii | 0.85 (1) | 2.06 (2) | 2.874 (5) | 163 (5) |
| O7—H7B···O1ii | 0.85 (3) | 2.08 (3) | 2.833 (4) | 148 (5) |
Symmetry codes: (i) x+1/2, y, −z+3/2; (ii) x−1/2, y, −z+3/2; (iii) −x+1/2, y+1/2, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BT5450).
References
- Bruker (2003). SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Eppley, H. J., Aubin, S. M., Streib, W. E., Bollinger, J. C., Hendrickson, D. N. & Christou, G. (1997). Inorg. Chem. 36, 109–115.
- Johnson, C. K. (1976). ORTEPII Report ORNL-5138. Oak Ridge National Laboratory, Tennessee, USA.
- Kukovec, B.-M., Popovic, Z., Kozlevcar, B. & Jaglicic, Z. (2008). Polyhedron, 27, 3631–3638.
- Martins, N. D., Ramos Silva, M., Silva, J. A., Matos Beja, A. & Sobral, A. J. F. N. (2008). Acta Cryst. E64, m829–m830. [DOI] [PMC free article] [PubMed]
- Martins, N. D., Silva, J. A., Ramos Silva, M., Matos Beja, A. & Sobral, A. J. F. N. (2008). Acta Cryst. E64, m394. [DOI] [PMC free article] [PubMed]
- Ramos Silva, M., Paixão, J. A., Matos Beja, A. & Alte da Veiga, L. (2001a). Acta Cryst. C57, 7–8. [DOI] [PubMed]
- Ramos Silva, M., Paixão, J. A., Matos Beja, A. & Alte da Veiga, L. (2001b). Acta Cryst. C57, 9–11. [DOI] [PubMed]
- Ramos Silva, M., Paixão, J. A., Matos Beja, A., da Veiga, L. A. & Martin-Gil, J. (2001c). J. Chem. Crystallogr. 31, 167–171.
- Ramos Silva, M., Matos Beja, A., Paixão, J. A. & Martin-Gil, J. (2005a). Acta Cryst. C61, m507–m509. [DOI] [PubMed]
- Ramos Silva, M., Matos Beja, A., Paixão, J. A. & Martin-Gil, J. (2005b). Acta Cryst. C61, m380–m382. [DOI] [PubMed]
- Sheldrick, G. M. (2000). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S160053681100064X/bt5450sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053681100064X/bt5450Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




