Abstract
In the crystal structure of the title compound, C10H9N2O2 +·Cl−·2H2O, the components are linked by O—H⋯O, N—H⋯O, O—H⋯Cl and N—H⋯Cl hydrogen bonds. In the cation, the imidazole ring is oriented at a dihedral angle of 13.67 (17)° with respect to the benzene ring. In the crystal, π–π stacking occurs between nearly parallel benzene rings, which are oriented at a dihedral angle of 3.4 (1)°, the centroid–centroid distance being 3.798 (3) Å.
Related literature
For related imidazole-containing compounds, see: Nyamori & Bala (2008 ▶); Nie et al. (2009 ▶).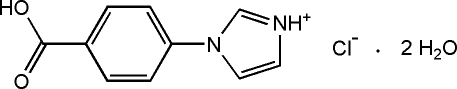
Experimental
Crystal data
C10H9N2O2 +·Cl−·2H2O
M r = 260.67
Orthorhombic,
a = 7.427 (5) Å
b = 17.708 (5) Å
c = 18.748 (5) Å
V = 2465.7 (19) Å3
Z = 8
Mo Kα radiation
μ = 0.32 mm−1
T = 298 K
0.30 × 0.20 × 0.20 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2001 ▶) T min = 0.912, T max = 0.940
14555 measured reflections
2165 independent reflections
1515 reflections with I > 2σ(I)
R int = 0.033
Refinement
R[F 2 > 2σ(F 2)] = 0.040
wR(F 2) = 0.146
S = 0.96
2165 reflections
175 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.20 e Å−3
Δρmin = −0.32 e Å−3
Data collection: SMART (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536811001838/xu5134sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811001838/xu5134Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H2⋯O1i | 0.86 | 2.21 | 2.822 (3) | 128 |
| N2—H2⋯Cl1ii | 0.86 | 2.62 | 3.231 (2) | 129 |
| O2—H7⋯O4iii | 0.82 | 1.77 | 2.594 (3) | 177 |
| O3—H10⋯Cl1iv | 0.83 (4) | 2.36 (4) | 3.150 (3) | 159 (3) |
| O3—H11⋯Cl1v | 0.94 (5) | 2.22 (5) | 3.141 (3) | 168 (3) |
| O4—H12⋯O3 | 0.76 (4) | 1.99 (4) | 2.718 (4) | 162 (4) |
| O4—H13⋯Cl1 | 0.86 (4) | 2.22 (4) | 3.066 (4) | 172 (3) |
Symmetry codes: (i) 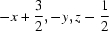 ; (ii)
; (ii) 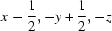 ; (iii)
; (iii) 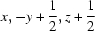 ; (iv)
; (iv) 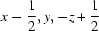 ; (v)
; (v) 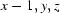 .
.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (21071001), the Education Committee of Anhui Province (KJ2009A52, KJ2010A30), the Team for Scientific Innovation Foundation of Anhui Province (2006 K J007TD), the Ministry of Education Person with Ability Foundation of Anhui University, the Science and Technological Fund of Anhui Province for Outstanding Youth (10040606Y22), and the 211 Project of Anhui University.
supplementary crystallographic information
Comment
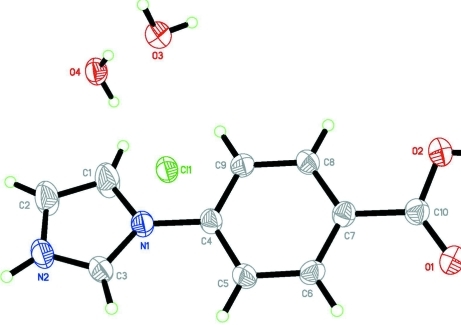
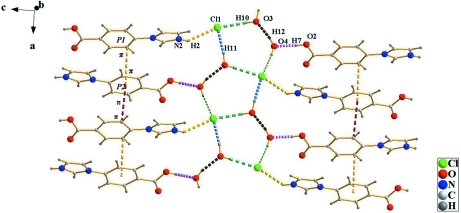
Imidazol - based materials had been investigated for their electrical and optical properties. Beside, the introduction about the structure of its complex has been reported (Nie et al., 2009). The title molecule that we has designed and synthesized is a good intermediate and penetratingly investigated. In the title molecule(I) (Fig.1), the bond lengths and angles show normal values. The imidazole ring is twisted out of the plane of the center benzene ring at a dihedral angle of 13.67 (17)°. In the title molecule (I) (Fig.2), the neighboring molecules connect through O—H···O, N—H···O, O—H···Cl and N—H···Cl hydrogen bonds (Table 1). π–π stacking is observed between nearly parallel benzene rings, the centroids distance being 3.798 (3) Å.
Experimental
A 150 ml round-bottom flask was charged with a magnetic stirrer and a reflux condenser, iminazole (44 mmol), K2CO3 (6.00 g, 43 mmol), 30 ml DMSO and a little Aliquat 336 were added. 4-Fluorobenzaldehyde (4.5 ml, 42 mmol) was added dropwise to the mixture at 363 K and stirred for 15 min. Then the reaction mixture was refluxed for 24 h at 353 K, cooled to room temperature, poured into 150 ml ice-water and filtered. The primrose yellow crude product was obtained, washed with distilled water, and dried in vacuo at room temperature, then purified by recrystallization with ethyl acetate to give the desired analytical pure intermediate products. Intermediate product (12.5 mmol) and 15 ml 20% (wt) NaOH (aq) were added to a round-bottom flask equipped with a magnetic stirrer and a reflux condenser at 333 K for 30 min. Then AgNO3 (4.00 g, 24 mmol) was added to the mixture group by group. The reaction mixture was refluxed for 24 h at 333 K, cooled to room temperature and filtered. Excessive HCl (1 M) was added to the filtrate and adjust pH to 2, a great deal of sediments were obtained and then filtered. The crude product was recrystallized from ethanol-water solution.
Refinement
Water H atoms were located in a difference Fourier map and refined isotropically. Other H atoms were placed in geometrically idealized positions and constrained to ride on their parent atoms, with O—H = 0.82 and C—H = 0.93 Å, Uiso(H) = 1.5Ueq(O) and 1.2Ueq(C).
Figures
Fig. 1.
The molecular structure of the title molecule(I) showing 30% probability displacement ellipsoids.
Fig. 2.
The H-bond and weak π–π interaction diagram of the title molecule(I).
Crystal data
| C10H9N2O2+·Cl−·2H2O | F(000) = 1088 |
| Mr = 260.67 | Dx = 1.404 Mg m−3 |
| Orthorhombic, Pbca | Mo Kα radiation, λ = 0.71069 Å |
| Hall symbol: -P 2ac 2ab | Cell parameters from 3445 reflections |
| a = 7.427 (5) Å | θ = 2.3–21.7° |
| b = 17.708 (5) Å | µ = 0.32 mm−1 |
| c = 18.748 (5) Å | T = 298 K |
| V = 2465.7 (19) Å3 | Block, yellow |
| Z = 8 | 0.30 × 0.20 × 0.20 mm |
Data collection
| Bruker SMART CCD area-detector diffractometer | 2165 independent reflections |
| Radiation source: fine-focus sealed tube | 1515 reflections with I > 2σ(I) |
| graphite | Rint = 0.033 |
| φ and ω scans | θmax = 25.0°, θmin = 2.3° |
| Absorption correction: multi-scan (SADABS; Bruker, 2001) | h = −8→7 |
| Tmin = 0.912, Tmax = 0.940 | k = −21→16 |
| 14555 measured reflections | l = −17→22 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.040 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.146 | H atoms treated by a mixture of independent and constrained refinement |
| S = 0.96 | w = 1/[σ2(Fo2) + (0.1P)2 + 0.3P] where P = (Fo2 + 2Fc2)/3 |
| 2165 reflections | (Δ/σ)max < 0.001 |
| 175 parameters | Δρmax = 0.20 e Å−3 |
| 0 restraints | Δρmin = −0.32 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.99581 (9) | 0.35765 (4) | 0.14987 (4) | 0.0637 (3) | |
| O3 | 0.4071 (3) | 0.34344 (12) | 0.18640 (13) | 0.0661 (6) | |
| N1 | 0.6552 (3) | 0.13527 (10) | 0.12446 (10) | 0.0467 (5) | |
| C4 | 0.6689 (3) | 0.11991 (13) | 0.19963 (12) | 0.0423 (6) | |
| O4 | 0.6325 (4) | 0.41591 (12) | 0.09501 (10) | 0.0628 (6) | |
| O2 | 0.6026 (3) | 0.11860 (11) | 0.46109 (9) | 0.0678 (6) | |
| H7 | 0.6154 | 0.1068 | 0.5031 | 0.102* | |
| O1 | 0.7751 (3) | 0.01818 (11) | 0.44419 (10) | 0.0717 (6) | |
| C10 | 0.6934 (3) | 0.07130 (14) | 0.42051 (13) | 0.0484 (6) | |
| C6 | 0.7468 (3) | 0.03739 (12) | 0.29454 (12) | 0.0473 (6) | |
| H6 | 0.7936 | −0.0083 | 0.3105 | 0.057* | |
| C7 | 0.6859 (3) | 0.09017 (12) | 0.34359 (12) | 0.0436 (6) | |
| C5 | 0.7388 (3) | 0.05180 (13) | 0.22262 (12) | 0.0481 (6) | |
| H5 | 0.7798 | 0.0163 | 0.1899 | 0.058* | |
| C8 | 0.6198 (3) | 0.15821 (13) | 0.31931 (13) | 0.0483 (6) | |
| H8 | 0.5805 | 0.1941 | 0.3520 | 0.058* | |
| C9 | 0.6110 (3) | 0.17379 (12) | 0.24755 (13) | 0.0469 (6) | |
| H9 | 0.5667 | 0.2199 | 0.2316 | 0.056* | |
| N2 | 0.6424 (3) | 0.12032 (12) | 0.01107 (11) | 0.0572 (6) | |
| H2 | 0.6454 | 0.0992 | −0.0302 | 0.069* | |
| C3 | 0.6691 (3) | 0.08596 (14) | 0.07173 (13) | 0.0532 (7) | |
| H3 | 0.6941 | 0.0348 | 0.0771 | 0.064* | |
| C2 | 0.6095 (6) | 0.19392 (17) | 0.02329 (16) | 0.0848 (11) | |
| C1 | 0.6184 (6) | 0.20415 (17) | 0.09362 (16) | 0.0934 (12) | |
| H1 | 0.6026 | 0.2496 | 0.1176 | 0.112* | |
| H12 | 0.554 (5) | 0.402 (2) | 0.117 (2) | 0.079 (13)* | |
| H13 | 0.734 (5) | 0.3965 (18) | 0.1065 (16) | 0.076 (11)* | |
| H11 | 0.283 (6) | 0.341 (2) | 0.179 (2) | 0.118 (15)* | |
| H10 | 0.421 (6) | 0.359 (2) | 0.228 (2) | 0.112 (15)* | |
| H4 | 0.597 (5) | 0.230 (2) | −0.017 (2) | 0.112 (12)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0640 (5) | 0.0782 (5) | 0.0489 (5) | −0.0013 (3) | 0.0008 (3) | −0.0005 (3) |
| O3 | 0.0661 (15) | 0.0778 (14) | 0.0545 (14) | 0.0097 (11) | 0.0031 (11) | −0.0011 (11) |
| N1 | 0.0583 (12) | 0.0435 (11) | 0.0383 (11) | −0.0004 (9) | −0.0028 (9) | −0.0008 (9) |
| C4 | 0.0480 (13) | 0.0421 (12) | 0.0370 (13) | −0.0030 (10) | −0.0038 (10) | −0.0021 (10) |
| O4 | 0.0735 (15) | 0.0696 (13) | 0.0452 (12) | 0.0004 (13) | 0.0000 (12) | 0.0038 (9) |
| O2 | 0.0905 (15) | 0.0737 (12) | 0.0393 (10) | 0.0220 (11) | 0.0038 (10) | 0.0017 (10) |
| O1 | 0.0959 (15) | 0.0695 (12) | 0.0497 (11) | 0.0272 (12) | −0.0011 (10) | 0.0107 (9) |
| C10 | 0.0514 (14) | 0.0493 (13) | 0.0444 (14) | 0.0007 (12) | −0.0044 (12) | −0.0011 (12) |
| C6 | 0.0571 (14) | 0.0351 (11) | 0.0497 (14) | 0.0043 (11) | −0.0047 (12) | 0.0016 (11) |
| C7 | 0.0460 (13) | 0.0431 (13) | 0.0416 (13) | −0.0026 (10) | −0.0050 (10) | 0.0009 (10) |
| C5 | 0.0623 (15) | 0.0406 (13) | 0.0413 (13) | 0.0032 (11) | 0.0000 (12) | −0.0057 (11) |
| C8 | 0.0602 (16) | 0.0449 (13) | 0.0397 (14) | 0.0045 (11) | −0.0018 (11) | −0.0059 (10) |
| C9 | 0.0583 (15) | 0.0374 (11) | 0.0450 (14) | 0.0064 (11) | −0.0015 (11) | 0.0012 (11) |
| N2 | 0.0716 (15) | 0.0631 (14) | 0.0369 (12) | −0.0002 (11) | −0.0019 (10) | −0.0056 (10) |
| C3 | 0.0677 (17) | 0.0500 (14) | 0.0420 (14) | 0.0050 (12) | −0.0007 (12) | −0.0075 (12) |
| C2 | 0.152 (3) | 0.0592 (19) | 0.0428 (17) | 0.011 (2) | −0.0073 (19) | 0.0062 (14) |
| C1 | 0.186 (4) | 0.0480 (16) | 0.0467 (17) | 0.017 (2) | −0.010 (2) | 0.0026 (13) |
Geometric parameters (Å, °)
| O3—H11 | 0.94 (5) | C6—C7 | 1.387 (3) |
| O3—H10 | 0.83 (4) | C6—H6 | 0.9300 |
| N1—C3 | 1.323 (3) | C7—C8 | 1.379 (3) |
| N1—C1 | 1.377 (3) | C5—H5 | 0.9300 |
| N1—C4 | 1.439 (3) | C8—C9 | 1.375 (3) |
| C4—C9 | 1.379 (3) | C8—H8 | 0.9300 |
| C4—C5 | 1.382 (3) | C9—H9 | 0.9300 |
| O4—H12 | 0.76 (4) | N2—C3 | 1.305 (3) |
| O4—H13 | 0.86 (3) | N2—C2 | 1.346 (4) |
| O2—C10 | 1.317 (3) | N2—H2 | 0.8600 |
| O2—H7 | 0.8200 | C3—H3 | 0.9300 |
| O1—C10 | 1.204 (3) | C2—C1 | 1.333 (4) |
| C10—C7 | 1.481 (3) | C2—H4 | 0.99 (4) |
| C6—C5 | 1.373 (3) | C1—H1 | 0.9300 |
| H11—O3—H10 | 106 (4) | C4—C5—H5 | 120.5 |
| C3—N1—C1 | 106.6 (2) | C9—C8—C7 | 121.0 (2) |
| C3—N1—C4 | 127.0 (2) | C9—C8—H8 | 119.5 |
| C1—N1—C4 | 126.3 (2) | C7—C8—H8 | 119.5 |
| C9—C4—C5 | 121.2 (2) | C8—C9—C4 | 118.9 (2) |
| C9—C4—N1 | 119.0 (2) | C8—C9—H9 | 120.5 |
| C5—C4—N1 | 119.8 (2) | C4—C9—H9 | 120.5 |
| H12—O4—H13 | 115 (3) | C3—N2—C2 | 109.3 (2) |
| C10—O2—H7 | 109.5 | C3—N2—H2 | 125.3 |
| O1—C10—O2 | 122.8 (2) | C2—N2—H2 | 125.3 |
| O1—C10—C7 | 123.6 (2) | N2—C3—N1 | 109.4 (2) |
| O2—C10—C7 | 113.6 (2) | N2—C3—H3 | 125.3 |
| C5—C6—C7 | 120.8 (2) | N1—C3—H3 | 125.3 |
| C5—C6—H6 | 119.6 | C1—C2—N2 | 106.9 (3) |
| C7—C6—H6 | 119.6 | C1—C2—H4 | 132 (2) |
| C8—C7—C6 | 119.1 (2) | N2—C2—H4 | 121 (2) |
| C8—C7—C10 | 122.1 (2) | C2—C1—N1 | 107.8 (3) |
| C6—C7—C10 | 118.8 (2) | C2—C1—H1 | 126.1 |
| C6—C5—C4 | 119.0 (2) | N1—C1—H1 | 126.1 |
| C6—C5—H5 | 120.5 | ||
| C3—N1—C4—C9 | −165.3 (2) | C6—C7—C8—C9 | 1.0 (4) |
| C1—N1—C4—C9 | 12.2 (4) | C10—C7—C8—C9 | −178.7 (2) |
| C3—N1—C4—C5 | 14.4 (4) | C7—C8—C9—C4 | 0.3 (4) |
| C1—N1—C4—C5 | −168.1 (3) | C5—C4—C9—C8 | −1.5 (4) |
| C5—C6—C7—C8 | −1.1 (4) | N1—C4—C9—C8 | 178.3 (2) |
| C5—C6—C7—C10 | 178.6 (2) | C2—N2—C3—N1 | −0.2 (3) |
| O1—C10—C7—C8 | −167.9 (3) | C1—N1—C3—N2 | −0.3 (3) |
| O2—C10—C7—C8 | 11.7 (3) | C4—N1—C3—N2 | 177.6 (2) |
| O1—C10—C7—C6 | 12.4 (4) | C3—N2—C2—C1 | 0.6 (4) |
| O2—C10—C7—C6 | −168.0 (2) | N2—C2—C1—N1 | −0.7 (4) |
| C7—C6—C5—C4 | 0.0 (4) | C3—N1—C1—C2 | 0.6 (4) |
| C9—C4—C5—C6 | 1.4 (4) | C4—N1—C1—C2 | −177.3 (3) |
| N1—C4—C5—C6 | −178.4 (2) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H2···O1i | 0.86 | 2.21 | 2.822 (3) | 128 |
| N2—H2···Cl1ii | 0.86 | 2.62 | 3.231 (2) | 129 |
| O2—H7···O4iii | 0.82 | 1.77 | 2.594 (3) | 177 |
| O3—H10···Cl1iv | 0.83 (4) | 2.36 (4) | 3.150 (3) | 159 (3) |
| O3—H11···Cl1v | 0.94 (5) | 2.22 (5) | 3.141 (3) | 168 (3) |
| O4—H12···O3 | 0.76 (4) | 1.99 (4) | 2.718 (4) | 162 (4) |
| O4—H13···Cl1 | 0.86 (4) | 2.22 (4) | 3.066 (4) | 172 (3) |
Symmetry codes: (i) −x+3/2, −y, z−1/2; (ii) x−1/2, −y+1/2, −z; (iii) x, −y+1/2, z+1/2; (iv) x−1/2, y, −z+1/2; (v) x−1, y, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: XU5134).
References
- Bruker (2001). SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2007). SAMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Nie, J.-J., Li, J.-H. & Xu, D.-J. (2009). Acta Cryst. E65, m822–m823. [DOI] [PMC free article] [PubMed]
- Nyamori, V. O. & Bala, M. D. (2008). Acta Cryst. E64, m1451. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536811001838/xu5134sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811001838/xu5134Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report