Abstract
The asymmetric unit of the title compound, C16H14F3N3OS·H2O, contains two independent molecules (A and B) and two water molecules, one of which is disordered over two positions in a 0.790 (8):0.210 (8) ratio. The molecular conformations are close, the benzimidazole mean plane and pyridine ring forming dihedral angles of 1.8 (3) and 0.1 (2)° in molecules A and B, respectively. The water molecules are involved in formation of two independent hydrogen-bonded chains via N—H⋯O and O—H⋯N hydrogen bonds. Chains propagating along the a axis are formed by molecule A and one independent water molecule, while chains propagating along the b axis are formed by molecule B and the other independent water molecule. The crystal packing exhibits π–π interactions, as indicated by short distances of 3.607 (3) and 3.701 (3) Å between the centroids of the imidazole and pyridine rings of neighbouring molecules.
Related literature
The title compound is an intermediate in the synthesis of the anti-ulcer drug lansoprazole [systematic name (RS)-2-([3-methyl-4-(2,2,2-trifluoroethoxy)pyridin-2-yl]methylsulfinyl)-1H-benzo[d]imidazole], see: Del Rio et al. (2007 ▶); Reddy et al. (2008 ▶); Iwahi et al. (1991 ▶). For related structures, see: Swamy & Ravikumar (2007 ▶); Hakim Al-arique et al. (2010 ▶).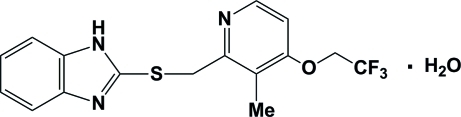
Experimental
Crystal data
C16H14F3N3OS·H2O
M r = 371.39
Triclinic,

a = 7.3526 (1) Å
b = 7.4702 (1) Å
c = 30.6500 (3) Å
α = 88.27°
β = 87.79°
γ = 89.13°
V = 1681.27 (4) Å3
Z = 4
Cu Kα radiation
μ = 2.15 mm−1
T = 296 K
0.28 × 0.12 × 0.10 mm
Data collection
Bruker SMART APEX diffractometer
12461 measured reflections
5446 independent reflections
5282 reflections with I > 2σ(I)
R int = 0.017
Refinement
R[F 2 > 2σ(F 2)] = 0.041
wR(F 2) = 0.117
S = 1.06
5446 reflections
462 parameters
H-atom parameters constrained
Δρmax = 0.41 e Å−3
Δρmin = −0.32 e Å−3
Data collection: SMART (Bruker, 2005 ▶); cell refinement: SAINT (Bruker, 2005 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: XP in SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810053730/cv5015sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810053730/cv5015Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2A—H2AA⋯OA | 0.86 | 1.95 | 2.771 (2) | 161 |
| OA—HA1⋯N1Ai | 0.83 | 2.00 | 2.806 (2) | 161 |
| N2B—H2BA⋯OB | 0.86 | 1.98 | 2.799 (3) | 160 |
| OB—HB1⋯N1Bii | 0.84 | 2.03 | 2.798 (3) | 152 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
This work was supported by the ‘New Drug Innovation 2009ZX09301–007’ project of the Ministry of Science and Technology of China.
supplementary crystallographic information
Comment
The title compound, (I), is important intermediate in the synthesis of lansoprazole (Del Rio et al., 2007; Reddy et al., 2008), which exhibits anti-ulcer effect (Iwahi et al.,1991). Herewith we present its crystal structure.
The asymmetric unit of (I) contains two independent molecules (Fig. 1), A and B, respectively, and two crystalline water molecules, one of which is disordered over two positions in a ratio 0.790 (8):0.210 (8). The molecular conformations of A and B are close - the benzimidazole mean plane and pyridine ring form the dihedral angles of 1.8 (3)° and 0.1 (2)° in A and B, respectively. The bond lengths and angles in A and B are normal and comparable with those observed in the related compounds (Swamy et al., 2007; Hakim,et al., 2010). The torsion angle of C7—S1—C8—C9 in A is 178.85 (12) ° (179.88 (14) ° in B).
The crystalline water molecules are involved in formation of two independent hydrogen-bonded chains via N—H···O and O—H···N hydrogen bonds (Table 1). The chains propagating along the axis a are formed by the molecule A and one independent water molecule, while the chains propagating along the axis b are formed by the molecule B and another independent water molecule. The crystal packing exhibits π-π interactions proved by short distances of 3.607 (3) and 3.701 (3) Å between the centroids of imidazole and pyridine rings from the neighbouring molecules.
Experimental
The raw material was kindly provided by Shanghai Enran Sci-Tech Investment Management Co., Ltd.The compound was dissolved in acetonitrile and suitable crystals of X-ray were obtained by slow evaporation at room temperature over a period of one week.
Refinement
Water H atoms were initially located in a difference Fourier map (O—H 0.80-0.85 Å), and refined as riding, with Uiso(H) = 1.5 Ueq(O). All other H atoms were constrained to an ideal geometry (C—H 0.93 - 0.97 Å; N—H 0.86 Å). All H atoms were refined as riding, with and Uiso(H) = 1.2 - 1.5 Ueq of the parent atom. One water molecule (OB) has been treated as disordered between two positions with the occupancies refined to 0.790 (8) and 0.210 (8), respectively.
Figures
Fig. 1.
The content of asymmetric unit (I) showing the atom-labelling scheme. Displacement ellipsoids are drawn at the 30% probability level. Dashed lines denote hydrogen bonds. Only major component of the disordered water molecule is shown. H atoms have been omitted for clarity.
Fig. 2.
A packing diagram.
Crystal data
| C16H14F3N3OS·H2O | Z = 4 |
| Mr = 371.39 | F(000) = 768 |
| Triclinic, P1 | Dx = 1.467 Mg m−3 |
| Hall symbol: -P 1 | Cu Kα radiation, λ = 1.54178 Å |
| a = 7.3526 (1) Å | Cell parameters from 9679 reflections |
| b = 7.4702 (1) Å | θ = 5.8–67.1° |
| c = 30.6500 (3) Å | µ = 2.15 mm−1 |
| α = 88.27° | T = 296 K |
| β = 87.79° | Prism, colourless |
| γ = 89.13° | 0.28 × 0.12 × 0.10 mm |
| V = 1681.27 (4) Å3 |
Data collection
| Bruker SMART APEX diffractometer | 5282 reflections with I > 2σ(I) |
| Radiation source: sealed tube | Rint = 0.017 |
| graphite | θmax = 67.4°, θmin = 4.3° |
| φ and ω scans | h = −7→8 |
| 12461 measured reflections | k = −8→8 |
| 5446 independent reflections | l = −33→36 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.041 | H-atom parameters constrained |
| wR(F2) = 0.117 | w = 1/[σ2(Fo2) + (0.0671P)2 + 0.6061P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.06 | (Δ/σ)max = 0.001 |
| 5446 reflections | Δρmax = 0.41 e Å−3 |
| 462 parameters | Δρmin = −0.32 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0062 (5) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| S1A | 0.20282 (6) | 0.80754 (6) | 0.034292 (14) | 0.04220 (15) | |
| F1A | −0.4248 (3) | 0.4400 (3) | −0.22095 (6) | 0.1133 (7) | |
| F2A | −0.5441 (2) | 0.6396 (2) | −0.18024 (5) | 0.0767 (4) | |
| F3A | −0.5417 (2) | 0.3690 (2) | −0.15787 (6) | 0.0898 (5) | |
| O1A | −0.28512 (19) | 0.57155 (19) | −0.11848 (4) | 0.0499 (3) | |
| N1A | −0.0279 (2) | 0.8860 (2) | 0.10435 (5) | 0.0449 (4) | |
| N2A | 0.2693 (2) | 0.9203 (2) | 0.11406 (5) | 0.0419 (3) | |
| H2AA | 0.3847 | 0.9196 | 0.1083 | 0.050* | |
| N3A | 0.1655 (2) | 0.6976 (2) | −0.04904 (5) | 0.0480 (4) | |
| C1A | 0.1826 (3) | 0.9679 (2) | 0.15275 (6) | 0.0414 (4) | |
| C2A | 0.2464 (3) | 1.0248 (3) | 0.19196 (7) | 0.0516 (5) | |
| H2AB | 0.3702 | 1.0347 | 0.1964 | 0.062* | |
| C3A | 0.1170 (3) | 1.0661 (3) | 0.22414 (7) | 0.0593 (6) | |
| H3AA | 0.1546 | 1.1059 | 0.2508 | 0.071* | |
| C4A | −0.0675 (3) | 1.0495 (3) | 0.21770 (7) | 0.0615 (6) | |
| H4AA | −0.1507 | 1.0806 | 0.2399 | 0.074* | |
| C5A | −0.1308 (3) | 0.9882 (3) | 0.17920 (7) | 0.0568 (5) | |
| H5AA | −0.2547 | 0.9751 | 0.1754 | 0.068* | |
| C6A | −0.0026 (3) | 0.9463 (2) | 0.14626 (6) | 0.0432 (4) | |
| C7A | 0.1375 (2) | 0.8745 (2) | 0.08674 (6) | 0.0390 (4) | |
| C8A | −0.0216 (2) | 0.7654 (2) | 0.01491 (6) | 0.0407 (4) | |
| H8AA | −0.0798 | 0.6726 | 0.0331 | 0.049* | |
| H8AB | −0.0961 | 0.8734 | 0.0167 | 0.049* | |
| C9A | −0.0052 (2) | 0.7070 (2) | −0.03177 (6) | 0.0384 (4) | |
| C10A | 0.1846 (3) | 0.6412 (3) | −0.08980 (7) | 0.0543 (5) | |
| H10A | 0.3019 | 0.6321 | −0.1021 | 0.065* | |
| C11A | 0.0431 (3) | 0.5958 (3) | −0.11481 (6) | 0.0504 (5) | |
| H11A | 0.0638 | 0.5563 | −0.1431 | 0.061* | |
| C12A | −0.1325 (3) | 0.6103 (2) | −0.09671 (6) | 0.0415 (4) | |
| C13A | −0.1595 (2) | 0.6663 (2) | −0.05382 (6) | 0.0388 (4) | |
| C14A | −0.2595 (3) | 0.5145 (3) | −0.16192 (6) | 0.0495 (5) | |
| H14A | −0.1914 | 0.4023 | −0.1624 | 0.059* | |
| H14B | −0.1921 | 0.6033 | −0.1794 | 0.059* | |
| C15A | −0.4423 (4) | 0.4908 (3) | −0.17966 (7) | 0.0618 (6) | |
| C16A | −0.3470 (3) | 0.6790 (3) | −0.03280 (7) | 0.0491 (5) | |
| H16A | −0.3389 | 0.7198 | −0.0035 | 0.074* | |
| H16B | −0.4021 | 0.5632 | −0.0321 | 0.074* | |
| H16C | −0.4198 | 0.7621 | −0.0494 | 0.074* | |
| OA | 0.6171 (2) | 0.9501 (2) | 0.07662 (6) | 0.0674 (4) | |
| HA1 | 0.7128 | 0.9071 | 0.0867 | 0.101* | |
| HA2 | 0.6392 | 1.0447 | 0.0630 | 0.101* | |
| S1B | 0.26211 (7) | 0.81787 (6) | 0.488240 (15) | 0.04885 (16) | |
| F1B | 0.0052 (3) | 1.4472 (3) | 0.75578 (5) | 0.1092 (7) | |
| F2B | 0.1852 (3) | 1.5488 (2) | 0.70486 (6) | 0.0992 (6) | |
| F3B | −0.0976 (3) | 1.5730 (2) | 0.69808 (6) | 0.1053 (6) | |
| O1B | 0.0650 (2) | 1.30722 (19) | 0.64771 (4) | 0.0576 (4) | |
| N1B | 0.3484 (2) | 1.0373 (2) | 0.41683 (5) | 0.0485 (4) | |
| N2B | 0.3662 (3) | 0.7438 (2) | 0.40584 (5) | 0.0506 (4) | |
| H2BA | 0.3605 | 0.6305 | 0.4114 | 0.061* | |
| N3B | 0.1553 (3) | 0.8612 (2) | 0.57285 (6) | 0.0578 (5) | |
| C1B | 0.4148 (3) | 0.8255 (3) | 0.36638 (6) | 0.0479 (4) | |
| C2B | 0.4657 (3) | 0.7591 (3) | 0.32581 (7) | 0.0604 (6) | |
| H2BB | 0.4733 | 0.6367 | 0.3212 | 0.072* | |
| C3B | 0.5043 (3) | 0.8839 (4) | 0.29259 (7) | 0.0652 (6) | |
| H3BA | 0.5386 | 0.8444 | 0.2649 | 0.078* | |
| C4B | 0.4932 (3) | 1.0663 (4) | 0.29952 (7) | 0.0648 (6) | |
| H4BA | 0.5205 | 1.1463 | 0.2765 | 0.078* | |
| C5B | 0.4428 (3) | 1.1314 (3) | 0.33978 (7) | 0.0605 (6) | |
| H5BA | 0.4354 | 1.2540 | 0.3442 | 0.073* | |
| C6B | 0.4032 (3) | 1.0091 (3) | 0.37361 (6) | 0.0480 (4) | |
| C7B | 0.3289 (3) | 0.8765 (3) | 0.43438 (6) | 0.0443 (4) | |
| C8B | 0.2354 (3) | 1.0420 (3) | 0.50903 (6) | 0.0473 (4) | |
| H8BA | 0.3498 | 1.1047 | 0.5058 | 0.057* | |
| H8BB | 0.1450 | 1.1088 | 0.4927 | 0.057* | |
| C9B | 0.1760 (3) | 1.0277 (3) | 0.55651 (6) | 0.0461 (4) | |
| C10B | 0.1024 (4) | 0.8433 (3) | 0.61464 (7) | 0.0637 (6) | |
| H10B | 0.0867 | 0.7280 | 0.6263 | 0.076* | |
| C11B | 0.0695 (3) | 0.9845 (3) | 0.64178 (7) | 0.0562 (5) | |
| H11B | 0.0327 | 0.9657 | 0.6709 | 0.067* | |
| C12B | 0.0929 (3) | 1.1550 (3) | 0.62422 (6) | 0.0479 (4) | |
| C13B | 0.1465 (3) | 1.1813 (3) | 0.58049 (6) | 0.0458 (4) | |
| C14B | 0.0293 (3) | 1.2837 (3) | 0.69304 (6) | 0.0558 (5) | |
| H14C | 0.1216 | 1.2065 | 0.7058 | 0.067* | |
| H14D | −0.0884 | 1.2288 | 0.6986 | 0.067* | |
| C15B | 0.0308 (4) | 1.4636 (4) | 0.71260 (8) | 0.0706 (7) | |
| C16B | 0.1682 (4) | 1.3666 (3) | 0.56065 (7) | 0.0620 (6) | |
| H16D | 0.1412 | 1.4535 | 0.5825 | 0.093* | |
| H16E | 0.0861 | 1.3836 | 0.5372 | 0.093* | |
| H16F | 0.2911 | 1.3811 | 0.5495 | 0.093* | |
| OB | 0.4154 (6) | 0.3918 (3) | 0.43764 (9) | 0.0886 (14) | 0.790 (8) |
| OB' | 0.251 (2) | 0.3984 (11) | 0.4323 (3) | 0.090 (5) | 0.210 (8) |
| HB1 | 0.3586 | 0.2971 | 0.4339 | 0.134* | 0.790 (8) |
| HB1' | 0.3361 | 0.3261 | 0.4312 | 0.134* | 0.210 (8) |
| HB2 | 0.5139 | 0.3549 | 0.4455 | 0.134* | 0.790 (8) |
| HB2' | 0.1581 | 0.3375 | 0.4272 | 0.134* | 0.210 (8) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1A | 0.0356 (3) | 0.0494 (3) | 0.0416 (3) | −0.00072 (17) | 0.00079 (18) | −0.00341 (18) |
| F1A | 0.0937 (13) | 0.179 (2) | 0.0719 (10) | 0.0062 (12) | −0.0171 (9) | −0.0662 (12) |
| F2A | 0.0676 (10) | 0.0855 (10) | 0.0770 (9) | 0.0115 (7) | −0.0118 (7) | 0.0009 (7) |
| F3A | 0.0722 (11) | 0.0777 (10) | 0.1210 (14) | −0.0239 (8) | −0.0155 (9) | −0.0013 (9) |
| O1A | 0.0464 (8) | 0.0642 (8) | 0.0394 (7) | −0.0068 (6) | 0.0000 (6) | −0.0081 (6) |
| N1A | 0.0361 (9) | 0.0526 (9) | 0.0458 (8) | −0.0014 (6) | −0.0003 (7) | −0.0009 (7) |
| N2A | 0.0322 (8) | 0.0490 (8) | 0.0445 (8) | −0.0006 (6) | −0.0006 (6) | −0.0029 (6) |
| N3A | 0.0378 (9) | 0.0584 (10) | 0.0476 (9) | −0.0039 (7) | 0.0042 (7) | −0.0056 (7) |
| C1A | 0.0427 (11) | 0.0390 (9) | 0.0423 (9) | 0.0002 (7) | 0.0001 (8) | 0.0014 (7) |
| C2A | 0.0512 (13) | 0.0535 (11) | 0.0506 (11) | −0.0028 (9) | −0.0058 (9) | −0.0042 (9) |
| C3A | 0.0729 (16) | 0.0612 (13) | 0.0440 (11) | −0.0026 (10) | 0.0011 (10) | −0.0059 (9) |
| C4A | 0.0666 (16) | 0.0698 (14) | 0.0469 (11) | 0.0042 (11) | 0.0141 (10) | −0.0032 (10) |
| C5A | 0.0449 (12) | 0.0698 (14) | 0.0547 (12) | 0.0005 (9) | 0.0086 (9) | 0.0013 (10) |
| C6A | 0.0416 (11) | 0.0453 (10) | 0.0424 (9) | 0.0005 (7) | 0.0013 (8) | 0.0016 (7) |
| C7A | 0.0359 (10) | 0.0379 (9) | 0.0431 (9) | 0.0007 (7) | −0.0021 (7) | 0.0024 (7) |
| C8A | 0.0368 (10) | 0.0440 (9) | 0.0411 (9) | −0.0010 (7) | −0.0004 (7) | −0.0010 (7) |
| C9A | 0.0375 (10) | 0.0368 (9) | 0.0404 (9) | −0.0013 (7) | 0.0023 (7) | 0.0012 (7) |
| C10A | 0.0404 (11) | 0.0708 (13) | 0.0513 (11) | −0.0056 (9) | 0.0109 (9) | −0.0083 (9) |
| C11A | 0.0508 (12) | 0.0587 (12) | 0.0414 (10) | −0.0043 (9) | 0.0089 (8) | −0.0065 (8) |
| C12A | 0.0425 (11) | 0.0407 (9) | 0.0410 (9) | −0.0049 (7) | −0.0005 (8) | 0.0010 (7) |
| C13A | 0.0398 (10) | 0.0361 (9) | 0.0400 (9) | −0.0011 (7) | 0.0036 (7) | 0.0010 (7) |
| C14A | 0.0578 (13) | 0.0492 (11) | 0.0416 (10) | 0.0000 (8) | 0.0010 (9) | −0.0073 (8) |
| C15A | 0.0673 (15) | 0.0666 (14) | 0.0526 (12) | 0.0008 (11) | −0.0056 (10) | −0.0176 (10) |
| C16A | 0.0382 (11) | 0.0578 (11) | 0.0512 (11) | −0.0022 (8) | 0.0025 (8) | −0.0053 (9) |
| OA | 0.0362 (9) | 0.0880 (12) | 0.0770 (11) | −0.0009 (7) | 0.0032 (7) | 0.0062 (9) |
| S1B | 0.0613 (3) | 0.0435 (3) | 0.0417 (3) | −0.0018 (2) | 0.0023 (2) | −0.00479 (19) |
| F1B | 0.184 (2) | 0.0958 (12) | 0.0464 (8) | 0.0074 (12) | 0.0272 (10) | −0.0194 (8) |
| F2B | 0.1211 (15) | 0.0910 (12) | 0.0870 (11) | −0.0297 (10) | 0.0105 (10) | −0.0319 (9) |
| F3B | 0.1288 (16) | 0.0833 (11) | 0.1016 (13) | 0.0405 (11) | 0.0145 (11) | −0.0096 (9) |
| O1B | 0.0808 (11) | 0.0539 (8) | 0.0378 (7) | 0.0024 (7) | 0.0050 (7) | −0.0076 (6) |
| N1B | 0.0569 (10) | 0.0439 (9) | 0.0445 (8) | 0.0010 (7) | 0.0025 (7) | −0.0065 (7) |
| N2B | 0.0636 (11) | 0.0399 (8) | 0.0485 (9) | −0.0004 (7) | 0.0033 (8) | −0.0091 (7) |
| N3B | 0.0814 (14) | 0.0469 (9) | 0.0449 (9) | −0.0054 (8) | 0.0032 (9) | −0.0030 (7) |
| C1B | 0.0470 (12) | 0.0527 (11) | 0.0443 (10) | 0.0000 (8) | −0.0010 (8) | −0.0089 (8) |
| C2B | 0.0628 (15) | 0.0654 (13) | 0.0539 (12) | −0.0007 (10) | 0.0010 (10) | −0.0197 (10) |
| C3B | 0.0583 (15) | 0.0947 (18) | 0.0430 (11) | −0.0019 (12) | 0.0022 (10) | −0.0127 (11) |
| C4B | 0.0622 (15) | 0.0843 (17) | 0.0470 (11) | −0.0021 (11) | 0.0016 (10) | 0.0063 (11) |
| C5B | 0.0670 (15) | 0.0596 (13) | 0.0545 (12) | −0.0006 (10) | 0.0006 (10) | 0.0029 (10) |
| C6B | 0.0473 (12) | 0.0528 (11) | 0.0438 (10) | 0.0017 (8) | −0.0009 (8) | −0.0043 (8) |
| C7B | 0.0458 (11) | 0.0443 (10) | 0.0430 (10) | −0.0001 (7) | −0.0016 (8) | −0.0080 (8) |
| C8B | 0.0554 (12) | 0.0438 (10) | 0.0428 (10) | −0.0037 (8) | 0.0031 (8) | −0.0070 (8) |
| C9B | 0.0480 (12) | 0.0484 (10) | 0.0420 (10) | −0.0038 (8) | −0.0018 (8) | −0.0031 (8) |
| C10B | 0.0935 (19) | 0.0489 (12) | 0.0483 (11) | −0.0076 (11) | 0.0039 (11) | 0.0012 (9) |
| C11B | 0.0701 (15) | 0.0587 (12) | 0.0393 (10) | −0.0053 (10) | 0.0018 (9) | 0.0002 (9) |
| C12B | 0.0508 (12) | 0.0532 (11) | 0.0404 (10) | −0.0020 (8) | −0.0022 (8) | −0.0087 (8) |
| C13B | 0.0499 (12) | 0.0472 (10) | 0.0407 (9) | −0.0037 (8) | −0.0012 (8) | −0.0037 (8) |
| C14B | 0.0641 (14) | 0.0643 (13) | 0.0386 (10) | 0.0024 (10) | 0.0058 (9) | −0.0062 (9) |
| C15B | 0.092 (2) | 0.0712 (15) | 0.0477 (12) | 0.0052 (14) | 0.0155 (12) | −0.0112 (11) |
| C16B | 0.0879 (18) | 0.0469 (11) | 0.0511 (12) | −0.0078 (10) | 0.0046 (11) | −0.0043 (9) |
| OB | 0.121 (4) | 0.0422 (11) | 0.1049 (19) | −0.0115 (12) | −0.0339 (17) | 0.0035 (11) |
| OB' | 0.119 (13) | 0.046 (4) | 0.103 (7) | 0.015 (5) | 0.006 (7) | −0.008 (4) |
Geometric parameters (Å, °)
| S1A—C7A | 1.7453 (18) | F1B—C15B | 1.332 (3) |
| S1A—C8A | 1.8107 (18) | F2B—C15B | 1.320 (3) |
| F1A—C15A | 1.333 (3) | F3B—C15B | 1.321 (3) |
| F2A—C15A | 1.331 (3) | O1B—C12B | 1.373 (2) |
| F3A—C15A | 1.324 (3) | O1B—C14B | 1.410 (2) |
| O1A—C12A | 1.367 (2) | N1B—C7B | 1.309 (3) |
| O1A—C14A | 1.415 (2) | N1B—C6B | 1.391 (2) |
| N1A—C7A | 1.314 (2) | N2B—C7B | 1.360 (2) |
| N1A—C6A | 1.394 (2) | N2B—C1B | 1.375 (3) |
| N2A—C7A | 1.359 (2) | N2B—H2BA | 0.8600 |
| N2A—C1A | 1.379 (2) | N3B—C10B | 1.328 (3) |
| N2A—H2AA | 0.8600 | N3B—C9B | 1.335 (3) |
| N3A—C10A | 1.332 (3) | C1B—C2B | 1.388 (3) |
| N3A—C9A | 1.344 (2) | C1B—C6B | 1.397 (3) |
| C1A—C2A | 1.387 (3) | C2B—C3B | 1.384 (4) |
| C1A—C6A | 1.396 (3) | C2B—H2BB | 0.9300 |
| C2A—C3A | 1.382 (3) | C3B—C4B | 1.386 (4) |
| C2A—H2AB | 0.9300 | C3B—H3BA | 0.9300 |
| C3A—C4A | 1.386 (4) | C4B—C5B | 1.376 (3) |
| C3A—H3AA | 0.9300 | C4B—H4BA | 0.9300 |
| C4A—C5A | 1.378 (3) | C5B—C6B | 1.386 (3) |
| C4A—H4AA | 0.9300 | C5B—H5BA | 0.9300 |
| C5A—C6A | 1.393 (3) | C8B—C9B | 1.504 (3) |
| C5A—H5AA | 0.9300 | C8B—H8BA | 0.9700 |
| C8A—C9A | 1.508 (2) | C8B—H8BB | 0.9700 |
| C8A—H8AA | 0.9700 | C9B—C13B | 1.391 (3) |
| C8A—H8AB | 0.9700 | C10B—C11B | 1.376 (3) |
| C9A—C13A | 1.385 (3) | C10B—H10B | 0.9300 |
| C10A—C11A | 1.369 (3) | C11B—C12B | 1.378 (3) |
| C10A—H10A | 0.9300 | C11B—H11B | 0.9300 |
| C11A—C12A | 1.389 (3) | C12B—C13B | 1.391 (3) |
| C11A—H11A | 0.9300 | C13B—C16B | 1.504 (3) |
| C12A—C13A | 1.398 (3) | C14B—C15B | 1.488 (3) |
| C13A—C16A | 1.502 (3) | C14B—H14C | 0.9700 |
| C14A—C15A | 1.485 (3) | C14B—H14D | 0.9700 |
| C14A—H14A | 0.9700 | C16B—H16D | 0.9600 |
| C14A—H14B | 0.9700 | C16B—H16E | 0.9600 |
| C16A—H16A | 0.9600 | C16B—H16F | 0.9600 |
| C16A—H16B | 0.9600 | OB—HB1 | 0.8399 |
| C16A—H16C | 0.9600 | OB—HB1' | 0.8013 |
| OA—HA1 | 0.8349 | OB—HB2 | 0.8132 |
| OA—HA2 | 0.8249 | OB'—HB1 | 1.0873 |
| S1B—C7B | 1.7490 (19) | OB'—HB1' | 0.8201 |
| S1B—C8B | 1.8146 (19) | OB'—HB2' | 0.8488 |
| C7A—S1A—C8A | 98.11 (8) | C7B—N2B—C1B | 106.89 (16) |
| C12A—O1A—C14A | 117.06 (15) | C7B—N2B—H2BA | 126.6 |
| C7A—N1A—C6A | 104.30 (16) | C1B—N2B—H2BA | 126.6 |
| C7A—N2A—C1A | 106.86 (15) | C10B—N3B—C9B | 117.15 (18) |
| C7A—N2A—H2AA | 126.6 | N2B—C1B—C2B | 132.8 (2) |
| C1A—N2A—H2AA | 126.6 | N2B—C1B—C6B | 105.28 (16) |
| C10A—N3A—C9A | 116.71 (17) | C2B—C1B—C6B | 122.0 (2) |
| N2A—C1A—C2A | 132.64 (19) | C3B—C2B—C1B | 116.8 (2) |
| N2A—C1A—C6A | 105.19 (16) | C3B—C2B—H2BB | 121.6 |
| C2A—C1A—C6A | 122.17 (18) | C1B—C2B—H2BB | 121.6 |
| C3A—C2A—C1A | 116.8 (2) | C2B—C3B—C4B | 121.6 (2) |
| C3A—C2A—H2AB | 121.6 | C2B—C3B—H3BA | 119.2 |
| C1A—C2A—H2AB | 121.6 | C4B—C3B—H3BA | 119.2 |
| C2A—C3A—C4A | 121.6 (2) | C5B—C4B—C3B | 121.4 (2) |
| C2A—C3A—H3AA | 119.2 | C5B—C4B—H4BA | 119.3 |
| C4A—C3A—H3AA | 119.2 | C3B—C4B—H4BA | 119.3 |
| C5A—C4A—C3A | 121.7 (2) | C4B—C5B—C6B | 118.1 (2) |
| C5A—C4A—H4AA | 119.2 | C4B—C5B—H5BA | 120.9 |
| C3A—C4A—H4AA | 119.2 | C6B—C5B—H5BA | 120.9 |
| C4A—C5A—C6A | 117.7 (2) | C5B—C6B—N1B | 130.08 (19) |
| C4A—C5A—H5AA | 121.2 | C5B—C6B—C1B | 120.15 (19) |
| C6A—C5A—H5AA | 121.2 | N1B—C6B—C1B | 109.77 (17) |
| C5A—C6A—N1A | 129.8 (2) | N1B—C7B—N2B | 113.33 (17) |
| C5A—C6A—C1A | 120.08 (19) | N1B—C7B—S1B | 127.96 (14) |
| N1A—C6A—C1A | 110.09 (16) | N2B—C7B—S1B | 118.72 (14) |
| N1A—C7A—N2A | 113.55 (16) | C9B—C8B—S1B | 108.66 (13) |
| N1A—C7A—S1A | 127.99 (14) | C9B—C8B—H8BA | 110.0 |
| N2A—C7A—S1A | 118.46 (13) | S1B—C8B—H8BA | 110.0 |
| C9A—C8A—S1A | 109.41 (12) | C9B—C8B—H8BB | 110.0 |
| C9A—C8A—H8AA | 109.8 | S1B—C8B—H8BB | 110.0 |
| S1A—C8A—H8AA | 109.8 | H8BA—C8B—H8BB | 108.3 |
| C9A—C8A—H8AB | 109.8 | N3B—C9B—C13B | 124.19 (18) |
| S1A—C8A—H8AB | 109.8 | N3B—C9B—C8B | 115.44 (17) |
| H8AA—C8A—H8AB | 108.2 | C13B—C9B—C8B | 120.37 (17) |
| N3A—C9A—C13A | 124.41 (16) | N3B—C10B—C11B | 124.2 (2) |
| N3A—C9A—C8A | 115.40 (16) | N3B—C10B—H10B | 117.9 |
| C13A—C9A—C8A | 120.19 (15) | C11B—C10B—H10B | 117.9 |
| N3A—C10A—C11A | 124.36 (18) | C10B—C11B—C12B | 117.57 (19) |
| N3A—C10A—H10A | 117.8 | C10B—C11B—H11B | 121.2 |
| C11A—C10A—H10A | 117.8 | C12B—C11B—H11B | 121.2 |
| C10A—C11A—C12A | 118.03 (18) | O1B—C12B—C11B | 123.44 (18) |
| C10A—C11A—H11A | 121.0 | O1B—C12B—C13B | 115.97 (17) |
| C12A—C11A—H11A | 121.0 | C11B—C12B—C13B | 120.59 (18) |
| O1A—C12A—C11A | 123.68 (17) | C9B—C13B—C12B | 116.33 (18) |
| O1A—C12A—C13A | 116.54 (16) | C9B—C13B—C16B | 122.53 (18) |
| C11A—C12A—C13A | 119.78 (18) | C12B—C13B—C16B | 121.12 (18) |
| C9A—C13A—C12A | 116.69 (16) | O1B—C14B—C15B | 107.66 (18) |
| C9A—C13A—C16A | 122.08 (16) | O1B—C14B—H14C | 110.2 |
| C12A—C13A—C16A | 121.23 (17) | C15B—C14B—H14C | 110.2 |
| O1A—C14A—C15A | 107.55 (17) | O1B—C14B—H14D | 110.2 |
| O1A—C14A—H14A | 110.2 | C15B—C14B—H14D | 110.2 |
| C15A—C14A—H14A | 110.2 | H14C—C14B—H14D | 108.5 |
| O1A—C14A—H14B | 110.2 | F2B—C15B—F3B | 105.5 (2) |
| C15A—C14A—H14B | 110.2 | F2B—C15B—F1B | 107.3 (2) |
| H14A—C14A—H14B | 108.5 | F3B—C15B—F1B | 107.3 (2) |
| F3A—C15A—F2A | 105.6 (2) | F2B—C15B—C14B | 113.3 (2) |
| F3A—C15A—F1A | 107.3 (2) | F3B—C15B—C14B | 113.3 (2) |
| F2A—C15A—F1A | 106.2 (2) | F1B—C15B—C14B | 109.8 (2) |
| F3A—C15A—C14A | 113.8 (2) | C13B—C16B—H16D | 109.5 |
| F2A—C15A—C14A | 113.79 (19) | C13B—C16B—H16E | 109.5 |
| F1A—C15A—C14A | 109.6 (2) | H16D—C16B—H16E | 109.5 |
| C13A—C16A—H16A | 109.5 | C13B—C16B—H16F | 109.5 |
| C13A—C16A—H16B | 109.5 | H16D—C16B—H16F | 109.5 |
| H16A—C16A—H16B | 109.5 | H16E—C16B—H16F | 109.5 |
| C13A—C16A—H16C | 109.5 | HB1—OB—HB1' | 19.7 |
| H16A—C16A—H16C | 109.5 | HB1—OB—HB2 | 102.8 |
| H16B—C16A—H16C | 109.5 | HB1'—OB—HB2 | 122.4 |
| HA1—OA—HA2 | 109.7 | HB1—OB'—HB1' | 5.6 |
| C7B—S1B—C8B | 98.24 (9) | HB1—OB'—HB2' | 102.8 |
| C12B—O1B—C14B | 116.91 (16) | HB1'—OB'—HB2' | 104.6 |
| C7B—N1B—C6B | 104.74 (16) | ||
| C7A—N2A—C1A—C2A | 179.6 (2) | C7B—N2B—C1B—C2B | −179.7 (2) |
| C7A—N2A—C1A—C6A | 0.33 (19) | C7B—N2B—C1B—C6B | −0.1 (2) |
| N2A—C1A—C2A—C3A | 178.4 (2) | N2B—C1B—C2B—C3B | 179.3 (2) |
| C6A—C1A—C2A—C3A | −2.4 (3) | C6B—C1B—C2B—C3B | −0.1 (3) |
| C1A—C2A—C3A—C4A | 0.7 (3) | C1B—C2B—C3B—C4B | 0.1 (4) |
| C2A—C3A—C4A—C5A | 1.2 (4) | C2B—C3B—C4B—C5B | −0.2 (4) |
| C3A—C4A—C5A—C6A | −1.3 (3) | C3B—C4B—C5B—C6B | 0.1 (4) |
| C4A—C5A—C6A—N1A | −178.5 (2) | C4B—C5B—C6B—N1B | −179.5 (2) |
| C4A—C5A—C6A—C1A | −0.3 (3) | C4B—C5B—C6B—C1B | −0.1 (3) |
| C7A—N1A—C6A—C5A | 177.7 (2) | C7B—N1B—C6B—C5B | 179.6 (2) |
| C7A—N1A—C6A—C1A | −0.6 (2) | C7B—N1B—C6B—C1B | 0.1 (2) |
| N2A—C1A—C6A—C5A | −178.36 (18) | N2B—C1B—C6B—C5B | −179.5 (2) |
| C2A—C1A—C6A—C5A | 2.2 (3) | C2B—C1B—C6B—C5B | 0.1 (3) |
| N2A—C1A—C6A—N1A | 0.2 (2) | N2B—C1B—C6B—N1B | 0.0 (2) |
| C2A—C1A—C6A—N1A | −179.23 (17) | C2B—C1B—C6B—N1B | 179.6 (2) |
| C6A—N1A—C7A—N2A | 0.8 (2) | C6B—N1B—C7B—N2B | −0.2 (2) |
| C6A—N1A—C7A—S1A | −179.74 (14) | C6B—N1B—C7B—S1B | 179.98 (16) |
| C1A—N2A—C7A—N1A | −0.8 (2) | C1B—N2B—C7B—N1B | 0.2 (2) |
| C1A—N2A—C7A—S1A | 179.75 (12) | C1B—N2B—C7B—S1B | −179.95 (15) |
| C8A—S1A—C7A—N1A | 0.36 (18) | C8B—S1B—C7B—N1B | −0.7 (2) |
| C8A—S1A—C7A—N2A | 179.76 (14) | C8B—S1B—C7B—N2B | 179.57 (16) |
| C7A—S1A—C8A—C9A | 178.84 (12) | C7B—S1B—C8B—C9B | 179.89 (14) |
| C10A—N3A—C9A—C13A | −1.6 (3) | C10B—N3B—C9B—C13B | 0.3 (3) |
| C10A—N3A—C9A—C8A | 178.03 (17) | C10B—N3B—C9B—C8B | −179.8 (2) |
| S1A—C8A—C9A—N3A | 0.16 (19) | S1B—C8B—C9B—N3B | −0.3 (2) |
| S1A—C8A—C9A—C13A | 179.79 (13) | S1B—C8B—C9B—C13B | 179.63 (16) |
| C9A—N3A—C10A—C11A | 1.0 (3) | C9B—N3B—C10B—C11B | −0.5 (4) |
| N3A—C10A—C11A—C12A | 0.4 (3) | N3B—C10B—C11B—C12B | 0.1 (4) |
| C14A—O1A—C12A—C11A | −0.3 (3) | C14B—O1B—C12B—C11B | 6.8 (3) |
| C14A—O1A—C12A—C13A | −179.93 (16) | C14B—O1B—C12B—C13B | −173.54 (19) |
| C10A—C11A—C12A—O1A | 179.03 (18) | C10B—C11B—C12B—O1B | −179.9 (2) |
| C10A—C11A—C12A—C13A | −1.4 (3) | C10B—C11B—C12B—C13B | 0.5 (3) |
| N3A—C9A—C13A—C12A | 0.7 (3) | N3B—C9B—C13B—C12B | 0.2 (3) |
| C8A—C9A—C13A—C12A | −178.93 (16) | C8B—C9B—C13B—C12B | −179.64 (18) |
| N3A—C9A—C13A—C16A | 179.90 (17) | N3B—C9B—C13B—C16B | −178.9 (2) |
| C8A—C9A—C13A—C16A | 0.3 (3) | C8B—C9B—C13B—C16B | 1.2 (3) |
| O1A—C12A—C13A—C9A | −179.52 (15) | O1B—C12B—C13B—C9B | 179.70 (18) |
| C11A—C12A—C13A—C9A | 0.8 (3) | C11B—C12B—C13B—C9B | −0.7 (3) |
| O1A—C12A—C13A—C16A | 1.2 (2) | O1B—C12B—C13B—C16B | −1.1 (3) |
| C11A—C12A—C13A—C16A | −178.39 (17) | C11B—C12B—C13B—C16B | 178.5 (2) |
| C12A—O1A—C14A—C15A | −176.73 (17) | C12B—O1B—C14B—C15B | 171.8 (2) |
| O1A—C14A—C15A—F3A | −60.9 (2) | O1B—C14B—C15B—F2B | −57.4 (3) |
| O1A—C14A—C15A—F2A | 60.1 (2) | O1B—C14B—C15B—F3B | 62.7 (3) |
| O1A—C14A—C15A—F1A | 178.9 (2) | O1B—C14B—C15B—F1B | −177.4 (2) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2A—H2AA···OA | 0.86 | 1.95 | 2.771 (2) | 161 |
| OA—HA1···N1Ai | 0.83 | 2.00 | 2.806 (2) | 161 |
| N2B—H2BA···OB | 0.86 | 1.98 | 2.799 (3) | 160 |
| OB—HB1···N1Bii | 0.84 | 2.03 | 2.798 (3) | 152 |
Symmetry codes: (i) x+1, y, z; (ii) x, y−1, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: CV5015).
References
- Bruker (2005). SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Del Rio, R. E., Wang, B., Achab, S. & Bohe, L. (2007). Org. Lett. 9, 2265–2268. [DOI] [PubMed]
- Hakim Al-arique, Q. N. M., Jasinski, J. P., Butcher, R. J., Yathirajan, H. S. & Narayana, B. (2010). Acta Cryst. E66, o1507–o1508. [DOI] [PMC free article] [PubMed]
- Iwahi, T., Satoh, H., Nakao, M., Iwasaki, T., Yamazaki, T., Kubo, K., Tamura, T. & Imada, A. (1991). Antimicrob. Agents Chemother. 35, 490–496. [DOI] [PMC free article] [PubMed]
- Reddy, G. M., Mukkanti, K., Kumar, T., Babu, J., Moses, M. & Reddy, P. P. (2008). Synth. Commun 38, 3477–3489.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Swamy, G. Y. S. K. & Ravikumar, K. (2007). J. Struct. Chem. 48, 715–718.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810053730/cv5015sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810053730/cv5015Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




