Abstract
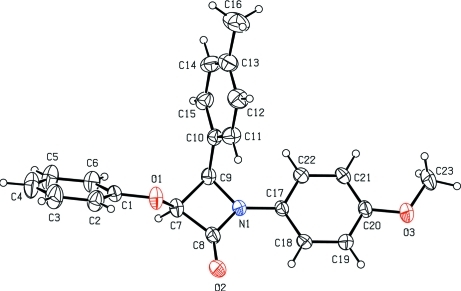
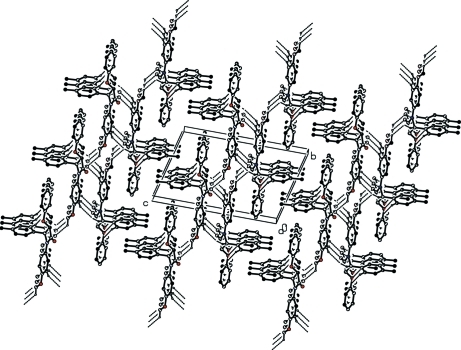
The central β-lactam ring of the title compound, C23H21NO3, is almost planar (r.m.s. deviation = 0.032Å). The methoxybenzene ring is almost coplanar with the β-lactam ring [dihedral angle = 1.87 (11)°], whereas the tolyl ring is almost normal to it [75.73 (12)°]. The dihedral angle between the β-lactam ring and the O-bonded phenyl ring is 51.95 (12)°. An intramolecular C—H⋯O interaction generates an S(6) ring. The crystal structure features intermolecular C—H⋯O hydrogen bonds, forming layers parallel to (011), and weak C—H⋯π interactions. Two aromatic π–π stacking interactions [centroid–centroid distances = 3.6744 (12) and 3.6799 (11) Å] are also observed.
Related literature
For the synthesis of the title compound and background to the biological properties of β-lactam compounds, see: Jarrahpour & Zarei (2010 ▶). For bond-length data, see: Allen et al. (1987 ▶). For hydrogen-bond motifs, see: Bernstein et al. (1995 ▶).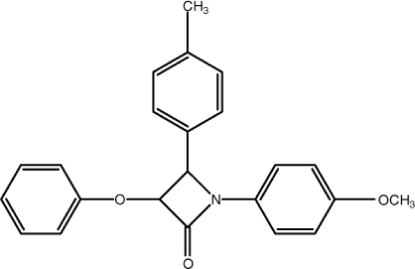
Experimental
Crystal data
C23H21NO3
M r = 359.41
Triclinic,
a = 6.0764 (3) Å
b = 9.9545 (5) Å
c = 16.4519 (10) Å
α = 104.360 (4)°
β = 91.261 (5)°
γ = 97.724 (4)°
V = 953.71 (9) Å3
Z = 2
Mo Kα radiation
μ = 0.08 mm−1
T = 296 K
0.60 × 0.37 × 0.12 mm
Data collection
Stoe IPDS 2 diffractometer
Absorption correction: integration (X-RED32; Stoe & Cie, 2002 ▶) T min = 0.964, T max = 0.990
13748 measured reflections
3961 independent reflections
2608 reflections with I > 2σ(I)
R int = 0.055
Refinement
R[F 2 > 2σ(F 2)] = 0.048
wR(F 2) = 0.119
S = 1.03
3961 reflections
246 parameters
H-atom parameters constrained
Δρmax = 0.13 e Å−3
Δρmin = −0.17 e Å−3
Data collection: X-AREA (Stoe & Cie, 2002 ▶); cell refinement: X-AREA; data reduction: X-RED32 (Stoe & Cie, 2002 ▶); program(s) used to solve structure: SIR97 (Altomare et al., 1999 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536811000663/hb5783sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811000663/hb5783Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg4 is the centroid of the C17–C22 benzene ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C18—H18⋯O2 | 0.93 | 2.47 | 3.093 (2) | 125 |
| C9—H9⋯O2i | 0.98 | 2.51 | 3.446 (2) | 160 |
| C15—H15⋯O1i | 0.93 | 2.54 | 3.435 (2) | 162 |
| C19—H19⋯O2ii | 0.93 | 2.54 | 3.415 (2) | 156 |
| C23—H23C⋯O2iii | 0.96 | 2.52 | 3.184 (2) | 126 |
| C5—H5⋯Cg4iv | 0.93 | 2.96 | 3.544 (3) | 122 |
Symmetry codes: (i) 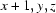 ; (ii)
; (ii) 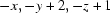 ; (iii)
; (iii) 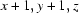 ; (iv)
; (iv) 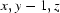 .
.
Acknowledgments
The authors acknowledge the Faculty of Arts and Sciences, Ondokuz Mayıs University, Turkey, for the use of the Stoe IPDS 2 diffractometer (purchased under grant F.279 of the University Research Fund). AJ and MR thank the Shiraz University Research Council for financial support (grant 88-GR—SC-23).
supplementary crystallographic information
Comment
The title compound, (I), is a β-lactam derivative with potential biological properties (Jarrahpour & Zarei, 2010); we now describe its crystal structure. In the title compound (I), (Fig. 1), the β-lactam ring (N1/C7–C9) is nearly planar [r.m.s. deviation = 0.032Å]. The dihedral angles between the planes of the rings in (I) are given in Table 2. The molecular dimensions are normal and lie within expected values for corresponding bond distances and angles (Allen et al., 1987).
The molecular structure is stabilized by intramolecular C—H···O hydrogen bonds, forming an S(6) graph-set motif (Bernstein et al., 1995) (Table 1). In the crystal structure, molecules are linked via intermolecular C—H···O hydrogen bonds (Table 1, Fig. 2), forming layers parallel to the bc plane. In addition, C—H···π interactions and two π-π stacking interactions [Cg1···Cg3(x, y, z) = 3.6744 (12) Å and Cg4···Cg4(1 - x, 2 - y, 1 - z) = 3.6799 (11) Å, where Cg1, Cg3 and Cg4 are the centroids of the N1/C7–C9 β-lactam, the C10–C15 and C17–C22 benzene rings, respectively] contribute to the stabilization of the structure.
Experimental
The title compound was prepared as described by Jarrahpour & Zarei (2010) and colourless prisms of (I) were recrystallized from ethyl acetate.
Refinement
All H atoms were placed at calculated positions and were treated as riding on their parent atoms with C—H = 0.93 (aromatic), 0.96(methyl) and 0.98 Å (methine), and with Uiso(H) = 1.5Ueq(C) for methyl and Uiso(H) = 1.2Ueq(C) for aromatic, methine.
Figures
Fig. 1.
The title compound with displacement ellipsoids for non-H atoms drawn at the 30% probability level.
Fig. 2.
View of the packing of (I) down the a axis. All H atoms are omitted for clarity.
Crystal data
| C23H21NO3 | Z = 2 |
| Mr = 359.41 | F(000) = 380 |
| Triclinic, P1 | Dx = 1.252 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 6.0764 (3) Å | Cell parameters from 19074 reflections |
| b = 9.9545 (5) Å | θ = 2.1–27.6° |
| c = 16.4519 (10) Å | µ = 0.08 mm−1 |
| α = 104.360 (4)° | T = 296 K |
| β = 91.261 (5)° | Prism, colourless |
| γ = 97.724 (4)° | 0.60 × 0.37 × 0.12 mm |
| V = 953.71 (9) Å3 |
Data collection
| Stoe IPDS 2 diffractometer | 3961 independent reflections |
| Radiation source: sealed X-ray tube, 12 x 0.4 mm long-fine focus | 2608 reflections with I > 2σ(I) |
| plane graphite | Rint = 0.055 |
| Detector resolution: 6.67 pixels mm-1 | θmax = 26.5°, θmin = 2.1° |
| ω scans | h = −7→7 |
| Absorption correction: integration (X-RED32; Stoe & Cie, 2002) | k = −12→12 |
| Tmin = 0.964, Tmax = 0.990 | l = −20→20 |
| 13748 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.048 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.119 | H-atom parameters constrained |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0548P)2 + 0.0685P] where P = (Fo2 + 2Fc2)/3 |
| 3961 reflections | (Δ/σ)max < 0.001 |
| 246 parameters | Δρmax = 0.13 e Å−3 |
| 0 restraints | Δρmin = −0.17 e Å−3 |
Special details
| Geometry. Bond distances, angles etc. have been calculated using the rounded fractional coordinates. All su's are estimated from the variances of the (full) variance-covariance matrix. The cell e.s.d.'s are taken into account in the estimation of distances, angles and torsion angles |
| Refinement. Refinement on F2 for ALL reflections except those flagged by the user for potential systematic errors. Weighted R-factors wR and all goodnesses of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The observed criterion of F2 > σ(F2) is used only for calculating -R-factor-obs etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R-factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.1189 (2) | 0.46929 (12) | 0.24918 (8) | 0.0593 (4) | |
| O2 | −0.0121 (2) | 0.69814 (13) | 0.39817 (9) | 0.0661 (5) | |
| O3 | 0.5557 (2) | 1.34079 (13) | 0.45143 (10) | 0.0691 (5) | |
| N1 | 0.3435 (2) | 0.76262 (13) | 0.35417 (9) | 0.0449 (4) | |
| C1 | 0.0980 (3) | 0.32558 (17) | 0.22621 (11) | 0.0466 (5) | |
| C2 | −0.0981 (3) | 0.2579 (2) | 0.18159 (14) | 0.0659 (7) | |
| C3 | −0.1326 (4) | 0.1146 (2) | 0.15480 (17) | 0.0855 (9) | |
| C4 | 0.0243 (5) | 0.0375 (2) | 0.17266 (18) | 0.0906 (10) | |
| C5 | 0.2170 (5) | 0.1043 (2) | 0.21619 (16) | 0.0841 (10) | |
| C6 | 0.2565 (3) | 0.2490 (2) | 0.24332 (13) | 0.0626 (7) | |
| C7 | 0.2563 (3) | 0.54632 (17) | 0.32009 (11) | 0.0493 (6) | |
| C8 | 0.1608 (3) | 0.67635 (17) | 0.36523 (11) | 0.0481 (6) | |
| C9 | 0.4601 (3) | 0.65118 (17) | 0.30514 (11) | 0.0456 (5) | |
| C10 | 0.5040 (3) | 0.66017 (17) | 0.21748 (11) | 0.0461 (5) | |
| C11 | 0.3614 (3) | 0.7126 (2) | 0.17006 (12) | 0.0590 (7) | |
| C12 | 0.4120 (4) | 0.7251 (2) | 0.09059 (14) | 0.0725 (8) | |
| C13 | 0.6024 (4) | 0.6850 (2) | 0.05514 (13) | 0.0711 (8) | |
| C14 | 0.7428 (3) | 0.6312 (2) | 0.10147 (15) | 0.0733 (8) | |
| C15 | 0.6962 (3) | 0.6196 (2) | 0.18176 (13) | 0.0603 (7) | |
| C16 | 0.6603 (5) | 0.7014 (3) | −0.03125 (17) | 0.1131 (13) | |
| C17 | 0.4029 (2) | 0.90918 (16) | 0.37839 (10) | 0.0417 (5) | |
| C18 | 0.2574 (3) | 0.99309 (17) | 0.42190 (11) | 0.0489 (6) | |
| C19 | 0.3149 (3) | 1.13583 (18) | 0.44533 (12) | 0.0523 (6) | |
| C20 | 0.5185 (3) | 1.19731 (17) | 0.42538 (11) | 0.0494 (6) | |
| C21 | 0.6635 (3) | 1.11455 (18) | 0.38189 (12) | 0.0514 (6) | |
| C22 | 0.6062 (3) | 0.97064 (18) | 0.35878 (11) | 0.0488 (6) | |
| C23 | 0.7547 (3) | 1.4105 (2) | 0.42925 (15) | 0.0691 (7) | |
| H2 | −0.20600 | 0.30950 | 0.16980 | 0.0790* | |
| H3 | −0.26370 | 0.06920 | 0.12420 | 0.1030* | |
| H4 | −0.00100 | −0.05980 | 0.15510 | 0.1090* | |
| H5 | 0.32400 | 0.05210 | 0.22790 | 0.1010* | |
| H6 | 0.38930 | 0.29390 | 0.27290 | 0.0750* | |
| H7 | 0.29430 | 0.48910 | 0.35750 | 0.0590* | |
| H9 | 0.59590 | 0.64290 | 0.33570 | 0.0550* | |
| H11 | 0.23000 | 0.73970 | 0.19210 | 0.0710* | |
| H12 | 0.31430 | 0.76160 | 0.06040 | 0.0870* | |
| H14 | 0.87190 | 0.60190 | 0.07840 | 0.0880* | |
| H15 | 0.79530 | 0.58410 | 0.21190 | 0.0720* | |
| H16A | 0.77970 | 0.77730 | −0.02580 | 0.1360* | |
| H16B | 0.53250 | 0.72090 | −0.05910 | 0.1360* | |
| H16C | 0.70560 | 0.61630 | −0.06360 | 0.1360* | |
| H18 | 0.12050 | 0.95250 | 0.43520 | 0.0590* | |
| H19 | 0.21710 | 1.19170 | 0.47470 | 0.0630* | |
| H21 | 0.79960 | 1.15550 | 0.36810 | 0.0620* | |
| H22 | 0.70480 | 0.91470 | 0.32990 | 0.0590* | |
| H23A | 0.75830 | 1.38950 | 0.36910 | 0.0830* | |
| H23B | 0.87980 | 1.37960 | 0.45200 | 0.0830* | |
| H23C | 0.76110 | 1.50980 | 0.45150 | 0.0830* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0676 (7) | 0.0352 (7) | 0.0707 (9) | −0.0008 (5) | −0.0211 (6) | 0.0116 (6) |
| O2 | 0.0539 (7) | 0.0533 (8) | 0.0853 (10) | −0.0075 (6) | 0.0196 (7) | 0.0132 (7) |
| O3 | 0.0758 (8) | 0.0372 (7) | 0.0866 (10) | −0.0080 (6) | 0.0139 (7) | 0.0091 (6) |
| N1 | 0.0449 (7) | 0.0359 (7) | 0.0502 (9) | −0.0019 (6) | 0.0055 (6) | 0.0079 (6) |
| C1 | 0.0499 (9) | 0.0379 (9) | 0.0504 (10) | 0.0010 (7) | 0.0033 (7) | 0.0112 (8) |
| C2 | 0.0555 (10) | 0.0520 (12) | 0.0820 (15) | 0.0035 (9) | −0.0070 (10) | 0.0048 (10) |
| C3 | 0.0791 (14) | 0.0564 (14) | 0.102 (2) | −0.0123 (11) | −0.0153 (13) | −0.0020 (12) |
| C4 | 0.125 (2) | 0.0410 (12) | 0.096 (2) | 0.0026 (13) | −0.0111 (16) | 0.0054 (12) |
| C5 | 0.1158 (19) | 0.0507 (13) | 0.0853 (17) | 0.0252 (12) | −0.0183 (14) | 0.0114 (12) |
| C6 | 0.0673 (11) | 0.0499 (11) | 0.0688 (13) | 0.0097 (9) | −0.0111 (10) | 0.0125 (10) |
| C7 | 0.0540 (9) | 0.0372 (9) | 0.0547 (11) | −0.0006 (7) | −0.0069 (8) | 0.0125 (8) |
| C8 | 0.0474 (9) | 0.0409 (9) | 0.0536 (11) | −0.0043 (7) | 0.0018 (8) | 0.0135 (8) |
| C9 | 0.0438 (8) | 0.0399 (9) | 0.0516 (10) | 0.0048 (7) | −0.0029 (7) | 0.0099 (8) |
| C10 | 0.0442 (8) | 0.0394 (9) | 0.0514 (11) | 0.0043 (7) | 0.0003 (7) | 0.0064 (8) |
| C11 | 0.0564 (10) | 0.0684 (13) | 0.0573 (12) | 0.0190 (9) | 0.0046 (9) | 0.0198 (10) |
| C12 | 0.0798 (14) | 0.0818 (16) | 0.0598 (13) | 0.0099 (11) | −0.0018 (11) | 0.0267 (11) |
| C13 | 0.0748 (13) | 0.0746 (15) | 0.0527 (13) | −0.0134 (11) | 0.0052 (10) | 0.0081 (11) |
| C14 | 0.0585 (11) | 0.0793 (15) | 0.0693 (15) | 0.0020 (10) | 0.0201 (11) | −0.0022 (12) |
| C15 | 0.0498 (9) | 0.0588 (12) | 0.0684 (14) | 0.0119 (8) | 0.0036 (9) | 0.0067 (10) |
| C16 | 0.127 (2) | 0.132 (3) | 0.0654 (17) | −0.0295 (19) | 0.0188 (15) | 0.0213 (16) |
| C17 | 0.0442 (8) | 0.0363 (9) | 0.0422 (9) | −0.0027 (6) | 0.0009 (7) | 0.0099 (7) |
| C18 | 0.0446 (8) | 0.0454 (10) | 0.0540 (11) | −0.0025 (7) | 0.0118 (7) | 0.0116 (8) |
| C19 | 0.0547 (9) | 0.0424 (10) | 0.0568 (11) | 0.0033 (8) | 0.0126 (8) | 0.0080 (8) |
| C20 | 0.0562 (9) | 0.0366 (9) | 0.0517 (11) | −0.0049 (7) | 0.0019 (8) | 0.0103 (8) |
| C21 | 0.0437 (8) | 0.0464 (10) | 0.0613 (12) | −0.0075 (7) | 0.0066 (8) | 0.0157 (8) |
| C22 | 0.0427 (8) | 0.0457 (10) | 0.0552 (11) | 0.0008 (7) | 0.0075 (7) | 0.0103 (8) |
| C23 | 0.0711 (12) | 0.0471 (11) | 0.0829 (15) | −0.0178 (9) | −0.0034 (10) | 0.0198 (10) |
Geometric parameters (Å, °)
| O1—C1 | 1.373 (2) | C18—C19 | 1.371 (3) |
| O1—C7 | 1.410 (2) | C19—C20 | 1.388 (3) |
| O2—C8 | 1.214 (2) | C20—C21 | 1.378 (3) |
| O3—C20 | 1.371 (2) | C21—C22 | 1.381 (3) |
| O3—C23 | 1.413 (2) | C2—H2 | 0.9300 |
| N1—C8 | 1.354 (2) | C3—H3 | 0.9300 |
| N1—C9 | 1.475 (2) | C4—H4 | 0.9300 |
| N1—C17 | 1.408 (2) | C5—H5 | 0.9300 |
| C1—C2 | 1.383 (3) | C6—H6 | 0.9300 |
| C1—C6 | 1.371 (3) | C7—H7 | 0.9800 |
| C2—C3 | 1.370 (3) | C9—H9 | 0.9800 |
| C3—C4 | 1.371 (4) | C11—H11 | 0.9300 |
| C4—C5 | 1.358 (4) | C12—H12 | 0.9300 |
| C5—C6 | 1.384 (3) | C14—H14 | 0.9300 |
| C7—C8 | 1.519 (3) | C15—H15 | 0.9300 |
| C7—C9 | 1.574 (3) | C16—H16A | 0.9600 |
| C9—C10 | 1.495 (2) | C16—H16B | 0.9600 |
| C10—C11 | 1.388 (3) | C16—H16C | 0.9600 |
| C10—C15 | 1.383 (3) | C18—H18 | 0.9300 |
| C11—C12 | 1.381 (3) | C19—H19 | 0.9300 |
| C12—C13 | 1.370 (3) | C21—H21 | 0.9300 |
| C13—C14 | 1.376 (3) | C22—H22 | 0.9300 |
| C13—C16 | 1.514 (3) | C23—H23A | 0.9600 |
| C14—C15 | 1.386 (3) | C23—H23B | 0.9600 |
| C17—C18 | 1.386 (2) | C23—H23C | 0.9600 |
| C17—C22 | 1.385 (2) | ||
| C1—O1—C7 | 120.21 (14) | C3—C2—H2 | 120.00 |
| C20—O3—C23 | 117.71 (15) | C2—C3—H3 | 120.00 |
| C8—N1—C9 | 96.00 (13) | C4—C3—H3 | 120.00 |
| C8—N1—C17 | 132.78 (13) | C3—C4—H4 | 120.00 |
| C9—N1—C17 | 131.20 (13) | C5—C4—H4 | 120.00 |
| O1—C1—C2 | 115.45 (16) | C4—C5—H5 | 120.00 |
| O1—C1—C6 | 124.67 (17) | C6—C5—H5 | 120.00 |
| C2—C1—C6 | 119.87 (17) | C1—C6—H6 | 120.00 |
| C1—C2—C3 | 119.70 (19) | C5—C6—H6 | 120.00 |
| C2—C3—C4 | 120.6 (2) | O1—C7—H7 | 113.00 |
| C3—C4—C5 | 119.5 (2) | C8—C7—H7 | 113.00 |
| C4—C5—C6 | 120.9 (2) | C9—C7—H7 | 113.00 |
| C1—C6—C5 | 119.4 (2) | N1—C9—H9 | 112.00 |
| O1—C7—C8 | 111.20 (14) | C7—C9—H9 | 112.00 |
| O1—C7—C9 | 117.37 (14) | C10—C9—H9 | 111.00 |
| C8—C7—C9 | 85.70 (13) | C10—C11—H11 | 120.00 |
| O2—C8—N1 | 132.64 (17) | C12—C11—H11 | 120.00 |
| O2—C8—C7 | 134.97 (17) | C11—C12—H12 | 119.00 |
| N1—C8—C7 | 92.39 (14) | C13—C12—H12 | 119.00 |
| N1—C9—C7 | 85.80 (12) | C13—C14—H14 | 119.00 |
| N1—C9—C10 | 115.03 (14) | C15—C14—H14 | 119.00 |
| C7—C9—C10 | 119.15 (15) | C10—C15—H15 | 120.00 |
| C9—C10—C11 | 122.47 (16) | C14—C15—H15 | 120.00 |
| C9—C10—C15 | 119.98 (17) | C13—C16—H16A | 109.00 |
| C11—C10—C15 | 117.52 (17) | C13—C16—H16B | 109.00 |
| C10—C11—C12 | 120.93 (18) | C13—C16—H16C | 109.00 |
| C11—C12—C13 | 121.6 (2) | H16A—C16—H16B | 110.00 |
| C12—C13—C14 | 117.7 (2) | H16A—C16—H16C | 109.00 |
| C12—C13—C16 | 121.6 (2) | H16B—C16—H16C | 109.00 |
| C14—C13—C16 | 120.7 (2) | C17—C18—H18 | 120.00 |
| C13—C14—C15 | 121.53 (19) | C19—C18—H18 | 120.00 |
| C10—C15—C14 | 120.74 (18) | C18—C19—H19 | 120.00 |
| N1—C17—C18 | 120.09 (13) | C20—C19—H19 | 120.00 |
| N1—C17—C22 | 120.40 (14) | C20—C21—H21 | 120.00 |
| C18—C17—C22 | 119.52 (16) | C22—C21—H21 | 120.00 |
| C17—C18—C19 | 120.12 (16) | C17—C22—H22 | 120.00 |
| C18—C19—C20 | 120.29 (17) | C21—C22—H22 | 120.00 |
| O3—C20—C19 | 114.80 (16) | O3—C23—H23A | 109.00 |
| O3—C20—C21 | 125.32 (16) | O3—C23—H23B | 109.00 |
| C19—C20—C21 | 119.88 (17) | O3—C23—H23C | 109.00 |
| C20—C21—C22 | 119.83 (17) | H23A—C23—H23B | 109.00 |
| C17—C22—C21 | 120.36 (16) | H23A—C23—H23C | 109.00 |
| C1—C2—H2 | 120.00 | H23B—C23—H23C | 109.00 |
| C7—O1—C1—C2 | 154.15 (17) | O1—C7—C8—N1 | −120.21 (15) |
| C7—O1—C1—C6 | −26.9 (3) | C9—C7—C8—O2 | 176.8 (2) |
| C1—O1—C7—C8 | −145.56 (15) | O1—C7—C9—C10 | −2.5 (2) |
| C1—O1—C7—C9 | 118.10 (17) | C8—C7—C9—N1 | 2.27 (12) |
| C23—O3—C20—C21 | 2.6 (3) | O1—C7—C8—O2 | 59.0 (3) |
| C23—O3—C20—C19 | −176.70 (17) | N1—C9—C10—C11 | −32.4 (2) |
| C8—N1—C9—C7 | −2.55 (13) | N1—C9—C10—C15 | 145.45 (17) |
| C17—N1—C8—O2 | 1.7 (3) | C7—C9—C10—C11 | 67.2 (2) |
| C9—N1—C8—O2 | −176.6 (2) | C7—C9—C10—C15 | −114.9 (2) |
| C9—N1—C8—C7 | 2.64 (14) | C9—C10—C15—C14 | −178.05 (18) |
| C17—N1—C8—C7 | −179.03 (17) | C15—C10—C11—C12 | −0.8 (3) |
| C17—N1—C9—C7 | 179.07 (16) | C9—C10—C11—C12 | 177.14 (18) |
| C9—N1—C17—C18 | 177.01 (16) | C11—C10—C15—C14 | −0.1 (3) |
| C8—N1—C17—C22 | 179.43 (17) | C10—C11—C12—C13 | 0.8 (3) |
| C9—N1—C17—C22 | −2.8 (3) | C11—C12—C13—C14 | 0.1 (3) |
| C8—N1—C17—C18 | −0.8 (3) | C11—C12—C13—C16 | −178.7 (2) |
| C8—N1—C9—C10 | 117.76 (16) | C12—C13—C14—C15 | −1.0 (3) |
| C17—N1—C9—C10 | −60.6 (2) | C16—C13—C14—C15 | 177.9 (2) |
| O1—C1—C6—C5 | −179.42 (19) | C13—C14—C15—C10 | 1.0 (3) |
| O1—C1—C2—C3 | 178.99 (19) | N1—C17—C22—C21 | 179.40 (16) |
| C6—C1—C2—C3 | 0.0 (3) | C18—C17—C22—C21 | −0.4 (3) |
| C2—C1—C6—C5 | −0.6 (3) | N1—C17—C18—C19 | −179.84 (16) |
| C1—C2—C3—C4 | 0.8 (4) | C22—C17—C18—C19 | −0.1 (3) |
| C2—C3—C4—C5 | −1.0 (4) | C17—C18—C19—C20 | 0.3 (3) |
| C3—C4—C5—C6 | 0.5 (4) | C18—C19—C20—C21 | 0.0 (3) |
| C4—C5—C6—C1 | 0.3 (4) | C18—C19—C20—O3 | 179.34 (17) |
| O1—C7—C9—N1 | 113.95 (15) | O3—C20—C21—C22 | −179.72 (17) |
| C8—C7—C9—C10 | −114.14 (17) | C19—C20—C21—C22 | −0.4 (3) |
| C9—C7—C8—N1 | −2.47 (13) | C20—C21—C22—C17 | 0.6 (3) |
Hydrogen-bond geometry (Å, °)
| Cg4 is the centroid of the C17–C22 benzene ring. |
| D—H···A | D—H | H···A | D···A | D—H···A |
| C18—H18···O2 | 0.93 | 2.47 | 3.093 (2) | 125 |
| C9—H9···O2i | 0.98 | 2.51 | 3.446 (2) | 160 |
| C15—H15···O1i | 0.93 | 2.54 | 3.435 (2) | 162 |
| C19—H19···O2ii | 0.93 | 2.54 | 3.415 (2) | 156 |
| C23—H23C···O2iii | 0.96 | 2.52 | 3.184 (2) | 126 |
| C5—H5···Cg4iv | 0.93 | 2.96 | 3.544 (3) | 122 |
Symmetry codes: (i) x+1, y, z; (ii) −x, −y+2, −z+1; (iii) x+1, y+1, z; (iv) x, y−1, z.
Table 2 The dihedral angles between the mean planes of the rings in (I) (°)
| Ring-2 | Ring-3 | Ring-4 | |
| Ring-1 | 51.95 (12) | 75.73 (12) | 1.87 (11) |
| Ring-2 | 86.93 (11) | 50.10 (10) | |
| Ring-3 | 76.54 (9) |
Ring-1 : N1/C7–C9 β-lactam ring, Ring-2 : C1–C6 phenyl ring, Ring-3 : C10–C15 benzene ring, Ring-4 : C17–C22 benzene ring.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HB5783).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Altomare, A., Burla, M. C., Camalli, M., Cascarano, G. L., Giacovazzo, C., Guagliardi, A., Moliterni, A. G. G., Polidori, G. & Spagna, R. (1999). J. Appl. Cryst. 32, 115–119.
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl. 34, 1555–1573.
- Farrugia, L. J. (1997). J. Appl. Cryst. 30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst. 32, 837–838.
- Jarrahpour, A. & Zarei, M. (2010). Tetrahedron, 66, 5017–5023.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Stoe & Cie (2002). X-AREA and X-RED32 Stoe & Cie, Darmstadt, Germany.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536811000663/hb5783sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811000663/hb5783Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report