Abstract
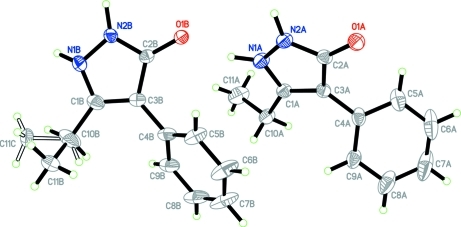
The asymmetric unit of the title compound, C11H12N2O, consists of two crystallographically independent molecules (A and B) with similar geometries. Both molecules exist in a keto form, the C=O bond length being 1.286 (2) Å in A and 1.283 (2) Å in B. The dihedral angles between the pyrazole ring and the attached phenyl ring are 43.28 (12) and 46.88 (11)°, respectively, for A and B. The ethyl unit in molecule B is disordered over two positions with a site-occupancy ratio of 0.508 (5):0.492 (5). In the crystal, each of the independent molecules forms a centrosymmetric dimer with an R 2 2(8) ring motif through a pair of N—H⋯O hydrogen bonds. These dimers are further connected into a three-dimensional network by intermolecular N—H⋯O and C—H⋯O hydrogen bonds. Intermolecular C—H⋯π interactions are also present.
Related literature
For background to pyrazole derivatives and their microbial activity, see: Ragavan et al. (2009 ▶, 2010 ▶). For bond-length data, see: Allen et al. (1987 ▶). For related structures, see: Loh et al. (2010 ▶, 2010a
▶,b
▶, 2011 ▶). For hydrogen-bond motifs, see: Bernstein et al. (1995 ▶). For the stability of the temperature controller used in the data collection, see: Cosier & Glazer (1986 ▶).
Experimental
Crystal data
C11H12N2O
M r = 188.23
Monoclinic,
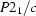
a = 11.0898 (3) Å
b = 13.2171 (4) Å
c = 15.0265 (5) Å
β = 114.539 (2)°
V = 2003.58 (11) Å3
Z = 8
Mo Kα radiation
μ = 0.08 mm−1
T = 100 K
0.60 × 0.16 × 0.13 mm
Data collection
Bruker SMART APEXII CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2009 ▶) T min = 0.953, T max = 0.989
22130 measured reflections
5845 independent reflections
3654 reflections with I > 2σ(I)
R int = 0.063
Refinement
R[F 2 > 2σ(F 2)] = 0.065
wR(F 2) = 0.166
S = 1.05
5845 reflections
284 parameters
2 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.38 e Å−3
Δρmin = −0.30 e Å−3
Data collection: APEX2 (Bruker, 2009 ▶); cell refinement: SAINT (Bruker, 2009 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536811001589/is2655sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811001589/is2655Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg1 and Cg2 are the centroids of the C4B–C9B and C4A–C9A rings, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1B—H1NB⋯O1A | 1.00 (2) | 1.73 (2) | 2.700 (2) | 161 (2) |
| N2B—H2NB⋯O1Bi | 1.02 (2) | 1.72 (2) | 2.738 (2) | 176 (2) |
| N2A—H2NA⋯O1Aii | 0.98 (3) | 1.74 (3) | 2.704 (2) | 171 (2) |
| N1A—H1NA⋯O1Biii | 0.98 (3) | 1.74 (3) | 2.691 (2) | 162 (2) |
| C8A—H8AA⋯O1Aiv | 0.93 | 2.47 | 3.370 (3) | 163 |
| C10A—H10C⋯Cg1iii | 0.97 | 2.61 | 3.464 (2) | 147 |
| C10B—H10E⋯Cg2 | 0.97 | 2.71 | 3.524 (3) | 142 |
Symmetry codes: (i) 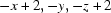 ; (ii)
; (ii) 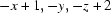 ; (iii)
; (iii) 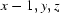 ; (iv)
; (iv) 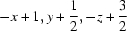 .
.
Acknowledgments
HKF and WSL thank Universiti Sains Malaysia (USM) for the Research University Grant (1001/PFIZIK/811160). WSL also thanks the Malaysian Government and USM for the award of a Research Fellowship. VV is grateful to the DST–India for funding through the Young Scientist Scheme (Fast Track Proposal).
supplementary crystallographic information
Comment
Antibacterial and antifungal activities of the azoles are most widely studied and some of them are in clinical practice as anti-microbial agents. However, the azole-resistant strains had led to the development of new anti-microbial compounds. In particular, pyrazole derivatives are extensively studied and used as anti-microbial agents. Pyrazole is an important class of heterocyclic compounds and many pyrazole derivatives are reported to have the broad spectrum of biological properties such as anti-inflammatory, antifungal, herbicidal, anti-tumour, cytotoxic, molecular modelling and antiviral activities. Pyrazole derivatives also act as anti-angiogenic agents, A3 adenosine receptor antagonists, neuropeptide YY5 receptor antagonists as well as kinase inhibitor for treatment of type 2 diabetes, hyperlipidemia, obesity and thrombopiotinmimetics. Recently urea derivatives of pyrazoles have been reported as potent inhibitors of p38 kinase. Since the high electronegativity of halogens (particularly chlorine and fluorine) in the aromatic part of the drug molecules play an important role in enhancing their biological activity, we are interested to have 4-fluoro or 4-chloro substitution in the aryls of 1,5-diaryl pyrazoles. As part of our on-going research aiming the synthesis of new anti-microbial compounds, we have reported the synthesis of novel pyrazole derivatives and their microbial activities (Ragavan et al., 2009, 2010).
The title compound (Fig. 1), consists of two crystallographically independent molecules, with similar geometries and exist in keto-form with the bond length of C═O being 1.286 (2) Å in molecule A and 1.283 (2) Å in molecule B. This indicates that the compound undergoes an enol-to-keto tautomerism during the crystallization process In molecule A, the pyrazole ring (N1A/N2A/C1A–C3A) is approximately planar [maximum deviation of 0.0262 (16) Å at N2A] and forms a dihedral angle of 43.28 (12)° with the attached phenyl ring (C4A–C9A). In molecule B, the pyrazole ring (N1B/N2B/C1B–C3B) is approximately planar with a maximum deviation of 0.0209 (15) Å at N1B and form a dihedral angle of 46.88 (11)° with the attached phenyl ring (C4B–C9B). The ethyl unit (C10B/C11B) in the molecule B is observed to be disordered over two positions with a site-occupancy ratio of 0.508 (5):0.492 (5). Bond lengths (Allen et al., 1987) and angles are within the normal ranges and are comparable to the related structures (Loh et al., 2010, 2011; Loh et al., 2010a,b).
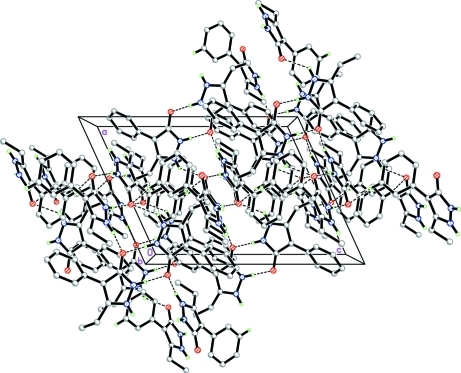
In the crystal packing (Fig. 2), intermolecular N2A—H2NA···O1A and N2B—H2NB···O1B hydrogen bonds (Table 1) link the neighbouring molecules to form dimers, generating R22(8) ring motifs (Bernstein et al., 1995) and are further packed into three-dimensional network by intermolecular N1B—H1NB···O1A, N1A—H1NA···O1B and C8A—H8AA···O1A hydrogen bonds (Table 1). The crystal structure is further stabilized by C—H···π interactions (Table 1) involving Cg1 (C4B–C9B) and Cg2 (C4A–C9A).
Experimental
The compound has been synthesized using the method available in the literature (Ragavan et al., 2010) and recrystallized using the ethanol-chloroform 1:1 mixture (yield 81%, m. p. 361.3–362.1 K).
Refinement
N-bound H atoms were located from a difference Fourier map and were refined freely [N—H = 0.97 (2) to 1.02 (2) Å]. The remaining H atoms were positioned geometrically with the bond length of C—H = 0.93 to 0.97 Å and were refined using a riding model, with Uiso(H) = 1.2 or 1.5Ueq(C). A rotating group model was applied to the methyl groups. The ethyl unit of molecule B was disordered over two positions with a site-occupancy of 0.508 (5):0.492 (5). Bond-distance restraints were applied for C10B—C11B and C10B—C11C.
Figures
Fig. 1.
The molecular structure of the title compound, showing 50% probability displacement ellipsoids and the atom-numbering scheme. Open bonds indicate the minor component.
Fig. 2.
The crystal packing of the title compound, showing the three-dimensional network. Only the major component is shown. H atoms not involved in the intermolecular interactions (dashed lines) have been omitted for clarity.
Crystal data
| C11H12N2O | F(000) = 800 |
| Mr = 188.23 | Dx = 1.248 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 4628 reflections |
| a = 11.0898 (3) Å | θ = 2.5–30.0° |
| b = 13.2171 (4) Å | µ = 0.08 mm−1 |
| c = 15.0265 (5) Å | T = 100 K |
| β = 114.539 (2)° | Needle, colourless |
| V = 2003.58 (11) Å3 | 0.60 × 0.16 × 0.13 mm |
| Z = 8 |
Data collection
| Bruker SMART APEXII CCD area-detector diffractometer | 5845 independent reflections |
| Radiation source: fine-focus sealed tube | 3654 reflections with I > 2σ(I) |
| graphite | Rint = 0.063 |
| φ and ω scans | θmax = 30.1°, θmin = 2.1° |
| Absorption correction: multi-scan (SADABS; Bruker, 2009) | h = −15→15 |
| Tmin = 0.953, Tmax = 0.989 | k = −18→18 |
| 22130 measured reflections | l = −20→21 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.065 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.166 | w = 1/[σ2(Fo2) + (0.0659P)2 + 0.5295P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.05 | (Δ/σ)max = 0.001 |
| 5845 reflections | Δρmax = 0.38 e Å−3 |
| 284 parameters | Δρmin = −0.30 e Å−3 |
| 2 restraints | Extinction correction: SHELXTL (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0163 (19) |
Special details
| Experimental. The crystal was placed in the cold stream of an Oxford Cryosystems Cobra open-flow nitrogen cryostat (Cosier & Glazer, 1986) operating at 100.0 (1) K. |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| O1A | 0.59450 (13) | 0.07629 (9) | 0.94578 (11) | 0.0314 (3) | |
| N1A | 0.29084 (17) | 0.16378 (11) | 0.92423 (13) | 0.0288 (4) | |
| N2A | 0.38633 (15) | 0.09048 (11) | 0.94491 (12) | 0.0271 (4) | |
| C1A | 0.32664 (18) | 0.24453 (13) | 0.88567 (14) | 0.0246 (4) | |
| C2A | 0.48806 (18) | 0.12736 (13) | 0.92718 (14) | 0.0244 (4) | |
| C3A | 0.44982 (17) | 0.22608 (12) | 0.88657 (14) | 0.0239 (4) | |
| C4A | 0.53044 (18) | 0.29233 (13) | 0.85342 (16) | 0.0298 (4) | |
| C5A | 0.6668 (2) | 0.30005 (15) | 0.90864 (18) | 0.0396 (5) | |
| H5AA | 0.7066 | 0.2645 | 0.9671 | 0.047* | |
| C6A | 0.7438 (2) | 0.36080 (18) | 0.8767 (3) | 0.0602 (8) | |
| H6AA | 0.8348 | 0.3652 | 0.9137 | 0.072* | |
| C7A | 0.6861 (3) | 0.41400 (18) | 0.7910 (3) | 0.0685 (10) | |
| H7AA | 0.7379 | 0.4549 | 0.7704 | 0.082* | |
| C8A | 0.5513 (3) | 0.40705 (19) | 0.7351 (2) | 0.0604 (8) | |
| H8AA | 0.5124 | 0.4430 | 0.6768 | 0.072* | |
| C9A | 0.4736 (2) | 0.34624 (16) | 0.76586 (18) | 0.0401 (5) | |
| H9AA | 0.3829 | 0.3415 | 0.7278 | 0.048* | |
| C10A | 0.23852 (19) | 0.33569 (14) | 0.85216 (16) | 0.0306 (4) | |
| H10C | 0.1769 | 0.3258 | 0.7845 | 0.037* | |
| H10D | 0.2927 | 0.3942 | 0.8546 | 0.037* | |
| C11A | 0.1600 (2) | 0.35798 (17) | 0.91215 (19) | 0.0450 (6) | |
| H11D | 0.1092 | 0.4187 | 0.8884 | 0.068* | |
| H11E | 0.2199 | 0.3668 | 0.9795 | 0.068* | |
| H11F | 0.1013 | 0.3025 | 0.9064 | 0.068* | |
| O1B | 1.07863 (12) | 0.06616 (9) | 0.92558 (9) | 0.0254 (3) | |
| N1B | 0.73764 (15) | 0.06391 (11) | 0.83821 (12) | 0.0275 (4) | |
| N2B | 0.86312 (14) | 0.04445 (11) | 0.90754 (12) | 0.0228 (3) | |
| C1B | 0.74826 (19) | 0.11417 (14) | 0.76342 (15) | 0.0303 (4) | |
| C2B | 0.95360 (17) | 0.07674 (12) | 0.87492 (13) | 0.0215 (4) | |
| C3B | 0.88097 (18) | 0.12138 (13) | 0.78120 (14) | 0.0251 (4) | |
| C4B | 0.9388 (2) | 0.16679 (13) | 0.71837 (14) | 0.0291 (4) | |
| C5B | 1.0386 (2) | 0.11721 (15) | 0.70194 (16) | 0.0361 (5) | |
| H5BA | 1.0694 | 0.0549 | 0.7315 | 0.043* | |
| C6B | 1.0928 (3) | 0.15955 (16) | 0.64204 (18) | 0.0505 (7) | |
| H6BA | 1.1588 | 0.1253 | 0.6312 | 0.061* | |
| C7B | 1.0483 (3) | 0.25308 (17) | 0.59830 (17) | 0.0541 (7) | |
| H7BA | 1.0840 | 0.2814 | 0.5579 | 0.065* | |
| C8B | 0.9506 (3) | 0.30381 (16) | 0.61513 (16) | 0.0459 (6) | |
| H8BA | 0.9209 | 0.3665 | 0.5862 | 0.055* | |
| C9B | 0.8973 (2) | 0.26163 (15) | 0.67475 (15) | 0.0360 (5) | |
| H9BA | 0.8326 | 0.2969 | 0.6862 | 0.043* | |
| C10B | 0.6280 (2) | 0.14952 (18) | 0.67717 (18) | 0.0499 (6) | |
| H10A | 0.5568 | 0.1581 | 0.6980 | 0.060* | 0.508 (5) |
| H10B | 0.6473 | 0.2157 | 0.6583 | 0.060* | 0.508 (5) |
| H10E | 0.6172 | 0.2209 | 0.6868 | 0.060* | 0.492 (5) |
| H10F | 0.6477 | 0.1442 | 0.6202 | 0.060* | 0.492 (5) |
| C11B | 0.5811 (4) | 0.0874 (3) | 0.5935 (3) | 0.0427 (13) | 0.508 (5) |
| H11A | 0.5027 | 0.1168 | 0.5442 | 0.064* | 0.508 (5) |
| H11B | 0.5608 | 0.0215 | 0.6104 | 0.064* | 0.508 (5) |
| H11C | 0.6481 | 0.0816 | 0.5689 | 0.064* | 0.508 (5) |
| C11C | 0.5046 (3) | 0.1031 (3) | 0.6533 (4) | 0.0395 (13) | 0.492 (5) |
| H11G | 0.4387 | 0.1358 | 0.5970 | 0.059* | 0.492 (5) |
| H11H | 0.4805 | 0.1090 | 0.7075 | 0.059* | 0.492 (5) |
| H11I | 0.5101 | 0.0329 | 0.6390 | 0.059* | 0.492 (5) |
| H1NB | 0.669 (2) | 0.0719 (16) | 0.8648 (17) | 0.043 (6)* | |
| H2NB | 0.881 (2) | 0.0033 (18) | 0.9694 (18) | 0.053 (7)* | |
| H2NA | 0.384 (2) | 0.029 (2) | 0.9802 (19) | 0.060 (8)* | |
| H1NA | 0.204 (3) | 0.1404 (18) | 0.9184 (19) | 0.052 (7)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1A | 0.0284 (7) | 0.0292 (7) | 0.0398 (9) | 0.0114 (5) | 0.0174 (7) | 0.0106 (6) |
| N1A | 0.0297 (9) | 0.0254 (8) | 0.0368 (10) | 0.0094 (6) | 0.0193 (8) | 0.0101 (6) |
| N2A | 0.0286 (8) | 0.0247 (8) | 0.0325 (9) | 0.0087 (6) | 0.0172 (7) | 0.0083 (6) |
| C1A | 0.0265 (9) | 0.0235 (8) | 0.0255 (10) | 0.0036 (7) | 0.0125 (8) | 0.0027 (7) |
| C2A | 0.0246 (9) | 0.0250 (8) | 0.0234 (10) | 0.0043 (7) | 0.0097 (7) | 0.0020 (7) |
| C3A | 0.0234 (9) | 0.0223 (8) | 0.0256 (10) | 0.0022 (6) | 0.0097 (7) | 0.0024 (7) |
| C4A | 0.0229 (9) | 0.0237 (9) | 0.0442 (13) | 0.0030 (7) | 0.0153 (9) | 0.0036 (8) |
| C5A | 0.0263 (10) | 0.0321 (10) | 0.0574 (15) | 0.0007 (8) | 0.0144 (10) | −0.0018 (10) |
| C6A | 0.0270 (12) | 0.0389 (13) | 0.116 (3) | −0.0031 (9) | 0.0309 (15) | −0.0020 (14) |
| C7A | 0.0510 (16) | 0.0381 (13) | 0.141 (3) | 0.0047 (11) | 0.064 (2) | 0.0232 (16) |
| C8A | 0.0510 (15) | 0.0508 (14) | 0.100 (2) | 0.0200 (11) | 0.0524 (16) | 0.0411 (14) |
| C9A | 0.0293 (11) | 0.0391 (11) | 0.0591 (16) | 0.0110 (8) | 0.0255 (11) | 0.0210 (10) |
| C10A | 0.0316 (10) | 0.0290 (9) | 0.0372 (12) | 0.0101 (7) | 0.0203 (9) | 0.0095 (8) |
| C11A | 0.0548 (15) | 0.0391 (12) | 0.0558 (16) | 0.0228 (10) | 0.0375 (13) | 0.0147 (10) |
| O1B | 0.0221 (6) | 0.0288 (6) | 0.0264 (7) | 0.0022 (5) | 0.0111 (6) | 0.0067 (5) |
| N1B | 0.0201 (8) | 0.0305 (8) | 0.0294 (9) | 0.0028 (6) | 0.0076 (7) | 0.0030 (6) |
| N2B | 0.0189 (7) | 0.0276 (7) | 0.0221 (8) | 0.0029 (5) | 0.0087 (6) | 0.0017 (6) |
| C1B | 0.0321 (10) | 0.0248 (9) | 0.0266 (10) | 0.0025 (7) | 0.0049 (8) | 0.0028 (7) |
| C2B | 0.0249 (9) | 0.0212 (8) | 0.0217 (9) | 0.0026 (6) | 0.0129 (7) | 0.0004 (6) |
| C3B | 0.0307 (10) | 0.0229 (8) | 0.0213 (9) | 0.0022 (7) | 0.0104 (8) | 0.0019 (7) |
| C4B | 0.0410 (11) | 0.0255 (9) | 0.0186 (9) | −0.0031 (7) | 0.0102 (8) | 0.0008 (7) |
| C5B | 0.0589 (14) | 0.0258 (9) | 0.0337 (12) | −0.0007 (9) | 0.0294 (11) | 0.0013 (8) |
| C6B | 0.092 (2) | 0.0353 (11) | 0.0465 (15) | −0.0071 (11) | 0.0504 (15) | −0.0031 (10) |
| C7B | 0.104 (2) | 0.0381 (12) | 0.0357 (13) | −0.0155 (13) | 0.0448 (15) | −0.0001 (10) |
| C8B | 0.0787 (18) | 0.0286 (10) | 0.0245 (11) | −0.0093 (10) | 0.0156 (12) | 0.0046 (8) |
| C9B | 0.0488 (13) | 0.0281 (10) | 0.0239 (10) | −0.0020 (8) | 0.0081 (9) | 0.0042 (7) |
| C10B | 0.0427 (13) | 0.0442 (13) | 0.0400 (14) | 0.0094 (10) | −0.0054 (11) | 0.0093 (10) |
| C11B | 0.032 (2) | 0.053 (3) | 0.033 (3) | 0.0001 (18) | 0.0032 (19) | 0.0136 (19) |
| C11C | 0.021 (2) | 0.047 (3) | 0.047 (3) | −0.0040 (17) | 0.0109 (19) | 0.014 (2) |
Geometric parameters (Å, °)
| O1A—C2A | 1.286 (2) | N1B—H1NB | 1.00 (2) |
| N1A—C1A | 1.350 (2) | N2B—C2B | 1.356 (2) |
| N1A—N2A | 1.372 (2) | N2B—H2NB | 1.02 (3) |
| N1A—H1NA | 0.98 (2) | C1B—C3B | 1.386 (3) |
| N2A—C2A | 1.353 (2) | C1B—C10B | 1.497 (3) |
| N2A—H2NA | 0.98 (3) | C2B—C3B | 1.427 (2) |
| C1A—C3A | 1.382 (2) | C3B—C4B | 1.471 (3) |
| C1A—C10A | 1.500 (2) | C4B—C5B | 1.394 (3) |
| C2A—C3A | 1.428 (2) | C4B—C9B | 1.401 (3) |
| C3A—C4A | 1.478 (2) | C5B—C6B | 1.391 (3) |
| C4A—C5A | 1.394 (3) | C5B—H5BA | 0.9300 |
| C4A—C9A | 1.396 (3) | C6B—C7B | 1.391 (3) |
| C5A—C6A | 1.395 (3) | C6B—H6BA | 0.9300 |
| C5A—H5AA | 0.9300 | C7B—C8B | 1.383 (4) |
| C6A—C7A | 1.371 (4) | C7B—H7BA | 0.9300 |
| C6A—H6AA | 0.9300 | C8B—C9B | 1.379 (3) |
| C7A—C8A | 1.381 (4) | C8B—H8BA | 0.9300 |
| C7A—H7AA | 0.9300 | C9B—H9BA | 0.9300 |
| C8A—C9A | 1.391 (3) | C10B—C11C | 1.403 (3) |
| C8A—H8AA | 0.9300 | C10B—C11B | 1.408 (3) |
| C9A—H9AA | 0.9300 | C10B—H10A | 0.9700 |
| C10A—C11A | 1.520 (3) | C10B—H10B | 0.9700 |
| C10A—H10C | 0.9700 | C10B—H10E | 0.9700 |
| C10A—H10D | 0.9700 | C10B—H10F | 0.9700 |
| C11A—H11D | 0.9600 | C11B—H11A | 0.9600 |
| C11A—H11E | 0.9600 | C11B—H11B | 0.9600 |
| C11A—H11F | 0.9600 | C11B—H11C | 0.9600 |
| O1B—C2B | 1.283 (2) | C11C—H11G | 0.9600 |
| N1B—C1B | 1.352 (2) | C11C—H11H | 0.9600 |
| N1B—N2B | 1.372 (2) | C11C—H11I | 0.9600 |
| C1A—N1A—N2A | 108.56 (15) | N1B—N2B—H2NB | 123.0 (14) |
| C1A—N1A—H1NA | 131.6 (14) | N1B—C1B—C3B | 109.01 (16) |
| N2A—N1A—H1NA | 115.8 (14) | N1B—C1B—C10B | 121.27 (19) |
| C2A—N2A—N1A | 109.22 (15) | C3B—C1B—C10B | 129.69 (19) |
| C2A—N2A—H2NA | 128.0 (15) | O1B—C2B—N2B | 122.08 (16) |
| N1A—N2A—H2NA | 121.5 (15) | O1B—C2B—C3B | 131.19 (16) |
| N1A—C1A—C3A | 108.78 (15) | N2B—C2B—C3B | 106.72 (16) |
| N1A—C1A—C10A | 120.80 (16) | C1B—C3B—C2B | 106.31 (16) |
| C3A—C1A—C10A | 130.40 (16) | C1B—C3B—C4B | 127.96 (17) |
| O1A—C2A—N2A | 122.23 (16) | C2B—C3B—C4B | 125.72 (17) |
| O1A—C2A—C3A | 130.95 (17) | C5B—C4B—C9B | 117.91 (18) |
| N2A—C2A—C3A | 106.82 (15) | C5B—C4B—C3B | 120.77 (16) |
| C1A—C3A—C2A | 106.39 (15) | C9B—C4B—C3B | 121.31 (18) |
| C1A—C3A—C4A | 128.76 (15) | C6B—C5B—C4B | 120.9 (2) |
| C2A—C3A—C4A | 124.86 (16) | C6B—C5B—H5BA | 119.5 |
| C5A—C4A—C9A | 118.53 (19) | C4B—C5B—H5BA | 119.5 |
| C5A—C4A—C3A | 120.08 (18) | C5B—C6B—C7B | 120.0 (2) |
| C9A—C4A—C3A | 121.37 (17) | C5B—C6B—H6BA | 120.0 |
| C4A—C5A—C6A | 120.4 (2) | C7B—C6B—H6BA | 120.0 |
| C4A—C5A—H5AA | 119.8 | C8B—C7B—C6B | 119.7 (2) |
| C6A—C5A—H5AA | 119.8 | C8B—C7B—H7BA | 120.1 |
| C7A—C6A—C5A | 120.3 (2) | C6B—C7B—H7BA | 120.1 |
| C7A—C6A—H6AA | 119.8 | C9B—C8B—C7B | 120.1 (2) |
| C5A—C6A—H6AA | 119.8 | C9B—C8B—H8BA | 119.9 |
| C6A—C7A—C8A | 120.2 (2) | C7B—C8B—H8BA | 119.9 |
| C6A—C7A—H7AA | 119.9 | C8B—C9B—C4B | 121.3 (2) |
| C8A—C7A—H7AA | 119.9 | C8B—C9B—H9BA | 119.3 |
| C7A—C8A—C9A | 120.0 (2) | C4B—C9B—H9BA | 119.3 |
| C7A—C8A—H8AA | 120.0 | C11C—C10B—C1B | 120.6 (3) |
| C9A—C8A—H8AA | 120.0 | C11B—C10B—C1B | 117.2 (2) |
| C8A—C9A—C4A | 120.6 (2) | C11B—C10B—H10A | 108.0 |
| C8A—C9A—H9AA | 119.7 | C1B—C10B—H10A | 108.0 |
| C4A—C9A—H9AA | 119.7 | C11B—C10B—H10B | 108.0 |
| C1A—C10A—C11A | 114.22 (16) | C1B—C10B—H10B | 108.0 |
| C1A—C10A—H10C | 108.7 | H10A—C10B—H10B | 107.2 |
| C11A—C10A—H10C | 108.7 | C11C—C10B—H10E | 107.2 |
| C1A—C10A—H10D | 108.7 | C1B—C10B—H10E | 107.2 |
| C11A—C10A—H10D | 108.7 | C11C—C10B—H10F | 107.2 |
| H10C—C10A—H10D | 107.6 | C1B—C10B—H10F | 107.2 |
| C10A—C11A—H11D | 109.5 | H10E—C10B—H10F | 106.8 |
| C10A—C11A—H11E | 109.5 | C10B—C11B—H11A | 109.5 |
| H11D—C11A—H11E | 109.5 | C10B—C11B—H11B | 109.5 |
| C10A—C11A—H11F | 109.5 | C10B—C11B—H11C | 109.5 |
| H11D—C11A—H11F | 109.5 | C10B—C11C—H11G | 109.5 |
| H11E—C11A—H11F | 109.5 | C10B—C11C—H11H | 109.5 |
| C1B—N1B—N2B | 108.18 (15) | H11G—C11C—H11H | 109.5 |
| C1B—N1B—H1NB | 128.5 (13) | C10B—C11C—H11I | 109.5 |
| N2B—N1B—H1NB | 114.6 (14) | H11G—C11C—H11I | 109.5 |
| C2B—N2B—N1B | 109.63 (15) | H11H—C11C—H11I | 109.5 |
| C2B—N2B—H2NB | 126.9 (14) | ||
| C1A—N1A—N2A—C2A | 5.0 (2) | N2B—N1B—C1B—C3B | 4.0 (2) |
| N2A—N1A—C1A—C3A | −3.5 (2) | N2B—N1B—C1B—C10B | −177.96 (18) |
| N2A—N1A—C1A—C10A | 177.71 (17) | N1B—N2B—C2B—O1B | −179.06 (15) |
| N1A—N2A—C2A—O1A | 175.93 (17) | N1B—N2B—C2B—C3B | 1.65 (18) |
| N1A—N2A—C2A—C3A | −4.4 (2) | N1B—C1B—C3B—C2B | −2.9 (2) |
| N1A—C1A—C3A—C2A | 0.8 (2) | C10B—C1B—C3B—C2B | 179.2 (2) |
| C10A—C1A—C3A—C2A | 179.43 (19) | N1B—C1B—C3B—C4B | 177.89 (17) |
| N1A—C1A—C3A—C4A | −179.05 (19) | C10B—C1B—C3B—C4B | 0.1 (3) |
| C10A—C1A—C3A—C4A | −0.4 (3) | O1B—C2B—C3B—C1B | −178.43 (18) |
| O1A—C2A—C3A—C1A | −178.1 (2) | N2B—C2B—C3B—C1B | 0.77 (19) |
| N2A—C2A—C3A—C1A | 2.2 (2) | O1B—C2B—C3B—C4B | 0.8 (3) |
| O1A—C2A—C3A—C4A | 1.7 (3) | N2B—C2B—C3B—C4B | 179.96 (16) |
| N2A—C2A—C3A—C4A | −177.92 (18) | C1B—C3B—C4B—C5B | −134.8 (2) |
| C1A—C3A—C4A—C5A | 138.1 (2) | C2B—C3B—C4B—C5B | 46.2 (3) |
| C2A—C3A—C4A—C5A | −41.7 (3) | C1B—C3B—C4B—C9B | 46.4 (3) |
| C1A—C3A—C4A—C9A | −43.5 (3) | C2B—C3B—C4B—C9B | −132.6 (2) |
| C2A—C3A—C4A—C9A | 136.7 (2) | C9B—C4B—C5B—C6B | −1.6 (3) |
| C9A—C4A—C5A—C6A | 0.2 (3) | C3B—C4B—C5B—C6B | 179.5 (2) |
| C3A—C4A—C5A—C6A | 178.6 (2) | C4B—C5B—C6B—C7B | 0.6 (4) |
| C4A—C5A—C6A—C7A | 0.4 (4) | C5B—C6B—C7B—C8B | 0.4 (4) |
| C5A—C6A—C7A—C8A | −0.6 (4) | C6B—C7B—C8B—C9B | −0.2 (4) |
| C6A—C7A—C8A—C9A | 0.2 (4) | C7B—C8B—C9B—C4B | −0.8 (3) |
| C7A—C8A—C9A—C4A | 0.4 (4) | C5B—C4B—C9B—C8B | 1.8 (3) |
| C5A—C4A—C9A—C8A | −0.6 (3) | C3B—C4B—C9B—C8B | −179.4 (2) |
| C3A—C4A—C9A—C8A | −179.0 (2) | N1B—C1B—C10B—C11C | −22.8 (4) |
| N1A—C1A—C10A—C11A | 33.3 (3) | C3B—C1B—C10B—C11C | 154.8 (3) |
| C3A—C1A—C10A—C11A | −145.2 (2) | N1B—C1B—C10B—C11B | −96.9 (3) |
| C1B—N1B—N2B—C2B | −3.53 (19) | C3B—C1B—C10B—C11B | 80.7 (3) |
Hydrogen-bond geometry (Å, °)
| Cg1 and Cg2 are the centroids of the C4B–C9B and C4A–C9A rings, respectively. |
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1B—H1NB···O1A | 1.00 (2) | 1.73 (2) | 2.700 (2) | 161 (2) |
| N2B—H2NB···O1Bi | 1.02 (2) | 1.72 (2) | 2.738 (2) | 176 (2) |
| N2A—H2NA···O1Aii | 0.98 (3) | 1.74 (3) | 2.704 (2) | 171 (2) |
| N1A—H1NA···O1Biii | 0.98 (3) | 1.74 (3) | 2.691 (2) | 162 (2) |
| C8A—H8AA···O1Aiv | 0.93 | 2.47 | 3.370 (3) | 163 |
| C10A—H10C···Cg1iii | 0.97 | 2.61 | 3.464 (2) | 147 |
| C10B—H10E···Cg2 | 0.97 | 2.71 | 3.524 (3) | 142 |
Symmetry codes: (i) −x+2, −y, −z+2; (ii) −x+1, −y, −z+2; (iii) x−1, y, z; (iv) −x+1, y+1/2, −z+3/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: IS2655).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl. 34, 1555–1573.
- Bruker (2009). APEX2, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Cosier, J. & Glazer, A. M. (1986). J. Appl. Cryst. 19, 105–107.
- Loh, W.-S., Fun, H.-K., Ragavan, R. V., Vijayakumar, V. & Sarveswari, S. (2010). Acta Cryst. E66, o2925. [DOI] [PMC free article] [PubMed]
- Loh, W.-S., Fun, H.-K., Ragavan, R. V., Vijayakumar, V. & Sarveswari, S. (2011). Acta Cryst. E67, o151–o152. [DOI] [PMC free article] [PubMed]
- Loh, W.-S., Fun, H.-K., Ragavan, R. V., Vijayakumar, V. & Venkatesh, M. (2010a). Acta Cryst. E66, o2563–o2564. [DOI] [PMC free article] [PubMed]
- Loh, W.-S., Fun, H.-K., Ragavan, R. V., Vijayakumar, V. & Venkatesh, M. (2010b). Acta Cryst. E66, o3050–o3051. [DOI] [PMC free article] [PubMed]
- Ragavan, R. V., Vijayakumar, V. & Sucheta Kumari, N. (2009). Eur. J. Med. Chem. 44, 3852–3857. [DOI] [PubMed]
- Ragavan, R. V., Vijayakumar, V. & Sucheta Kumari, N. (2010). Eur. J. Med. Chem. 45, 1173–1180. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536811001589/is2655sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811001589/is2655Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report