Abstract
In the title molecule, C16H9NO5S, there is an intramolecular O—H⋯O hydrogen bond involving the quinolone carbonyl O atom and a carboxyl OH group. In the crystal, intermolecular O—H⋯O hydrogen bonds between the carbonyl group of the quinolone carboxyl group, and a second carboxyl group on the thiazeto moiety lead to the formation of chains propagating along [201] and perpendicular to the π-stacks of molecules.
Related literature
For background to the biological importance of thiazetoquinoline antibiotics, see: Ozaki et al. (1991 ▶). For similar work using different procedures, see: Ito et al. (1992 ▶, 1994 ▶); Matsuoka et al. (1999 ▶).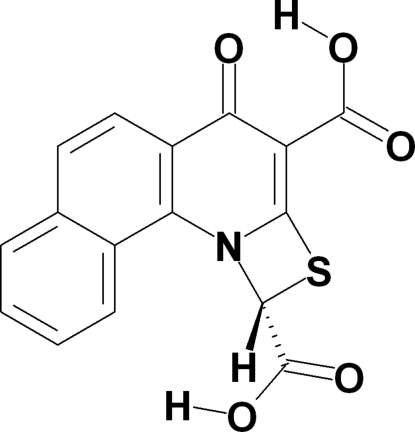
Experimental
Crystal data
C16H9NO5S
M r = 327.31
Monoclinic,

a = 7.237 (2) Å
b = 16.171 (5) Å
c = 11.929 (4) Å
β = 106.081 (8)°
V = 1341.5 (7) Å3
Z = 4
Mo Kα radiation
μ = 0.27 mm−1
T = 153 K
0.18 × 0.04 × 0.04 mm
Data collection
Rigaku Saturn diffractometer
Absorption correction: numerical (ABSCOR; Higashi, 1999 ▶) T min = 0.974, T max = 0.996
17300 measured reflections
2769 independent reflections
2614 reflections with I > 2σ(I)
R int = 0.074
Refinement
R[F 2 > 2σ(F 2)] = 0.088
wR(F 2) = 0.164
S = 1.30
2769 reflections
214 parameters
2 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.31 e Å−3
Δρmin = −0.31 e Å−3
Data collection: CrystalClear (Rigaku, 2005 ▶); cell refinement: CrystalClear; data reduction: CrystalClear; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: Mercury (Macrae et al., 2006 ▶); software used to prepare material for publication: publCIF (Westrip, 2010 ▶).
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536811003333/su2249sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811003333/su2249Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O5—H5A⋯O1 | 0.96 (4) | 1.57 (4) | 2.504 (4) | 161 (4) |
| O3—H3⋯O4i | 0.97 (3) | 1.62 (3) | 2.569 (4) | 166 (3) |
Symmetry code: (i)  .
.
Table 2. π⋯π interactions (Å, °).
Angle of elevation defined as the angle of the Cg(I)→Cg(J) vector and the normal to plane J. Cg1, Cg2 and Cg3 are the centroids of the C7–C12, N1/C1–C4/C13 and C4–C7/C12/C13 rings, respectively.
| π⋯π | Distance | Angle of Elevation |
|---|---|---|
| Cg1⋯Cg2i | 3.560 (2) | 19.56 |
| Cg3⋯Cg2i | 3.644 (2) | 22.75 |
| Cg3⋯Cg3i | 3.688 (2) | 24.39 |
Symmetry code: (i) −x + 1, −y, −z.
supplementary crystallographic information
Comment
4-oxo-1,4-dihydroquinoline-3-carboxylic acid derivatives (quinolones) are an important class of antibacterial agents, and a significant market exists for thiazetoquinoline antibiotics (Matsuoka et al.,1999; Ito et al., 1992; Ito et al., 1994; Ozaki et al., 1991). To this end, the title comound was obtained from the reaction of ethyl 2-{[2- ethoxy-2-oxoethyl)thio)-4-hydroxybenzo[h]quinoline-3-carboxylate with 1,2-dibromopropane in the presence of a catalytic amount of KI, followed by saponification using sodium hydroxide.
The molecular structure of the title molecule is shown in Fig. 1. It exhibits intra- (O5—H5a···O1) and intermolecular (O3—H3···O4i) hydrogen bonding (Table 1 and Fig. 2) leading to a chain-like arrangement of molecules which run along [201] and perpendicular to the π stacks (Fig. 2). Centroid-centroid distances range from 3.560 (2) to 3.688 (2) Å with angles of elevation between 19.56 and 24.39° (Table 2), while the inter-planar distance, as defined by the adjacent 14-atom (N1,C1—C13) ring system is 3.34 (1) Å.
Experimental
To a mixture of ethyl 2-{[2- ethoxy-2-oxoethyl)thio)-4-hydroxybenzo[h]quinoline-3-carboxylate (1 mmol) and K2CO3 (2.8 mmol) in dry DMF (25 ml) under a nitrogen atmosphere was added 1,2-dibromopropane (2.8 mmol) along with a catlytic amount of KI. The reaction mixture was heated at 343 K for 24 h, and then poured into ice-H2O. The resulting thiazetoquinoline derivative was collected by filtration. The separated product was reacted with sodium hydroxide (2.2 mmol) in water (20 ml) and heated at 373 K for 3–4 h. After being cooled, the reaction mixture was neutralized with hydrochloric acid (1 mol/L), extracted with CH2Cl2, dried over MgSO4, and then evaporated. The obtained solid was purified by recrystallization from ethanol to afford the title compound as a yellowish white powder. Mp. 508 K, yield = 39%. 1H-NMR and 13C-NMR data are given in the archived CIF.
Refinement
The OH H-atoms, H3 and H5a, were located from difference Fourier maps, and were refined with distance restraints: O-H = 0.96 (3) Å, with Uiso(H) = 1.2Ueq(O). The C-bound H-atoms were included in calculated positions and treated as riding atoms: C-H = 0.95, 0.98, 0.99 and 1.0 Å for H-aromatic, H-methyl, H-methylene and methine H-atoms, respectively, with Uiso(H) = k × Ueq(parent C-atom), where k = 1.5 for H-methyl and k = 1.2 for all other H-atoms.
Figures
Fig. 1.
A view of the molecular structure of the title molecule, with displacement ellipsoids drawn at the 50% probability level.
Fig. 2.
A partial view of the crystal packing of the title compound. Both the hydrogen bonding [symmetry codes: (i) x-1, y, z; (ii) x, -y+1/2, z+1/2; (iii) x+1, y, z+1] and π···π interactions [symmetry codes: (ii) x, -y+1/2, z+1/2; (iv) -x+1, y+1/2, -z+1/2] are shown as dashed lines; ring centroids are marked by small spheres. See Tables 1 and 2 for details.
Crystal data
| C16H9NO5S | F(000) = 672 |
| Mr = 327.31 | Dx = 1.621 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71075 Å |
| Hall symbol: -P 2ybc | Cell parameters from 4915 reflections |
| a = 7.237 (2) Å | θ = 2.2–30.6° |
| b = 16.171 (5) Å | µ = 0.27 mm−1 |
| c = 11.929 (4) Å | T = 153 K |
| β = 106.081 (8)° | Needle, colourless |
| V = 1341.5 (7) Å3 | 0.18 × 0.04 × 0.04 mm |
| Z = 4 |
Data collection
| Rigaku Saturn diffractometer | 2769 independent reflections |
| Radiation source: fine-focus sealed tube | 2614 reflections with I > 2σ(I) |
| graphite - Rigaku SHINE | Rint = 0.074 |
| Detector resolution: 14.63 pixels mm-1 | θmax = 26.5°, θmin = 2.5° |
| ω scans | h = −9→9 |
| Absorption correction: numerical (ABSCOR; Higashi, 1999) | k = −20→20 |
| Tmin = 0.974, Tmax = 0.996 | l = −14→14 |
| 17300 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.088 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.164 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.30 | w = 1/[σ2(Fo2) + (0.0366P)2 + 2.1946P] where P = (Fo2 + 2Fc2)/3 |
| 2769 reflections | (Δ/σ)max < 0.001 |
| 214 parameters | Δρmax = 0.31 e Å−3 |
| 2 restraints | Δρmin = −0.31 e Å−3 |
Special details
| Experimental. Spectroscopic data:1H-NMR: (500 MHz, DMSO-d6): δ= 8.27(1H, d, J=8.8), 8.25(1H, d, J=8.4), 8.17(1H, d, J=7.5), 8.02(1H, d, J=8.80), 7.83(1H, dd, J=11.0, 4.0), 7.81–7.76(1H, m), 7.73(1H, s)}. 13C-NMR: (500 MHz, DMSO-d6): δ= 175.76, 165.64, 165.25, 164.26, 136.09, 135.26, 129.58, 128.97, 127.58, 126.05, 122.67, 122.33, 121.53, 121.15, 103.64, 70.43. |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.60834 (14) | 0.26465 (6) | 0.14885 (8) | 0.0347 (3) | |
| O1 | 0.5029 (4) | −0.01520 (17) | 0.3384 (2) | 0.0387 (7) | |
| O2 | 0.2537 (4) | 0.33079 (19) | −0.0999 (3) | 0.0512 (8) | |
| O3 | 0.1270 (4) | 0.25356 (17) | 0.0178 (2) | 0.0370 (6) | |
| O4 | 0.7870 (4) | 0.20609 (17) | 0.3966 (2) | 0.0379 (7) | |
| O5 | 0.7226 (4) | 0.08587 (19) | 0.4695 (2) | 0.0439 (7) | |
| N1 | 0.4277 (4) | 0.14703 (17) | 0.0711 (2) | 0.0257 (6) | |
| C1 | 0.5397 (5) | 0.1656 (2) | 0.1790 (3) | 0.0274 (7) | |
| C2 | 0.5731 (5) | 0.1139 (2) | 0.2722 (3) | 0.0289 (8) | |
| C3 | 0.4829 (5) | 0.0354 (2) | 0.2544 (3) | 0.0303 (8) | |
| C4 | 0.3696 (5) | 0.0139 (2) | 0.1369 (3) | 0.0277 (8) | |
| C5 | 0.2892 (5) | −0.0668 (2) | 0.1153 (3) | 0.0318 (8) | |
| H5 | 0.3105 | −0.1057 | 0.1774 | 0.038* | |
| C6 | 0.1828 (5) | −0.0887 (2) | 0.0074 (3) | 0.0317 (8) | |
| H6 | 0.1297 | −0.1428 | −0.0048 | 0.038* | |
| C7 | 0.1484 (5) | −0.0329 (2) | −0.0882 (3) | 0.0281 (8) | |
| C8 | 0.0343 (5) | −0.0566 (2) | −0.2002 (3) | 0.0327 (8) | |
| H8 | −0.0180 | −0.1108 | −0.2118 | 0.039* | |
| C9 | −0.0019 (5) | −0.0027 (3) | −0.2920 (3) | 0.0364 (9) | |
| H9 | −0.0819 | −0.0190 | −0.3661 | 0.044* | |
| C10 | 0.0790 (6) | 0.0762 (2) | −0.2765 (3) | 0.0359 (9) | |
| H10 | 0.0552 | 0.1130 | −0.3410 | 0.043* | |
| C11 | 0.1927 (5) | 0.1015 (2) | −0.1696 (3) | 0.0324 (8) | |
| H11 | 0.2475 | 0.1553 | −0.1611 | 0.039* | |
| C12 | 0.2288 (5) | 0.0480 (2) | −0.0719 (3) | 0.0275 (8) | |
| C13 | 0.3401 (5) | 0.0702 (2) | 0.0439 (3) | 0.0254 (7) | |
| C14 | 0.4427 (5) | 0.2249 (2) | 0.0104 (3) | 0.0298 (8) | |
| H14 | 0.5090 | 0.2174 | −0.0521 | 0.036* | |
| C15 | 0.2618 (5) | 0.2759 (2) | −0.0304 (3) | 0.0327 (8) | |
| C16 | 0.7025 (5) | 0.1379 (2) | 0.3852 (3) | 0.0332 (9) | |
| H5A | 0.649 (6) | 0.039 (2) | 0.432 (4) | 0.052* | |
| H3 | 0.008 (4) | 0.277 (2) | −0.029 (3) | 0.044* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0343 (5) | 0.0303 (5) | 0.0351 (5) | −0.0046 (4) | 0.0023 (4) | −0.0016 (4) |
| O1 | 0.0403 (16) | 0.0408 (16) | 0.0313 (14) | 0.0000 (12) | 0.0036 (12) | 0.0104 (12) |
| O2 | 0.0460 (17) | 0.0450 (17) | 0.062 (2) | 0.0053 (14) | 0.0138 (15) | 0.0244 (16) |
| O3 | 0.0304 (14) | 0.0391 (15) | 0.0396 (15) | 0.0034 (12) | 0.0065 (12) | 0.0059 (12) |
| O4 | 0.0335 (14) | 0.0420 (16) | 0.0337 (14) | −0.0013 (13) | 0.0022 (11) | −0.0073 (12) |
| O5 | 0.0438 (17) | 0.0558 (19) | 0.0260 (14) | −0.0055 (14) | −0.0006 (12) | 0.0038 (13) |
| N1 | 0.0257 (15) | 0.0240 (15) | 0.0263 (15) | −0.0017 (12) | 0.0053 (12) | 0.0024 (12) |
| C1 | 0.0220 (17) | 0.0300 (18) | 0.0289 (18) | −0.0005 (14) | 0.0051 (14) | −0.0049 (15) |
| C2 | 0.0265 (18) | 0.033 (2) | 0.0273 (18) | −0.0005 (15) | 0.0080 (14) | −0.0024 (15) |
| C3 | 0.0285 (18) | 0.036 (2) | 0.0268 (18) | 0.0060 (16) | 0.0076 (14) | 0.0044 (15) |
| C4 | 0.0243 (17) | 0.0300 (19) | 0.0294 (18) | 0.0035 (15) | 0.0087 (14) | 0.0009 (15) |
| C5 | 0.0285 (19) | 0.0283 (19) | 0.040 (2) | 0.0039 (15) | 0.0111 (16) | 0.0066 (16) |
| C6 | 0.0278 (18) | 0.0252 (19) | 0.042 (2) | 0.0007 (15) | 0.0092 (16) | 0.0008 (16) |
| C7 | 0.0237 (17) | 0.0275 (18) | 0.0326 (19) | 0.0035 (14) | 0.0072 (14) | −0.0029 (15) |
| C8 | 0.0259 (18) | 0.033 (2) | 0.039 (2) | −0.0002 (15) | 0.0076 (16) | −0.0087 (17) |
| C9 | 0.0282 (19) | 0.045 (2) | 0.031 (2) | 0.0022 (17) | 0.0004 (15) | −0.0106 (17) |
| C10 | 0.039 (2) | 0.038 (2) | 0.0280 (19) | 0.0004 (18) | 0.0058 (16) | 0.0025 (17) |
| C11 | 0.034 (2) | 0.0300 (19) | 0.0323 (19) | −0.0032 (16) | 0.0082 (16) | −0.0041 (16) |
| C12 | 0.0220 (17) | 0.0298 (19) | 0.0303 (18) | 0.0010 (14) | 0.0065 (14) | −0.0007 (15) |
| C13 | 0.0224 (16) | 0.0250 (17) | 0.0302 (18) | 0.0013 (14) | 0.0094 (14) | 0.0003 (15) |
| C14 | 0.0265 (18) | 0.0290 (19) | 0.0329 (19) | 0.0002 (15) | 0.0065 (15) | 0.0027 (15) |
| C15 | 0.035 (2) | 0.0271 (19) | 0.0325 (19) | −0.0016 (16) | 0.0029 (16) | −0.0013 (16) |
| C16 | 0.0300 (19) | 0.044 (2) | 0.0267 (19) | 0.0051 (17) | 0.0093 (15) | −0.0021 (17) |
Geometric parameters (Å, °)
| S1—C1 | 1.744 (4) | C5—C6 | 1.351 (5) |
| S1—C14 | 1.866 (4) | C5—H5 | 0.9500 |
| O1—C3 | 1.271 (4) | C6—C7 | 1.421 (5) |
| O2—C15 | 1.205 (4) | C6—H6 | 0.9500 |
| O3—C15 | 1.314 (5) | C7—C8 | 1.416 (5) |
| O3—H3 | 0.963 (19) | C7—C12 | 1.423 (5) |
| O4—C16 | 1.250 (5) | C8—C9 | 1.366 (5) |
| O5—C16 | 1.288 (5) | C8—H8 | 0.9500 |
| O5—H5A | 0.965 (19) | C9—C10 | 1.394 (6) |
| N1—C1 | 1.350 (4) | C9—H9 | 0.9500 |
| N1—C13 | 1.392 (4) | C10—C11 | 1.375 (5) |
| N1—C14 | 1.472 (4) | C10—H10 | 0.9500 |
| C1—C2 | 1.358 (5) | C11—C12 | 1.416 (5) |
| C2—C3 | 1.416 (5) | C11—H11 | 0.9500 |
| C2—C16 | 1.464 (5) | C12—C13 | 1.438 (5) |
| C3—C4 | 1.456 (5) | C14—C15 | 1.509 (5) |
| C4—C13 | 1.406 (5) | C14—H14 | 1.0000 |
| C4—C5 | 1.423 (5) | ||
| C1—S1—C14 | 73.49 (16) | C7—C8—H8 | 119.5 |
| C15—O3—H3 | 107 (3) | C8—C9—C10 | 119.8 (3) |
| C16—O5—H5A | 103 (3) | C8—C9—H9 | 120.1 |
| C1—N1—C13 | 122.3 (3) | C10—C9—H9 | 120.1 |
| C1—N1—C14 | 99.9 (3) | C11—C10—C9 | 121.1 (4) |
| C13—N1—C14 | 137.7 (3) | C11—C10—H10 | 119.5 |
| N1—C1—C2 | 124.6 (3) | C9—C10—H10 | 119.5 |
| N1—C1—S1 | 97.9 (2) | C10—C11—C12 | 120.5 (3) |
| C2—C1—S1 | 137.5 (3) | C10—C11—H11 | 119.7 |
| C1—C2—C3 | 117.2 (3) | C12—C11—H11 | 119.7 |
| C1—C2—C16 | 121.0 (3) | C11—C12—C7 | 118.3 (3) |
| C3—C2—C16 | 121.8 (3) | C11—C12—C13 | 124.3 (3) |
| O1—C3—C2 | 120.8 (3) | C7—C12—C13 | 117.3 (3) |
| O1—C3—C4 | 121.0 (3) | N1—C13—C4 | 115.7 (3) |
| C2—C3—C4 | 118.2 (3) | N1—C13—C12 | 123.0 (3) |
| C13—C4—C5 | 119.1 (3) | C4—C13—C12 | 121.3 (3) |
| C13—C4—C3 | 121.8 (3) | N1—C14—C15 | 116.8 (3) |
| C5—C4—C3 | 119.1 (3) | N1—C14—S1 | 88.6 (2) |
| C6—C5—C4 | 120.6 (3) | C15—C14—S1 | 112.6 (3) |
| C6—C5—H5 | 119.7 | N1—C14—H14 | 112.3 |
| C4—C5—H5 | 119.7 | C15—C14—H14 | 112.3 |
| C5—C6—C7 | 121.7 (3) | S1—C14—H14 | 112.3 |
| C5—C6—H6 | 119.2 | O2—C15—O3 | 127.1 (4) |
| C7—C6—H6 | 119.2 | O2—C15—C14 | 119.8 (4) |
| C8—C7—C6 | 120.8 (3) | O3—C15—C14 | 113.1 (3) |
| C8—C7—C12 | 119.2 (3) | O4—C16—O5 | 123.0 (3) |
| C6—C7—C12 | 120.0 (3) | O4—C16—C2 | 120.2 (3) |
| C9—C8—C7 | 121.0 (4) | O5—C16—C2 | 116.8 (4) |
| C9—C8—H8 | 119.5 |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O5—H5A···O1 | 0.96 (4) | 1.57 (4) | 2.504 (4) | 161 (4) |
| O3—H3···O4i | 0.97 (3) | 1.62 (3) | 2.569 (4) | 166 (3) |
Symmetry codes: (i) x−1, −y+1/2, z−1/2.
Table 2 π···π interactions (Å, °)
Angle of elevation defined as the angle of the Cg(I)→Cg(J) vector and the normal to plane J. Cg1, Cg2 and Cg3 are the centroids of the C7–C12, N1/C1–C4/C13 and C4–C7/C12/C13 rings, respectively.
| π···π | Distance | Angle of Elevation |
| Cg1···Cg2i | 3.560 (2) | 19.56 |
| Cg3···Cg2i | 3.644 (2) | 22.75 |
| Cg3···Cg3i | 3.688 (2) | 24.39 |
Symmetry code: (i) -x+1, -y, -z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: SU2249).
References
- Higashi, T. (1999). ABSCOR Rigaku Corporation, Tokyo, Japan.
- Ito, Y., Kato, H., Yasuda, S., Yoshida, T. & Yamamoto, Y. (1992). JP 92-21664 (July 27, 1992).
- Ito, Y., Kato, H., Yasuda, S., Yoshida, T. & Yamamoto, Y. (1994). JP 06016677 A (Jan 25, 1994).
- Macrae, C. F., Edgington, P. R., McCabe, P., Pidcock, E., Shields, G. P., Taylor, R., Towler, M. & van de Streek, J. (2006). J. Appl. Cryst. 39, 453–457.
- Matsuoka, M., Segawa, J., Amimoto, I., Masui, Y., Tomii, Y., Kitano, M. & Kise, M. (1999). Chem. Pharm. Bull. (Tokyo), 47, 1765–1773. [DOI] [PubMed]
- Ozaki, M., Matsuda, M., Tomii, Y., Kimura, K., Segawa, J., Kitano, M., Kise, M., Shibata, K., Otsuki, M. & Nishino, T. (1991). Antimicrob. Agents Chemother. 35, 2496–2499. [DOI] [PMC free article] [PubMed]
- Rigaku (2005). CrystalClear Rigaku Corporation, Tokyo, Japan.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536811003333/su2249sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811003333/su2249Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




