Abstract
In the title compound, C20H20Cl2N4O2, the nitroimidazole ring makes dihedral angles of 17.00 (1) and 60.45 (11)° with the phenyl and chlorophenyl rings, respectively. The three-coordinate N atom connected to two methylene and one methine C atoms shows pyramidal coordination.
Related literature
For the use of nitrogen mustards containing the β-chloroethylamine unit as antitumor drugs, see: Zhuang et al. (2008 ▶). Nitroimidazole compounds are also used extensively in the treatment of various cancers as clinical radiosensitizers, see: Cai et al. (2009 ▶). For the synthesis, see: Fang et al. (2010 ▶); Gan et al. (2010 ▶).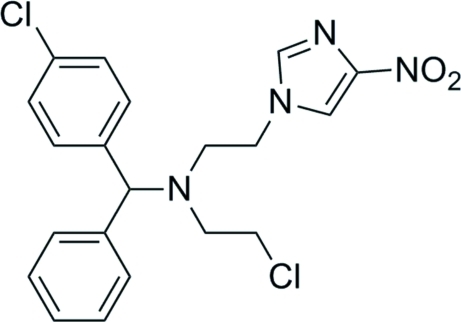
Experimental
Crystal data
C20H20Cl2N4O2
M r = 419.30
Monoclinic,

a = 8.8206 (16) Å
b = 25.005 (5) Å
c = 9.0450 (17) Å
β = 100.802 (3)°
V = 1959.6 (6) Å3
Z = 4
Mo Kα radiation
μ = 0.36 mm−1
T = 298 K
0.32 × 0.24 × 0.18 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.903, T max = 0.938
9838 measured reflections
3449 independent reflections
2464 reflections with I > 2σ(I)
R int = 0.037
Refinement
R[F 2 > 2σ(F 2)] = 0.053
wR(F 2) = 0.139
S = 1.03
3449 reflections
253 parameters
H-atom parameters constrained
Δρmax = 0.49 e Å−3
Δρmin = −0.24 e Å−3
Data collection: SMART (Bruker, 2000 ▶); cell refinement: SAINT (Bruker, 2000 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S160053681100256X/ng5107sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053681100256X/ng5107Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
We thank Southwest University (grant Nos. SWUB2006018 and XSGX0602), the Natural Science Foundation of Chongqing (grant No. 2009BB5296) and the Research Funds for the Central Universities (XDJK2009c092) for financial support.
supplementary crystallographic information
Comment
Nitrogen mustards as anticancer agents containing typical β-chloroethylamine moiety with easy synthesis and inexpensive expense are one of the most important antitumor drugs (Zhuang et al., 2008). Nitroimidazole compounds are also extensively used in the treatment of various cancers as clinical radiosensitizer (Cai et al., 2009). In view of this, it is of great interest for us to investigate the nitrogen mustard-based nitroimidazoles as new potential anticancer agents. Herein we would like to report the crystal structure of the title compound (I).
The title compound, C20H20Cl2N4O2, crystallized in non-chiral monoclinic crystal system of P2(1)/n space group, including a racemic chiral isomers. In the molecule, the nitroimidazole ring makes dihedral angles of 17.00 (1) and 60.45 (11)°, respectively, with the benzene and chlorophenyl ring.
Experimental
The intermediate 2-chloro-N-(2-chloroethyl)-N-((4-chlorophenyl(phenyl)methyl)ethanamine (0.85 g, 2.5 mmol), which was prepared according to the procedure of Fang et al.(2010) and Gan et al.(2010), reacted with 4-nitroimidazole (0.34 g, 3.0 mmol) in the presence of weak base in acetonitrile at 60 °C for 12 h to produce the title compound (I) 0.30 g as white solid via silica gel column chromatography (ethyl acetate/petroleum ether, 1/2, V/V). A crystal of (I) suitable for X-ray analysis was grown from a mixture solution of ethyl acetate and petroleum ether by slow evaporation at room temperature.
Refinement
Hydrogen atoms were placed in idealized positions and treated as riding, with C—H = 0.93Å (CH), 0.98Å (CH) or 0.98Å (CH2) and Uiso(H) = 1.2 Ueq(C).
Figures
Fig. 1.
Ellipsoid plot.
Crystal data
| C20H20Cl2N4O2 | Z = 4 |
| Mr = 419.30 | F(000) = 872 |
| Monoclinic, P21/n | Dx = 1.421 Mg m−3 |
| Hall symbol: -P 2yn | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.8206 (16) Å | θ = 2.4–21.3° |
| b = 25.005 (5) Å | µ = 0.36 mm−1 |
| c = 9.0450 (17) Å | T = 298 K |
| β = 100.802 (3)° | Block, colourless |
| V = 1959.6 (6) Å3 | 0.32 × 0.24 × 0.18 mm |
Data collection
| Bruker SMART CCD area-detector diffractometer | 3449 independent reflections |
| Radiation source: fine-focus sealed tube | 2464 reflections with I > 2σ(I) |
| graphite | Rint = 0.037 |
| phi and ω scans | θmax = 25.0°, θmin = 1.6° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | h = −10→7 |
| Tmin = 0.903, Tmax = 0.938 | k = −29→29 |
| 9838 measured reflections | l = −10→10 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.053 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.139 | H-atom parameters constrained |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0576P)2 + 0.904P] where P = (Fo2 + 2Fc2)/3 |
| 3449 reflections | (Δ/σ)max = 0.001 |
| 253 parameters | Δρmax = 0.49 e Å−3 |
| 0 restraints | Δρmin = −0.24 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.3270 (3) | 0.05952 (13) | 0.0927 (3) | 0.0487 (7) | |
| C2 | 0.4173 (4) | 0.01834 (13) | 0.1569 (4) | 0.0615 (9) | |
| H2 | 0.3972 | −0.0165 | 0.1231 | 0.074* | |
| C3 | 0.5385 (4) | 0.02915 (12) | 0.2723 (3) | 0.0538 (8) | |
| H3 | 0.5994 | 0.0009 | 0.3161 | 0.065* | |
| C4 | 0.5734 (3) | 0.07989 (11) | 0.3257 (3) | 0.0402 (6) | |
| C5 | 0.4771 (3) | 0.12070 (12) | 0.2588 (3) | 0.0498 (7) | |
| H5 | 0.4957 | 0.1556 | 0.2933 | 0.060* | |
| C6 | 0.3544 (3) | 0.11081 (12) | 0.1423 (3) | 0.0514 (8) | |
| H6 | 0.2917 | 0.1386 | 0.0985 | 0.062* | |
| C7 | 0.7143 (3) | 0.08861 (11) | 0.4496 (3) | 0.0401 (6) | |
| H7 | 0.7628 | 0.0533 | 0.4654 | 0.048* | |
| C8 | 0.6827 (3) | 0.10452 (10) | 0.6032 (3) | 0.0400 (6) | |
| C9 | 0.8038 (3) | 0.10174 (11) | 0.7245 (3) | 0.0456 (7) | |
| H9 | 0.8999 | 0.0902 | 0.7091 | 0.055* | |
| C10 | 0.7859 (4) | 0.11550 (12) | 0.8666 (3) | 0.0536 (8) | |
| H10 | 0.8695 | 0.1137 | 0.9463 | 0.064* | |
| C11 | 0.6439 (4) | 0.13203 (13) | 0.8913 (4) | 0.0592 (8) | |
| H11 | 0.6313 | 0.1417 | 0.9875 | 0.071* | |
| C12 | 0.5226 (4) | 0.13412 (14) | 0.7746 (4) | 0.0657 (9) | |
| H12 | 0.4263 | 0.1448 | 0.7912 | 0.079* | |
| C13 | 0.5415 (3) | 0.12044 (12) | 0.6309 (3) | 0.0538 (8) | |
| H13 | 0.4573 | 0.1220 | 0.5518 | 0.065* | |
| C14 | 0.9049 (3) | 0.10277 (13) | 0.2887 (3) | 0.0547 (8) | |
| H14A | 0.9671 | 0.1308 | 0.2559 | 0.066* | |
| H14B | 0.8231 | 0.0941 | 0.2046 | 0.066* | |
| C15 | 1.0037 (4) | 0.05433 (13) | 0.3272 (4) | 0.0591 (8) | |
| H15A | 1.0457 | 0.0436 | 0.2400 | 0.071* | |
| H15B | 0.9407 | 0.0252 | 0.3526 | 0.071* | |
| C16 | 0.7954 (3) | 0.18032 (11) | 0.3913 (3) | 0.0498 (7) | |
| H16A | 0.7132 | 0.1884 | 0.4456 | 0.060* | |
| H16B | 0.7574 | 0.1876 | 0.2855 | 0.060* | |
| C17 | 0.9333 (4) | 0.21643 (13) | 0.4485 (3) | 0.0557 (8) | |
| H17A | 1.0205 | 0.2049 | 0.4051 | 0.067* | |
| H17B | 0.9081 | 0.2528 | 0.4158 | 0.067* | |
| C18 | 1.1030 (3) | 0.19238 (12) | 0.6938 (4) | 0.0523 (8) | |
| H18 | 1.1775 | 0.1748 | 0.6516 | 0.063* | |
| C19 | 0.9802 (3) | 0.22472 (11) | 0.8478 (3) | 0.0479 (7) | |
| C20 | 0.8953 (3) | 0.23602 (11) | 0.7104 (4) | 0.0521 (8) | |
| H20 | 0.8017 | 0.2542 | 0.6891 | 0.063* | |
| Cl1 | 0.17478 (11) | 0.04618 (4) | −0.05458 (11) | 0.0835 (3) | |
| Cl2 | 1.15787 (10) | 0.06636 (4) | 0.48090 (12) | 0.0777 (3) | |
| N1 | 0.8356 (2) | 0.12352 (9) | 0.4102 (2) | 0.0412 (5) | |
| N2 | 0.9760 (3) | 0.21524 (9) | 0.6115 (3) | 0.0458 (6) | |
| N3 | 1.1106 (3) | 0.19743 (10) | 0.8388 (3) | 0.0538 (6) | |
| N4 | 0.9403 (4) | 0.23746 (13) | 0.9901 (4) | 0.0692 (8) | |
| O1 | 1.0250 (3) | 0.22177 (12) | 1.1050 (3) | 0.0897 (9) | |
| O2 | 0.8228 (4) | 0.26290 (15) | 0.9864 (4) | 0.1142 (12) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0397 (16) | 0.0603 (19) | 0.0430 (17) | −0.0055 (14) | −0.0002 (13) | −0.0095 (14) |
| C2 | 0.059 (2) | 0.0473 (18) | 0.073 (2) | −0.0065 (16) | −0.0015 (18) | −0.0138 (16) |
| C3 | 0.0534 (18) | 0.0446 (17) | 0.060 (2) | 0.0017 (14) | 0.0013 (16) | −0.0014 (14) |
| C4 | 0.0360 (14) | 0.0425 (15) | 0.0416 (16) | 0.0020 (12) | 0.0059 (12) | 0.0013 (12) |
| C5 | 0.0451 (16) | 0.0416 (16) | 0.0565 (19) | 0.0023 (13) | −0.0061 (14) | −0.0055 (14) |
| C6 | 0.0427 (17) | 0.0503 (18) | 0.0561 (19) | 0.0063 (14) | −0.0040 (14) | 0.0038 (14) |
| C7 | 0.0361 (14) | 0.0399 (15) | 0.0417 (16) | 0.0039 (12) | 0.0005 (12) | 0.0038 (12) |
| C8 | 0.0383 (15) | 0.0369 (14) | 0.0433 (16) | −0.0022 (12) | 0.0041 (12) | 0.0054 (12) |
| C9 | 0.0347 (15) | 0.0561 (18) | 0.0450 (17) | 0.0012 (13) | 0.0049 (13) | 0.0102 (14) |
| C10 | 0.0498 (18) | 0.068 (2) | 0.0392 (17) | −0.0077 (16) | −0.0004 (14) | 0.0072 (15) |
| C11 | 0.063 (2) | 0.068 (2) | 0.0475 (18) | −0.0007 (17) | 0.0118 (16) | −0.0073 (16) |
| C12 | 0.0505 (19) | 0.087 (3) | 0.059 (2) | 0.0159 (18) | 0.0107 (17) | −0.0067 (18) |
| C13 | 0.0397 (16) | 0.069 (2) | 0.0493 (18) | 0.0076 (15) | −0.0010 (14) | −0.0013 (15) |
| C14 | 0.0442 (17) | 0.076 (2) | 0.0419 (17) | −0.0036 (16) | 0.0032 (14) | 0.0063 (15) |
| C15 | 0.0513 (19) | 0.072 (2) | 0.057 (2) | −0.0036 (17) | 0.0181 (16) | −0.0093 (17) |
| C16 | 0.0454 (17) | 0.0521 (18) | 0.0463 (17) | −0.0048 (14) | −0.0055 (14) | 0.0111 (14) |
| C17 | 0.0549 (19) | 0.0574 (19) | 0.0500 (18) | −0.0139 (15) | −0.0022 (15) | 0.0171 (15) |
| C18 | 0.0396 (16) | 0.0621 (19) | 0.054 (2) | 0.0032 (15) | 0.0060 (14) | 0.0025 (15) |
| C19 | 0.0413 (16) | 0.0467 (17) | 0.0552 (19) | −0.0076 (14) | 0.0074 (14) | −0.0049 (14) |
| C20 | 0.0404 (17) | 0.0441 (17) | 0.067 (2) | 0.0010 (14) | −0.0030 (16) | −0.0054 (15) |
| Cl1 | 0.0626 (6) | 0.0995 (7) | 0.0751 (6) | −0.0043 (5) | −0.0216 (5) | −0.0236 (5) |
| Cl2 | 0.0537 (5) | 0.0773 (6) | 0.0937 (7) | 0.0113 (4) | −0.0080 (5) | 0.0054 (5) |
| N1 | 0.0341 (12) | 0.0473 (13) | 0.0411 (13) | 0.0014 (10) | 0.0038 (10) | 0.0070 (10) |
| N2 | 0.0381 (13) | 0.0465 (13) | 0.0494 (15) | −0.0064 (11) | −0.0001 (11) | 0.0047 (11) |
| N3 | 0.0452 (15) | 0.0627 (16) | 0.0507 (16) | −0.0013 (13) | 0.0017 (12) | 0.0051 (13) |
| N4 | 0.0560 (19) | 0.080 (2) | 0.072 (2) | −0.0215 (16) | 0.0145 (17) | −0.0256 (17) |
| O1 | 0.094 (2) | 0.117 (2) | 0.0570 (16) | −0.0273 (18) | 0.0125 (16) | −0.0085 (15) |
| O2 | 0.078 (2) | 0.151 (3) | 0.117 (3) | 0.012 (2) | 0.0262 (18) | −0.065 (2) |
Geometric parameters (Å, °)
| C1—C2 | 1.364 (4) | C13—H13 | 0.9300 |
| C1—C6 | 1.365 (4) | C14—N1 | 1.450 (4) |
| C1—Cl1 | 1.737 (3) | C14—C15 | 1.495 (4) |
| C2—C3 | 1.374 (4) | C14—H14A | 0.9700 |
| C2—H2 | 0.9300 | C14—H14B | 0.9700 |
| C3—C4 | 1.372 (4) | C15—Cl2 | 1.779 (3) |
| C3—H3 | 0.9300 | C15—H15A | 0.9700 |
| C4—C5 | 1.392 (4) | C15—H15B | 0.9700 |
| C4—C7 | 1.524 (4) | C16—N1 | 1.466 (3) |
| C5—C6 | 1.384 (4) | C16—C17 | 1.525 (4) |
| C5—H5 | 0.9300 | C16—H16A | 0.9700 |
| C6—H6 | 0.9300 | C16—H16B | 0.9700 |
| C7—N1 | 1.475 (3) | C17—N2 | 1.452 (4) |
| C7—C8 | 1.520 (4) | C17—H17A | 0.9700 |
| C7—H7 | 0.9800 | C17—H17B | 0.9700 |
| C8—C13 | 1.375 (4) | C18—N3 | 1.307 (4) |
| C8—C9 | 1.382 (4) | C18—N2 | 1.350 (4) |
| C9—C10 | 1.368 (4) | C18—H18 | 0.9300 |
| C9—H9 | 0.9300 | C19—N3 | 1.353 (4) |
| C10—C11 | 1.376 (4) | C19—C20 | 1.355 (4) |
| C10—H10 | 0.9300 | C19—N4 | 1.432 (4) |
| C11—C12 | 1.356 (4) | C20—N2 | 1.348 (4) |
| C11—H11 | 0.9300 | C20—H20 | 0.9300 |
| C12—C13 | 1.384 (4) | N4—O2 | 1.211 (4) |
| C12—H12 | 0.9300 | N4—O1 | 1.225 (4) |
| C2—C1—C6 | 121.1 (3) | N1—C14—H14A | 108.5 |
| C2—C1—Cl1 | 119.2 (2) | C15—C14—H14A | 108.5 |
| C6—C1—Cl1 | 119.8 (2) | N1—C14—H14B | 108.5 |
| C1—C2—C3 | 119.0 (3) | C15—C14—H14B | 108.5 |
| C1—C2—H2 | 120.5 | H14A—C14—H14B | 107.5 |
| C3—C2—H2 | 120.5 | C14—C15—Cl2 | 111.8 (2) |
| C4—C3—C2 | 122.8 (3) | C14—C15—H15A | 109.2 |
| C4—C3—H3 | 118.6 | Cl2—C15—H15A | 109.2 |
| C2—C3—H3 | 118.6 | C14—C15—H15B | 109.2 |
| C3—C4—C5 | 116.5 (3) | Cl2—C15—H15B | 109.2 |
| C3—C4—C7 | 119.3 (2) | H15A—C15—H15B | 107.9 |
| C5—C4—C7 | 124.2 (2) | N1—C16—C17 | 112.0 (2) |
| C6—C5—C4 | 121.8 (3) | N1—C16—H16A | 109.2 |
| C6—C5—H5 | 119.1 | C17—C16—H16A | 109.2 |
| C4—C5—H5 | 119.1 | N1—C16—H16B | 109.2 |
| C1—C6—C5 | 118.9 (3) | C17—C16—H16B | 109.2 |
| C1—C6—H6 | 120.6 | H16A—C16—H16B | 107.9 |
| C5—C6—H6 | 120.6 | N2—C17—C16 | 111.8 (2) |
| N1—C7—C8 | 109.3 (2) | N2—C17—H17A | 109.3 |
| N1—C7—C4 | 115.8 (2) | C16—C17—H17A | 109.3 |
| C8—C7—C4 | 116.5 (2) | N2—C17—H17B | 109.3 |
| N1—C7—H7 | 104.6 | C16—C17—H17B | 109.3 |
| C8—C7—H7 | 104.6 | H17A—C17—H17B | 107.9 |
| C4—C7—H7 | 104.6 | N3—C18—N2 | 113.2 (3) |
| C13—C8—C9 | 117.5 (3) | N3—C18—H18 | 123.4 |
| C13—C8—C7 | 124.7 (3) | N2—C18—H18 | 123.4 |
| C9—C8—C7 | 117.8 (2) | N3—C19—C20 | 112.3 (3) |
| C10—C9—C8 | 121.7 (3) | N3—C19—N4 | 121.4 (3) |
| C10—C9—H9 | 119.2 | C20—C19—N4 | 126.2 (3) |
| C8—C9—H9 | 119.2 | N2—C20—C19 | 105.0 (3) |
| C9—C10—C11 | 119.8 (3) | N2—C20—H20 | 127.5 |
| C9—C10—H10 | 120.1 | C19—C20—H20 | 127.5 |
| C11—C10—H10 | 120.1 | C14—N1—C16 | 112.7 (2) |
| C12—C11—C10 | 119.7 (3) | C14—N1—C7 | 113.5 (2) |
| C12—C11—H11 | 120.2 | C16—N1—C7 | 115.5 (2) |
| C10—C11—H11 | 120.2 | C20—N2—C18 | 106.5 (2) |
| C11—C12—C13 | 120.4 (3) | C20—N2—C17 | 126.7 (3) |
| C11—C12—H12 | 119.8 | C18—N2—C17 | 126.8 (3) |
| C13—C12—H12 | 119.8 | C18—N3—C19 | 103.0 (3) |
| C8—C13—C12 | 121.0 (3) | O2—N4—O1 | 125.0 (3) |
| C8—C13—H13 | 119.5 | O2—N4—C19 | 116.4 (3) |
| C12—C13—H13 | 119.5 | O1—N4—C19 | 118.5 (3) |
| N1—C14—C15 | 115.1 (2) | ||
| C6—C1—C2—C3 | −0.6 (5) | N1—C14—C15—Cl2 | −58.3 (3) |
| Cl1—C1—C2—C3 | 179.4 (2) | N1—C16—C17—N2 | −70.4 (3) |
| C1—C2—C3—C4 | −0.4 (5) | N3—C19—C20—N2 | −0.3 (3) |
| C2—C3—C4—C5 | 1.3 (4) | N4—C19—C20—N2 | −178.3 (3) |
| C2—C3—C4—C7 | −177.3 (3) | C15—C14—N1—C16 | 156.5 (3) |
| C3—C4—C5—C6 | −1.3 (4) | C15—C14—N1—C7 | −69.7 (3) |
| C7—C4—C5—C6 | 177.2 (3) | C17—C16—N1—C14 | −83.4 (3) |
| C2—C1—C6—C5 | 0.6 (5) | C17—C16—N1—C7 | 143.8 (2) |
| Cl1—C1—C6—C5 | −179.4 (2) | C8—C7—N1—C14 | 163.5 (2) |
| C4—C5—C6—C1 | 0.4 (5) | C4—C7—N1—C14 | −62.6 (3) |
| C3—C4—C7—N1 | 118.5 (3) | C8—C7—N1—C16 | −64.1 (3) |
| C5—C4—C7—N1 | −60.0 (3) | C4—C7—N1—C16 | 69.8 (3) |
| C3—C4—C7—C8 | −110.9 (3) | C19—C20—N2—C18 | 0.6 (3) |
| C5—C4—C7—C8 | 70.6 (3) | C19—C20—N2—C17 | 179.6 (3) |
| N1—C7—C8—C13 | 121.7 (3) | N3—C18—N2—C20 | −0.7 (3) |
| C4—C7—C8—C13 | −11.9 (4) | N3—C18—N2—C17 | −179.7 (3) |
| N1—C7—C8—C9 | −59.8 (3) | C16—C17—N2—C20 | −70.0 (4) |
| C4—C7—C8—C9 | 166.6 (2) | C16—C17—N2—C18 | 108.8 (3) |
| C13—C8—C9—C10 | −1.6 (4) | N2—C18—N3—C19 | 0.4 (3) |
| C7—C8—C9—C10 | 179.7 (3) | C20—C19—N3—C18 | −0.1 (3) |
| C8—C9—C10—C11 | 0.7 (4) | N4—C19—N3—C18 | 178.0 (3) |
| C9—C10—C11—C12 | 0.5 (5) | N3—C19—N4—O2 | 178.6 (3) |
| C10—C11—C12—C13 | −0.9 (5) | C20—C19—N4—O2 | −3.6 (5) |
| C9—C8—C13—C12 | 1.2 (4) | N3—C19—N4—O1 | −2.0 (4) |
| C7—C8—C13—C12 | 179.8 (3) | C20—C19—N4—O1 | 175.8 (3) |
| C11—C12—C13—C8 | 0.0 (5) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: NG5107).
References
- Bruker (2000). SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Cai, J. L., Li, S., Zhou, C. H., Gan, L. L. & Wu, J. (2009). Chin J New Drugs, 18, 598–652.
- Fang, B., Zhou, C. H. & Rao, X. C. (2010). Eur J Med Chem 45, 4388–4398. [DOI] [PubMed]
- Gan, L. L., Fang, B. & Zhou, C. H. (2010). Bull Kor. Chem Soc 31, 3684–3693.
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Zhuang, Y. Y., Zhou, C. H., Wang, Y. F. & Li, D. H. (2008). Chin Pharm J 43, 1281–1287.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S160053681100256X/ng5107sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S160053681100256X/ng5107Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



