Abstract
Although the nonrandom nature of interphase chromosome arrangement is widely accepted, how nuclear organization relates to genomic function remains unclear. Nuclear subcompartments may play a role by offering rich microenvironments that regulate chromatin state and ensure optimal transcriptional efficiency. Technological advances now provide genome-wide and four-dimensional analyses, permitting global characterizations of nuclear order. These approaches will help uncover how seemingly separate nuclear processes may be coupled and aid in the effort to understand the role of nuclear organization in development and disease.
Introduction
A striking feature of the eukaryotic cell nucleus is the packing of DNA into highly folded chromatin that fits into a very limited space. However, chromatin occupies only half of the available nuclear volume, and the remaining interchromatin space harbors nuclear subcompartments and soluble components involved in dynamic structural changes to chromatin domains. Increasing evidence suggests that the arrangement of chromosomes, gene loci, and nuclear bodies is nonrandom and exhibits features of self-organization in space and time. In the context of a highly ordered environment, the nucleus thus supports the efficient and precise coordination of diverse processes, including transcription, DNA repair, and replication.
Chromosomes and individual genes occupy preferred locations relative to one another and to landmarks within the interphase nucleus. Furthermore, localization has been associated with transcriptional activity. In metazoans, the lamina lining the inside of the nuclear envelope is considered to be a repressive environment, harboring transcriptionally inactive facultative heterochromatin. In contrast, particularly in yeast, subsets of actively transcribed loci can be found at nuclear pores as well as specific interior compartments. Thus, specific architecture may create functional microenvironments to coordinate transcriptional activity. Furthermore, changes in the transcriptional state have been associated with the movement of loci into and out of nuclear subcompartments.
In this review, we examine the evidence for spatial organization affecting gene expression and how changes in transcriptional status are related to the selective localization of chromatin to specific nuclear subcompartments, including the nuclear lamina, the nuclear pore, transcription factories, nucleoli and perinucleolar regions, and polycomb bodies (Fig. 1). We discuss how spatial architecture is relevant to disease states, such as cancer, that disrupt genome integrity. We also discuss the capabilities and limitations of the most pertinent techniques for investigating spatial architecture at the population level and in single cells as well as novel approaches that will allow a better evaluation of the relationship between the architecture of the nucleus and features of the linear genome, such as gene expression, histone modification, or binding of transcription factors. Finally, the development of mathematical approaches has permitted a more complete picture of the dynamic nucleus, which can further our understanding of critical developmental processes, such as cell specification and differentiation.
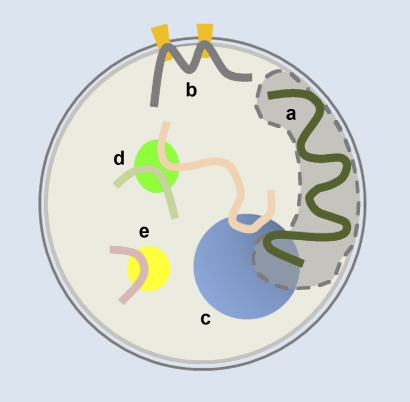
Figure 1.
Chromosome conformation and transcriptional activity are affected by the association of chromosomal regions with peripheral or central subcompartments. (a) The nuclear lamina and adjacent nuclear space, (b) nuclear pore complexes, (c) a nucleolus, where ribosomal DNA loci from different chromosomes cluster, (d) a transcription factory, where coregulated genes preferentially colocalize, and (e) a polycomb body. The filled gray region represents a CT that is interacting with both a and c.
Interphase chromosome territories (CTs)
The spatial organization of whole chromosomes has emerged in recent decades as an important factor in gene regulation and genome stability. Using Giemsa staining and light and electron microscopy, Stack et al. (1977) observed that chromosomes occupy distinct domains in the eukaryotic interphase nucleus. Cremer et al. (1982) were the first to provide experimental evidence for the existence of such interphase CTs (Cremer and Cremer, 2006, 2010; Heard and Bickmore, 2007). Targeting specific regions of the nucleus with a microlaser showed that damage was not randomly distributed across many chromosomes but limited to a few locations, and the investigators surmised that chromosomes must, therefore, be constrained to CTs. Arrangement of these CTs is defined relative to each other as well as by proximity to the nuclear periphery. Nonrandom positioning is demonstrated in the segregation of gene-rich and gene-poor chromosomes, which tend to localize toward the nuclear interior or periphery, respectively. This nonrandom organization of CTs is evident across many different cell types and appears to be conserved through eukaryotic evolution (Croft et al., 1999; Boyle et al., 2001; Cremer et al., 2001; Neusser et al., 2007). It is thought that this high level of organization contributes to inter- and intrachromosomal interactions and a coordinated expression among sets of genes. Dynamic activity in CTs complements the highly organized system. Long-range movements of gene loci to and from CTs have been reported (with distances ≤5 µm) and have been linked to gene activation and silencing, presumably as access to the transcriptional machinery changes (Chuang et al., 2006; Dundr et al., 2007; Meister et al., 2010). However, it is yet unclear whether gene looping is the primary mechanism for colocalization of distant loci or whether a higher order rearrangement of chromatin mediates the interaction (Strickfaden et al., 2010).
The 3D architecture of chromosomes can compartmentalize the nucleus and reflect regional gene expression (Kosak and Groudine, 2004; Bolzer et al., 2005; Misteli, 2007; Dekker, 2008), but the analysis of nuclear architecture has been limited by methods that focus on interactions between specific loci rather than an unbiased genome-wide analysis (Dostie et al., 2006; Simonis et al., 2006; Zhao et al., 2006). The chromosome conformation capture (3C) technique identifies chromatin interactions between two regions of interest by cross-linking, restriction enzyme digestion, intermolecular ligation, and PCR analysis of the resulting linked DNA fragments (Dekker et al., 2002). Recently described variants of the 3C technique have been used to investigate nuclear organization on a more global level (Lieberman-Aiden et al., 2009). Using one of these, Hi-C, which probes the 3D architecture of whole genomes by coupling proximity-based ligation with massively parallel sequencing, Lieberman-Aiden et al. (2009) and van Berkum et al. (2010) constructed proximity maps of the human genome in B cell and erythroid cell lines and confirmed the presence of CTs, proximity of small, gene-rich chromosomes, and the spatial segregation of open and closed chromatin.
The arrangement of interphase chromosomes into distinct territories is now generally accepted as a basic principle of nuclear organization (Cremer and Cremer, 2010). There are two prevalent models of CT organization in the interphase nucleus. In the CT–interchromatin compartment (IC) model, nuclei contain CTs and the IC. Interconnected higher order chromatin networks define the predominant structure within individual CTs (Visser et al., 2000; Albiez et al., 2006). The IC is a 3D contiguous spatial network that forms channels between CTs (Albiez et al., 2006; Rouquette et al., 2009). Actively transcribed genes lie at the surface of CTs, in the perichromatin region, where transcription, pre-mRNA processing, and DNA replication occur. Because of the variable width of ICs, the CT-IC model does not exclude the possibility of interactions between CTs, in which inter- and intrachromosomal rearrangements may occur (Markaki et al., 2011; Rouquette et al., 2010). In contrast, in the interchromatin network model, the interchromatin space acts as a commons for frequent intermingling of chromatin loops in cis and trans (Branco and Pombo, 2006). Chromatin looping out of CTs is only constrained by immediate steric interactions, such as intrachromosomal higher order conformation, adjacent chromosomes, nuclear subcompartments, and the nuclear envelope. Significant interchromosomal contact revealed by Hi-C data suggests regular intermingling, which is consistent with the interchromatin network model (Lieberman-Aiden et al., 2009). Intermingling of chromosomal regions may influence whole chromosome organization as well, potentially promoting chromosome-wide and genome-wide spatial repositioning by influencing the behavior of adjacent chromatin in cis and trans. Actively transcribed and repressed genes may frequently cluster into distinct functional subcompartments (Cremer and Cremer, 2010).
It is also important to consider two limitations of microscopy assays in the investigation of CTs and derivative models. First, repetitive sequences are eliminated during the generation of chromosome-specific paints. Although the common assumption is that repeats are buried within the CT, this has not been formally shown, and it is possible that repeats may coat a territory or extend from it. Second, although there are numerous studies of specific loci looping from their respective CTs to active or repressive subcompartments, the number of loops from any given CT is unknown (Volpi et al., 2000; Ragoczy et al., 2003). Moreover, by visualization of the CT and a specific looped locus, the surrounding sequences connecting the locus to the CT are generally not detected. Thus, the true extent of the CT (CT boundaries) and the extent of intermingling have not been very accurately defined given the limitations of current reagents and methodologies.
Peripheral dynamics
Nuclear organization will depend on a variety of cellular cues influenced by environment, developmental state, and cell cycle stage. Thus, models of nuclear organization must encompass the dynamic flexibility of chromosomes required by an adaptable and responsive nuclear environment, while at the same time allowing for a high level of order as reflected in the functional specialization of nuclear subcompartments. The relationship between spatial arrangement and gene expression is illustrated by the dynamic association of chromosomes with peripheral structures at the nuclear envelope as well as with more central nuclear subcompartments. In general, peripheral localization correlates with attenuated transcription and heterochromatin, whereas movement toward the nuclear center correlates with higher transcription and euchromatin. However, two competing domains may be influencing gene expression at the nuclear periphery.
One important component is the nuclear lamina, a network of intermediate filaments (lamins, LAP2β, emerin, etc.) lining the inner nuclear membrane, providing both a mechanical and signaling scaffold for the nucleus. The nuclear lamina has been shown to interact with chromatin, and loci at this interface tend to exhibit reduced expression. Moreover, tethering experiments in which loci are anchored to the nuclear lamina have revealed silencing upon peripheral localization (Finlan et al., 2008; Reddy et al., 2008). Genome-wide scanning for loci in close association with the nuclear lamina, using DNA adenine methyltransferase identification (DamID; a method that uses fusions to a bacterial methyltransferase to identify genomic binding sites of a particular protein), has revealed numerous lamin-associated domains (LADs) in largely silent heterochromatic regions of human, mouse, and Drosophila melanogaster nuclei (Pickersgill et al., 2006; Guelen et al., 2008). Regions in the tens of kilobases to >10 Mb have been identified, but the exact lamin-binding motifs have not been defined (Peric-Hupkes et al., 2010). The ubiquitous nature of these interactions was recently demonstrated by Peric-Hupkes et al. (2010), who identified LADs during spatial reorganization during the differentiation of embryonic stem cells (ESCs) into neural progenitor cells and astrocytes. The study revealed that in each of the three differentiation stages as well as in 3T3 mouse embryonic fibroblasts ∼40% of the genome is comprised of LADs and that LADs in the different cell types overlap by ∼70–90%. Notably, during differentiation, some LADs reposition away from the periphery, suggesting impending or actual transcriptional activity (Meister et al., 2010). Thus, although overlapping sets of LADs are present in different cell types regardless of lineage or developmental stage, LAD identity is dynamic.
Chromatin also interacts with components of the nuclear pore complex. Unlike peripheral association with the nuclear lamina, loci near nuclear pore complexes tend to be euchromatic and more actively transcribed (Blobel, 1985; Krull et al., 2010). The association between nuclear pore proteins and genomic DNA has been documented in yeast, Drosophila, and mammalian systems (Akhtar and Gasser, 2007; Luthra et al., 2007; Brown et al., 2008). In yeast, chromatin immunoprecipitation (ChIP) of nuclear pore–associated proteins has revealed interactions with actively transcribed genes (Casolari et al., 2004). Additionally, nuclear porins in the inner nuclear pore basket have been shown to associate with particular DNA sequences in active promoters, which may facilitate optimal gene expression (Schmid et al., 2006; Taddei et al., 2006; Ahmed et al., 2010). It is important to note that interactions between promoters and soluble nucleoplasmic nuclear porins have been documented; therefore, localization to the nuclear pore cannot be determined using ChIP and DamID alone and should be confirmed by FISH (Capelson et al., 2010; Kalverda et al., 2010). The localization of chromatin to the nuclear pore microenvironment could expedite transport of transcripts into the cytoplasm as a result of proximity, lower concentrations of obstructive condensed DNA, and higher concentrations of pre-mRNA–processing machinery (Blobel, 1985; Ishii et al., 2002; Kylberg et al., 2008; Krull et al., 2010; Strambio-De-Castillia et al., 2010).
Although mechanisms governing spatial positioning and how specific nuclear subcompartments at the periphery influence transcriptional state are not well understood, they appear to be important regulatory events in development. In a study in human ESCs, repressed chromatin, marked by H3K27 trimethylation, was enriched at the nuclear periphery, but a small percentage of active chromatin at the periphery was preserved. Differentiated cells contained the same percentage of active chromatin but had significantly less peripheral repressed chromatin than ESCs (Luo et al., 2009). In addition, some histone deacetylases associate with lamin-associated proteins at the periphery (Somech et al., 2005), suggesting an actively maintained repressive environment. Thus, targeting certain loci to the peripheral microenvironment may ensure the persistence of repression until appropriate cues signal the release and activation of developmentally required genes. During differentiation, loci containing up-regulated genes move from a repressive to an active nuclear compartment, whereas loci containing down-regulated genes move in the opposite direction (Brown et al., 1997; Skok et al., 2001; Kosak et al., 2002; Ragoczy et al., 2006). On a local level, movement of loci may be accompanied by the looping of loci from their CTs (Williams et al., 2006). Furthermore, investigation of the monoallelically expressed GFAP gene showed that the active allele is found in a more internal radial position than the nonexpressed allele (Takizawa et al., 2008).
A notable inversion of the predominant pattern of nuclear organization is found in retinal rod cells of nocturnal mammals, where, though the same high level of order is present, heterochromatin is internal and euchromatin is peripheral, an arrangement that minimizes light scattering (Solovei et al., 2009). Therefore, rod cells exemplify the broad flexibility of biological systems and also illustrate that the functional requirements of a specific cell type may closely guide nuclear architecture.
Central dynamics
Many other nuclear subcompartments participate in the dynamic regulation of the genome and likely impose a particular spatial configuration inside the interphase nucleus. For example, nucleoli, the most prominent nuclear bodies, emerge from the congregation of multiple tandem repeats of ribosomal DNA from several chromosomes. They are the sites of ribosomal RNA (rRNA) transcription by RNA polymerase I, posttranscriptional processing of the rRNA, and assembly of ribosomal subunits. Spatial clustering of these distant genomic loci is critical for their function, making nucleoli a prominent example of specialized nuclear compartmentalization (Boisvert et al., 2007).
The perinucleolar region is another nuclear domain with distinct characteristics, exhibiting low RNA polymerase II transcriptional activity (Németh et al., 2010). Sequencing, genome-wide microarray analysis, 3D FISH, and immunofluorescence have revealed that nucleolar-associated domains comprise ∼4% of the human genome, are largely transcriptionally repressed, and are enriched in repressive histone modifications and depleted of those associated with transcriptional activity (Németh et al., 2010). A notable exception to transcriptional repression in the perinucleolar region is the RNA polymerase III–transcribed tRNA genes (Bertrand et al., 1998). tRNA genes are enriched in nucleolar-associated domains, suggesting a possible common mechanism for the spatial positioning of the many genomic regions carrying tRNA genes (Németh et al., 2010). This positioning is dependent on RNA polymerase III occupancy and transcriptional activity, as promoter point mutations eliminate association with the nucleolus (Thompson et al., 2003). The close spatial proximity of tRNA and rRNA synthesis could be important for the coordinated expression of translational machinery.
Polycomb bodies provide another example of spatial positioning in the interior of the nucleus that results in gene repression. Polycomb bodies are discrete foci that consist of polycomb group (PcG) proteins, which attenuate gene expression in many cell types, and target loci that contain PcG response elements (Saurin et al., 1998). PcG proteins and response elements have been shown to facilitate long-range interchromosomal interactions of multiple loci even when loci are artificially inserted into ectopic sites (Bantignies et al., 2003; Vazquez et al., 2006). Genes associated with polycomb bodies undergo chromatin modifications catalyzed by the PcG complex proteins PRC1 and PRC2 (mechanisms of PcG repression are reviewed in Morey and Helin, 2010). Polycomb-mediated repression appears to be critical for maintaining the appropriate temporal repression of loci during development, as their disruption can induce the activation of differentiation pathways (O’Carroll et al., 2001; Pasini et al., 2007; Ezhkova et al., 2009).
Transcription factories
Dynamic movements of genes into and out of a CT may permit specific loci or sets of coregulated genes to rapidly engage the transcription machinery. Indeed, transcription of coordinately regulated genes that rely on the same transcription factors and cofactors would be much more efficient if this machinery were preassembled at high local concentrations in compartments. The observation that labeled nascent transcripts localize to discrete transcription foci led to the concept of such “transcription factories” (Jackson et al., 1993; Wansink et al., 1993), and it is now known that RNA polymerase II accumulates in transcription factories at ≤1,000-fold higher concentrations than elsewhere in the nucleoplasm (Cook, 2002). In addition, the observation that RNA polymerases are relatively immobile and anchor to nuclear structures supports the transcription factory model, as actively transcribing genes can move efficiently through clustered polymerase complexes rather than the polymerase tracking along DNA (Sexton et al., 2007; Papantonis et al., 2010).
Transcription factories specialize depending on the type of polymerase, the transcription factors, and the chromosomal regions present. For example, RNA polymerase I localizes to nucleoli for ribosome synthesis, whereas RNA polymerase II and III localize to dedicated, mutually exclusive nucleoplasmic regions (Pombo et al., 1999). Genomic regions and cell type may also determine the transcription factory function reviewed in Bartlett et al. (2006). In a recent study, Schoenfelder et al. (2010) used a variation of 3C to screen the genome for loci that colocalize with active α- and β-globin genes (Hba and Hbb) in erythroid cells. Many erythroid-specific genes, both on the same and different chromosomes, were found associated with the globin genes. Each globin gene colocalized with sets of loci with very little overlap. Moreover, a subset of genes identified by colocalization with Hbb shared binding sites for Krüppel-like factor 1 (Klf1), an erythroid lineage transcription factor. Klf1 was found in a subset of transcription factories along with the identified Klf1-regulated genes, suggesting the existence of lineage-specific factories optimized for the expression of coregulated genes. Consistent with this hypothesis, in the absence of Klf1, the coregulated genes failed to colocalize.
However, many questions remain, such as whether transcription factories are stable or whether they spontaneously and transiently self-organize and whether the developmental state influences the status and dynamics of transcription factories. Confirming the validity of transcription factor dynamics will require further investigation of loci on a genomic scale as well as development of higher resolution detection methods and tracking transcription factory formation and localization over time.
Methods for exploring the 3D genome
A variety of biochemical methods have been developed to investigate the conformation of chromatin (or chromosomal) regions, providing insight into nuclear subcompartments and local gene expression patterns. Initially, such characterizations only analyzed one or a few loci at most (Dostie et al., 2006; Simonis et al., 2006; Zhao et al., 2006), but the recent development of unbiased high throughput genome-wide spatial analyses has the potential to reveal critical global characteristics of the nucleus that may participate in the regulation of transcriptional programs.
As previously stated, the recently developed Hi-C method probes the 3D architecture of whole genomes (Fig. 2 A) by coupling proximity-based ligation with massively parallel sequencing (Fig. 2 B). Spatial proximity maps of the human genome have been constructed using Hi-C at a resolution of 0.1–1 Mb (Lieberman-Aiden et al., 2009), and the use of a similar method to Hi-C has provided a model of the 3D architecture of the yeast genome (Duan et al., 2010). Maps constructed using the human genome data support the presence of CTs and the spatial proximity of small, gene-rich chromosomes. The maps also identified an additional level of genome organization that is characterized by the spatial segregation of open and closed chromatin (based on DNase I hypersensitivity) into two genome-wide compartments. Although the compartment patterns appear to be similar between different cell types, the composition of individual loci differs, and there is a strong correlation between the compartment pattern and chromatin accessibility in the same cell type (Lieberman-Aiden et al., 2009). These results demonstrate the power of Hi-C to map the conformations of whole genomes and, furthermore, that open and closed chromatin domains throughout the genome occupy different spatial compartments in the nucleus. These patterns may distinguish specific cell types or states. The predecessor method to Hi-C, 3C, only probes small genomic regions. Advances in this technique, such as 4C or 5C, allow analysis of whole genomic regions or long-range interactions but are still biased in the sense that they rely on PCR amplification with specific bait primer sets and application to microarrays (for more information on these techniques see Naumova and Dekker, 2010; van Steensel and Dekker, 2010). Another recent method, chromatin interaction analysis with paired-end ditag sequencing, uses a combination of ChIP with a specific antibody, intramolecular ligation of enriched sequences, and sequencing, allowing the detection of protein factor binding as well as long-range interactions (Fullwood et al., 2009).
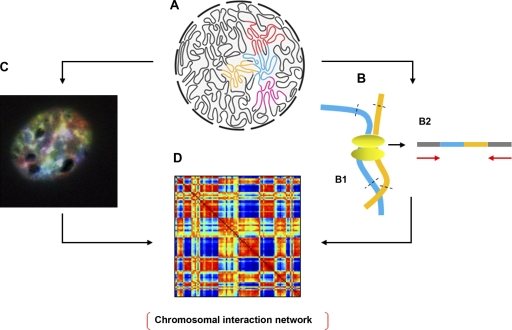
Figure 2.
Methods for exploring the 3D genome. (A) A schematic representation of the 3D genome (courtesy of J. Dekker and N.L. van Berkum, University of Massachusetts Medical School, Worcester, MA). (B) The Hi-C technique. (B1) DNA is cross-linked and digested with restriction enzymes. (B2) High throughput sequencing (8.5 million reads) is used to determine the spatial proximity of sequences, including those on the same or different chromosomes using paired-end sequencing. (C) Image of a murine hematopoietic progenitor interphase nucleus labeled by SKY. All chromosomes are labeled with a unique color to visualize their territories. (D) Once algorithms are developed to characterize the spatial relationships among all CTs in the interphase nucleus simultaneously from SKY data, spatial proximity maps can be generated that integrate both SKY and Hi-C data to more precisely define inter- and intrachromosomal interactions.
As described for CT analyses using FISH whole chromosome probes, one major issue with these approaches is that they exclude many repetitive and nonprotein-coding sequences. Because identified sequences are mapped back to the genome, any sequences from nonannotated genomic regions and repeats are not considered in the analysis (Shapiro and von Sternberg, 2005; Alexander et al., 2010). Repetitive elements are found throughout the genome, and it is possible that they influence expressed loci through long-range regulatory regions or by having other structural roles in higher order chromatin arrangements (O’Sullivan et al., 2009). For example, macrosatellite D4Z4 repeats in chromosome 4 appear to have an epigenetic role in the expression of a locus that acts in a dominant fashion to cause facioscapulohumeral muscular dystrophy. With >10 repeats, the entire region is heterochromatic and methylated, whereas with <10 repeats, the region is less methylated and less heterochromatic, allowing expression of the “toxic” DUX4 transcript (Lemmers et al., 2010). Spatial analysis has yet to be conducted, but an attractive explanation is that architecture and long-range interactions are critical for activation or silencing in this region.
Current biochemical chromatin conformation analyses measure the probability of any given interaction by the number of sequencing reads obtained from a population of cells. However, these techniques do not reveal which interactions are functionally relevant, which must be confirmed experimentally. Furthermore, these techniques require millions of cells to improve the likelihood of capturing a particular interaction, making it difficult to determine which interactions/configurations occur simultaneously in any given cell (Simonis et al., 2007). High resolution live-cell imaging, with gene loci and nuclear bodies labeled using bright fluorophores and low photobleaching or novel multiplex labeling systems, will likely be necessary to validate many findings about the genomic organization at the single-cell level. Microscopy approaches more effectively reveal individual nuclear components and their topography with respect to each other. Multicolor DNA and RNA FISH (M-FISH or 3D-FISH) allows visualization of interphase chromosomes in their entirety as well as transcription events. The spectral karyotyping (SKY) technique uses 3D-FISH as well as specialized equipment and computational analyses to allow capture of the entire emission spectrum in a single image (Fig. 2 C). These techniques facilitate comparisons that may help identify cell-to-cell variability of chromatin arrangements and cell type–specific arrangements of certain chromatin domains.
Bolzer et al. (2005) were the first to use 3D-FISH to simultaneously detect all chromosomes in single diploid human fibroblasts in interphase and in prometaphase rosettes. They discovered that particular CTs consistently bordered only select neighbors, and their analysis suggested that the intermingling of chromatin loops between CTs was not extensive, as this would have made the CTs much less discrete. However, this does not exclude the possibility of nonrandom intermingling between CTs or colocalization of genes in the IC. In addition, investigation of relative positioning of all CTs has not been revisited or explored in other cell types. If significant intermingling does occur, accurate measurements of chromosome shape and relative position will be difficult to determine by microscopy (Cremer and Cremer, 2010).
One difficulty with any 3D characterization using labeling methods and microscopy is the shape of nuclei, which depends on cell type. Human fibroblasts, for example, have flat ellipsoid nuclei, in which the orientation of structures within can be well defined using major and minor axes. Spherical nuclei, however, present a challenge in that it is difficult to assign a spatial coordinate system and polarity. Geometric computational techniques can assist in defining specific features, such as a central point or centroid, and help to characterize relationships between CTs by determining the distance between centroids and the closest distance between any two CTs. Development of more sophisticated computational tools may permit further investigation of complex CT characteristics, such as shared volume and contact area (Eils et al., 1996; Roix et al., 2003; Cremer and Cremer, 2010). Furthermore, microscopy techniques that provide a higher resolution in three dimensions, such as laser-based microscopy and focused ion beam with scanning electron microscopy, will be critical to refining existing models of chromatin organization (Hell, 2007; Schroeder-Reiter et al., 2009).
Although the single time points in recent Hi-C and M-FISH studies have provided important clues about how genomes organize (Bolzer et al., 2005; Lieberman-Aiden et al., 2009), extending this to multiple time points is required to understand the dynamic flexibility of chromosome organization. Across a time course of differentiation, for example, much can be learned about the importance of gene positioning in the context of development and transcriptional regulation. An investigation of two time points during differentiation in the murine hematopoietic lineage suggests that dramatic changes in total genomic order take place (Rajapakse et al., 2009). In this study, prometaphase rosettes were examined using SKY. Although this arrangement was validated by pairwise comparisons of a few interphase CTs (Kosak et al., 2007), it is not yet known whether all prometaphase chromosome arrangements are reflected in the interphase nucleus. Moreover, additional time points during the course of differentiation may provide a more detailed picture of chromosome reorganization. An additional consideration in whole-genome 3D analyses is the use of a nontransformed primary cell system with a normal karyotype.
Technical advances also have facilitated a genome-wide study of specific interactions and processes in the linear genome. Replication timing during S phase can be tracked using BrdU pulse labeling, microarray analysis, and deep sequencing (Hiratani et al., 2008); transcription factor–DNA interactions or histone modifications can be globally profiled using ChIP combined with microarray analysis or sequencing (ChIP-chip or ChIP-seq); and chromatin-interacting proteins can be tracked using DamID. Heterochromatin and euchromatin can also be profiled using DNase I treatments that target open chromatin coupled with ligation-mediated PCR amplification and microarrays or sequencing (Naumova and Dekker, 2010). Comparisons between such profiles and spatial proximity maps may provide additional information about how seemingly separate nuclear processes coincide and influence each other. Ryba et al. (2010) showed that G1 replication timing decision points were related to spatial architecture. Many other potential relationships await investigation. For example, transcription factors that act as master regulators, such as MyoD in differentiating skeletal muscle cells, may influence nuclear architecture. MyoD, in addition to its expected binding of muscle-specific regulatory regions, also binds broadly throughout the genome at sites associated with histone acetylation before muscle cell differentiation. Thus, MyoD may also play a role in regulating epigenetic features and genomic organization (Cao et al., 2010).
Spatial positioning and cancer
Various cancers are associated with specific translocations. It has been shown that the high frequency of certain translocations reflects prevalent tissue-specific spatial positioning of particular chromosomes, as these translocations occur when the chromosome partners are in close proximity (Misteli, 2004; Soutoglou and Misteli, 2008). Moreover, translocations affect spatial positioning as well as gene expression (Croft et al., 1999; Taslerová et al., 2003; Harewood et al., 2010) and may play a role in tumorigenesis through disruption of the coupling between spatial arrangements and expression. For example, Harewood et al. (2010) studied the effect of a reciprocal translocation between chromosome 11 and 22 (t(11;22)(q23;q11)) that is associated with an increased risk of breast cancer and has been shown to affect the spatial positioning of chromosomes. In a transcriptome analysis, cell lines from individuals with the translocation had higher numbers of differentially expressed genes compared with the expected variation in cell lines from individuals without it (Harewood et al., 2010). However, not every translocation, even when associated with tumorigenesis, results in changes in spatial positioning or gene expression (Snow et al., 2011). This is not a surprising result if chromosome arrangement is a key feature in gene regulation. The time or path to reach an important global configuration may change after a translocation or mutation, but only major disruptions in chromosome topology that alter localization patterns of individual genes may increase the likelihood of disease states.
Repositioning in cancer cells, compared with normal tissue, also appears to be gene specific and has been linked to disease-specific 3D rearrangements. Meaburn et al. (2009) identified several genes with altered localization in cells derived from invasive breast cancer tissue compared with cells from normal tissue. Changes in copy number did not correlate with chromosome rearrangement, suggesting that spatial repositioning was not a result of genome instability. In addition, breast cancer cells could be distinguished from noncancer cells based on gene repositioning alone with nearly 100% accuracy using the gene HES5 (Meaburn et al., 2009). Thus, spatial organization appears to be an important feature related to nuclear function that has a potential diagnostic utility for identifying cancerous tissue.
Perspective
Self-organization and optimum configurations.
It has been suggested that the positioning of genes and chromosomes as well as formation of nuclear subcompartments is the result of self-organization (Misteli, 2001; Kosak and Groudine, 2004). Self-organization in a system is a process by which the global level pattern emerges solely from many interactions among lower level components; the pattern is an emergent property of the system rather than a property imposed on the system by an external ordering influence (Ashby, 1947; Camazine et al., 2003). The rules for behavior in such systems are nonlinear, and the whole of a nonlinear system is not simply an additive function of its parts (Anderson, 1972; Strogatz, 1994, 2001, 2003). A more refined view of self-organization is that the global pattern, although not in control of the local interactions, can feed back to influence those local components (Fig. 3 A). The resulting changes in local behavior may then change the global pattern, and the self-organized system fine tunes over time. Thus, self-organized systems have local to global and global to local feedback that leads to increasing order, and over time, the system exhibits a continual interplay of bottom-up and top-down processes. Therefore, the coordination of the activities of individual complex elements enables a system to develop, sustain complexity at a higher level, and change over time (Fig. 3 B).
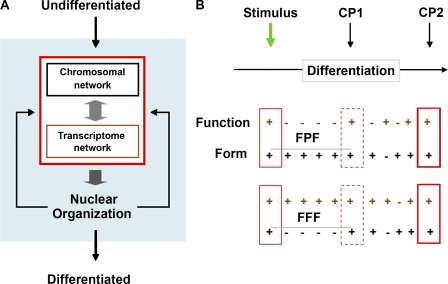
Figure 3.
The mechanics of self-organization and differentiation. (A) Local interactions that make up the transcriptome network (gene coregulation) and spatial architecture that makes up the chromosomal network mutually influence each other and lead to the emergence of cell-specific nuclear organization. In turn, this order feeds back to strengthen the local associations, and the self-organized system fine tunes over time. (B) Two possible models, form precedes function (FPF) and form follows function (FFF), representing the relationship between chromosomal (form) and transcriptome (function) networks over time during differentiation (horizontal black arrow). At the green arrow, a stimulus initiates the process of differentiation. CP1 and CP2 are critical points 1 and 2 or defined states in nuclear architecture. The solid, dotted, and bold boxes represent nuclear organization as determined by the interaction between the networks. Plus and minus symbols show changing or unchanging architecture, respectively, within each network. In the undifferentiated state in both models, the transcription and space networks are changing and mutually influence each other. After the stimulus in the form precedes function model, only the space network changes over time until CP1. At CP1, when the space network has reached a particular configuration, the transcription network begins to change, and again they become mutually related. In the form follows function model, only the transcription network changes over time until CP1, and the system behaves in the opposite fashion as the form precedes function model, with the transcription network leading the space network. Between CP1 and CP2 in both models, the system fine tunes, or feedback between the networks leads to the optimum configuration for terminal differentiation at CP2.
An intriguing example of self-organization in biology is the slime mold Physarum polycephalum, which has a talent for finding the most efficient configuration in its quest for food. When the slime mold is placed near an array of different-sized oat flakes, it makes contact with all flakes within reach. Next, it begins erasing connections between smaller oat flakes and strengthening the connections between bigger and central flakes. Eventually, it will converge to the optimal solution in which the most robust connections are established between the most important/central nodes and the redundant connections slowly disappear (Tero et al., 2010). In many scientific disciplines, optimality has long been an organizing principle of a system. We can argue the same for the living cell. The cell is a self-organizing, self-replicating, environmentally responsive machine of staggering complexity. The instructions for this complexity are contained within the cell’s genetic code, but how this information is accessed, read, and interpreted is influenced by the environment and epigenetic factors during development and differentiation.
Networks.
The basic mechanisms underlying self-organization in complex biological networks are still far from clear. However, self-organizing systems can be simplified, while retaining complex information, by the deconstruction of their elements into well-defined networks. If we think of the nucleus as a system composed of such networks, studying the dynamics between networks may provide a useful framework for investigating complex 4D nuclear organization. A network—a graph in the mathematics literature—is a collection of points, or nodes, joined in some fashion by lines, or edges. In recent years, there has been a strong upsurge in the study of networks in many disciplines, ranging from computer science and communications to sociology and epidemiology (Newman et al., 2006). For characterizing nuclear function, both gene regulation and spatial configuration can be defined in terms of networks (Rajapakse et al., 2010). The configuration of a gene relative to the host chromosome as well as to genes on other chromosomes will be referred to as the spatial network, subsequently shown in Fig. 4 A. Fig. 4 A shows the structural organization of the nucleus. Fig. 4 B shows the transcriptome network, which represents the functional organization of the nucleus. In Fig. 4 A, nodes are chromosomes or genes in which the edges are computed based on the proximity of chromosomes. In Fig. 4 B, nodes are chromosomes or genes, and the edges are computed based on gene coregulation. One of the measures of gene coregulation is the relative entropy between gene expression profiles. It has been shown that the two cell networks shown in Fig. 4 (A and B) are not only related during differentiation but also highly correlated (Rajapakse et al., 2009). This intriguing connection has been established by defining and showing the collective similarity between the coregulated gene regulatory network and the chromosomal interaction network in the nucleus during in vitro differentiation of murine hematopoietic progenitors (Bruno et al., 2004; Kosak et al., 2007; Rajapakse et al., 2009). These studies suggest that, in fact, gene coregulation leads to chromosomal associations, which in turn feed back to strengthen local associations. Thus, the nucleus reorganizes itself functionally and structurally. Although the global reorganization of chromosomes occurs during differentiation (Kim et al., 2004; Parada et al., 2004; Kosak et al., 2007), it is unclear whether local changes in positioning, such as the looping of loci from CTs to active or repressive compartments, drive global reorganization on the whole chromosome level or vice versa.
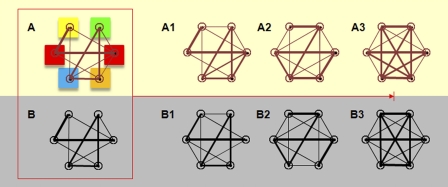
Figure 4.
A network view of differentiation showing spatial and transcriptome networks in which nodes are genes or chromosomes. (A–B) Networks representing the signature of the undifferentiated state, in which some genes are connected, and some are not. The connectivity is weighted; that is, the strength of the connection depends on the gene pair, which is shown by edge thickness. In A, the colored boxes indicate their subnuclear location as described in Fig. 1, which defines node dynamics and, therefore, network edges (spatial distances). In B, the connectivity is also weighted, which defines gene coregulation. A1–A3 and B1–B3 represent the organization of the spatial and transcriptome networks during differentiation. The networks mutually influence each other, and fine tuning over time gives the differentiated system a unique network signature.
Within the context of these recent results, the major question of how the structural organization of the nucleus relates to its function remains unanswered. By using a combination of Hi-C and interphase 3D-FISH along with more sophisticated computational and imaging techniques, we may be able to move toward a more complete map of local and global spatial proximities with which to construct the chromosomal interaction network (Fig. 2 D), allowing more accurate snapshots of the dynamics of a cell’s functional and structural networks. One important question is how to identify the fundamental processes that help establish stable transitions between cellular states during development. For example, we can address on a global scale whether lineage determination patterns a specific nuclear architecture that shapes the expression of differentiation genes or whether the transcription of differentiation genes initiates transitions in nuclear architecture. We suggest that investigating the relationships between nuclear architecture (form) and expression (function) will be critical to improve our understanding of cell fate (Fig. 3 B), including missteps that can propel normal cells into an unstable state that leads to cancer. By studying disruptions in networks that globally represent the nucleus of any cell type, we can potentially predict instabilities and ultimately determine how to redirect cells from a differentiated state to pluripotency or from a pathological to a benign state. Emerging technologies in microscopy, high throughput biochemical assays, and computational tools invite an integrated approach that puts a more refined understanding of nuclear organization within reach.
Acknowledgments
We thank Lindsey Muir, Tobias Ragoczy, David Scalzo, Hector Rincon, Michael Morrison, and Stephen Tapscott for discussions and critical reading of the manuscript. We also thank Job Dekker and Nynke L. van Berkum for providing Fig. 2 A.
I. Rajapakse is supported by the Mentored Quantitative Research Career Development Award (K25) from the National Institutes of Health (NIH; grant 1K25DK082791-01A109), and M. Groudine is supported by the NIH (grants R37 DK44746 and RO1 HL65440).
Footnotes
Abbreviations used in this paper:
- ChIP
- chromatin immunoprecipitation
- CT
- chromosome territory
- DamID
- DNA adenine methyltransferase identification
- ESC
- embryonic stem cell
- IC
- interchromatin compartment
- LAD
- lamin-associated domain
- PcG
- polycomb group
- rRNA
- ribosomal RNA
- SKY
- spectral karyotyping
References
- Ahmed S., Brickner D.G., Light W.H., Cajigas I., McDonough M., Froyshteter A.B., Volpe T., Brickner J.H. 2010. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat. Cell Biol. 12:111–118 10.1038/ncb2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar A., Gasser S.M. 2007. The nuclear envelope and transcriptional control. Nat. Rev. Genet. 8:507–517 10.1038/nrg2122 [DOI] [PubMed] [Google Scholar]
- Albiez H., Cremer M., Tiberi C., Vecchio L., Schermelleh L., Dittrich S., Küpper K., Joffe B., Thormeyer T., von Hase J., et al. 2006. Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res. 14:707–733 10.1007/s10577-006-1086-x [DOI] [PubMed] [Google Scholar]
- Alexander R.P., Fang G., Rozowsky J., Snyder M., Gerstein M.B. 2010. Annotating non-coding regions of the genome. Nat. Rev. Genet. 11:559–571 10.1038/nrg2814 [DOI] [PubMed] [Google Scholar]
- Anderson P.W. 1972. More is different. Science. 177:393–396 10.1126/science.177.4047.393 [DOI] [PubMed] [Google Scholar]
- Ashby W.R. 1947. Principles of the self-organizing dynamic system. J. Gen. Psychol. 37:125–128 10.1080/00221309.1947.9918144 [DOI] [PubMed] [Google Scholar]
- Bantignies F., Grimaud C., Lavrov S., Gabut M., Cavalli G. 2003. Inheritance of Polycomb-dependent chromosomal interactions in Drosophila. Genes Dev. 17:2406–2420 10.1101/gad.269503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett J., Blagojevic J., Carter D., Eskiw C., Fromaget M., Job C., Shamsher M., Trindade I.F., Xu M., Cook P.R. 2006. Specialized transcription factories. Biochem. Soc. Symp. 73:67–75 [DOI] [PubMed] [Google Scholar]
- Bertrand E., Houser-Scott F., Kendall A., Singer R.H., Engelke D.R. 1998. Nucleolar localization of early tRNA processing. Genes Dev. 12:2463–2468 10.1101/gad.12.16.2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G. 1985. Gene gating: a hypothesis. Proc. Natl. Acad. Sci. USA. 82:8527–8529 10.1073/pnas.82.24.8527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert F.-M., van Koningsbruggen S., Navascués J., Lamond A.I. 2007. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8:574–585 10.1038/nrm2184 [DOI] [PubMed] [Google Scholar]
- Bolzer A., Kreth G., Solovei I., Koehler D., Saracoglu K., Fauth C., Müller S., Eils R., Cremer C., Speicher M.R., Cremer T. 2005. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 3:e157 10.1371/journal.pbio.0030157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle S., Gilchrist S., Bridger J.M., Mahy N.L., Ellis J.A., Bickmore W.A. 2001. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum. Mol. Genet. 10:211–219 10.1093/hmg/10.3.211 [DOI] [PubMed] [Google Scholar]
- Branco M.R., Pombo A. 2006. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 4:e138 10.1371/journal.pbio.0040138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.R., Kennedy C.J., Delmar V.A., Forbes D.J., Silver P.A. 2008. Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev. 22:627–639 10.1101/gad.1632708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K.E., Guest S.S., Smale S.T., Hahm K., Merkenschlager M., Fisher A.G. 1997. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 91:845–854 10.1016/S0092-8674(00)80472-9 [DOI] [PubMed] [Google Scholar]
- Bruno L., Hoffmann R., McBlane F., Brown J., Gupta R., Joshi C., Pearson S., Seidl T., Heyworth C., Enver T. 2004. Molecular signatures of self-renewal, differentiation, and lineage choice in multipotential hemopoietic progenitor cells in vitro. Mol. Cell. Biol. 24:741–756 10.1128/MCB.24.2.741-756.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camazine S., Deneubourg J.-L., Franks N.R., Sneyd J., Theraulaz G., Bonabeau E. 2003. Self-Organization in Biological Systems. Princeton University Press, Princeton, NJ: 538 pp [Google Scholar]
- Cao Y., Yao Z., Sarkar D., Lawrence M., Sanchez G.J., Parker M.H., MacQuarrie K.L., Davison J., Morgan M.T., Ruzzo W.L., et al. 2010. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev. Cell. 18:662–674 10.1016/j.devcel.2010.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M., Liang Y., Schulte R., Mair W., Wagner U., Hetzer M.W. 2010. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 140:372–383 10.1016/j.cell.2009.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari J.M., Brown C.R., Komili S., West J., Hieronymus H., Silver P.A. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 117:427–439 10.1016/S0092-8674(04)00448-9 [DOI] [PubMed] [Google Scholar]
- Chuang C.-H., Carpenter A.E., Fuchsova B., Johnson T., de Lanerolle P., Belmont A.S. 2006. Long-range directional movement of an interphase chromosome site. Curr. Biol. 16:825–831 10.1016/j.cub.2006.03.059 [DOI] [PubMed] [Google Scholar]
- Cook P.R. 2002. Predicting three-dimensional genome structure from transcriptional activity. Nat. Genet. 32:347–352 10.1038/ng1102-347 [DOI] [PubMed] [Google Scholar]
- Cremer M., von Hase J., Volm T., Brero A., Kreth G., Walter J., Fischer C., Solovei I., Cremer C., Cremer T. 2001. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res. 9:541–567 10.1023/A:1012495201697 [DOI] [PubMed] [Google Scholar]
- Cremer T., Cremer C. 2006. Rise, fall and resurrection of chromosome territories: a historical perspective. Part I. The rise of chromosome territories. Eur. J. Histochem. 50:161–176 [PubMed] [Google Scholar]
- Cremer T., Cremer M. 2010. Chromosome territories. Cold Spring Harbor Perspect. Biol. 2:a003889 10.1101/cshperspect.a003889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T., Cremer C., Baumann H., Luedtke E.K., Sperling K., Teuber V., Zorn C. 1982. Rabl’s model of the interphase chromosome arrangement tested in Chinese hamster cells by premature chromosome condensation and laser-UV-microbeam experiments. Hum. Genet. 60:46–56 10.1007/BF00281263 [DOI] [PubMed] [Google Scholar]
- Croft J.A., Bridger J.M., Boyle S., Perry P., Teague P., Bickmore W.A. 1999. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 145:1119–1131 10.1083/jcb.145.6.1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J. 2008. Gene regulation in the third dimension. Science. 319:1793–1794 10.1126/science.1152850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J., Rippe K., Dekker M., Kleckner N. 2002. Capturing chromosome conformation. Science. 295:1306–1311 10.1126/science.1067799 [DOI] [PubMed] [Google Scholar]
- Dostie J., Richmond T.A., Arnaout R.A., Selzer R.R., Lee W.L., Honan T.A., Rubio E.D., Krumm A., Lamb J., Nusbaum C., et al. 2006. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 16:1299–1309 10.1101/gr.5571506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z., Andronescu M., Schutz K., McIlwain S., Kim Y.J., Lee C., Shendure J., Fields S., Blau C.A., Noble W.S. 2010. A three-dimensional model of the yeast genome. Nature. 465:363–367 10.1038/nature08973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M., Ospina J.K., Sung M.-H., John S., Upender M., Ried T., Hager G.L., Matera A.G. 2007. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J. Cell Biol. 179:1095–1103 10.1083/jcb.200710058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eils R., Dietzel S., Bertin E., Schröck E., Speicher M.R., Ried T., Robert-Nicoud M., Cremer C., Cremer T. 1996. Three-dimensional reconstruction of painted human interphase chromosomes: active and inactive X chromosome territories have similar volumes but differ in shape and surface structure. J. Cell Biol. 135:1427–1440 10.1083/jcb.135.6.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E., Pasolli H.A., Parker J.S., Stokes N., Su I.H., Hannon G., Tarakhovsky A., Fuchs E. 2009. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 136:1122–1135 10.1016/j.cell.2008.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlan L.E., Sproul D., Thomson I., Boyle S., Kerr E., Perry P., Ylstra B., Chubb J.R., Bickmore W.A. 2008. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 4:e1000039 10.1371/journal.pgen.1000039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood M.J., Liu M.H., Pan Y.F., Liu J., Xu H., Mohamed Y.B., Orlov Y.L., Velkov S., Ho A., Mei P.H., et al. 2009. An oestrogen-receptor-α-bound human chromatin interactome. Nature. 462:58–64 10.1038/nature08497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L., Pagie L., Brasset E., Meuleman W., Faza M.B., Talhout W., Eussen B.H., de Klein A., Wessels L., de Laat W., van Steensel B. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 453:948–951 10.1038/nature06947 [DOI] [PubMed] [Google Scholar]
- Harewood L., Schütz F., Boyle S., Perry P., Delorenzi M., Bickmore W.A., Reymond A. 2010. The effect of translocation-induced nuclear reorganization on gene expression. Genome Res. 20:554–564 10.1101/gr.103622.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E., Bickmore W. 2007. The ins and outs of gene regulation and chromosome territory organisation. Curr. Opin. Cell Biol. 19:311–316 10.1016/j.ceb.2007.04.016 [DOI] [PubMed] [Google Scholar]
- Hell S.W. 2007. Far-field optical nanoscopy. Science. 316:1153–1158 10.1126/science.1137395 [DOI] [PubMed] [Google Scholar]
- Hiratani I., Ryba T., Itoh M., Yokochi T., Schwaiger M., Chang C.-W., Lyou Y., Townes T.M., Schübeler D., Gilbert D.M. 2008. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 6:e245 10.1371/journal.pbio.0060245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K., Arib G., Lin C., Van Houwe G., Laemmli U.K. 2002. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell. 109:551–562 10.1016/S0092-8674(02)00756-0 [DOI] [PubMed] [Google Scholar]
- Jackson D.A., Hassan A.B., Errington R.J., Cook P.R. 1993. Visualization of focal sites of transcription within human nuclei. EMBO J. 12:1059–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalverda B., Pickersgill H., Shloma V.V., Fornerod M. 2010. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 140:360–371 10.1016/j.cell.2010.01.011 [DOI] [PubMed] [Google Scholar]
- Kim S.H., McQueen P.G., Lichtman M.K., Shevach E.M., Parada L.A., Misteli T. 2004. Spatial genome organization during T-cell differentiation. Cytogenet. Genome Res. 105:292–301 10.1159/000078201 [DOI] [PubMed] [Google Scholar]
- Kosak S.T., Groudine M. 2004. Form follows function: The genomic organization of cellular differentiation. Genes Dev. 18:1371–1384 10.1101/gad.1209304 [DOI] [PubMed] [Google Scholar]
- Kosak S.T., Skok J.A., Medina K.L., Riblet R., Le Beau M.M., Fisher A.G., Singh H. 2002. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 296:158–162 10.1126/science.1068768 [DOI] [PubMed] [Google Scholar]
- Kosak S.T., Scalzo D., Alworth S.V., Li F., Palmer S., Enver T., Lee J.S.J., Groudine M. 2007. Coordinate gene regulation during hematopoiesis is related to genomic organization. PLoS Biol. 5:e309 10.1371/journal.pbio.0050309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull S., Dörries J., Boysen B., Reidenbach S., Magnius L., Norder H., Thyberg J., Cordes V.C. 2010. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J. 29:1659–1673 10.1038/emboj.2010.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kylberg K., Björkroth B., Ivarsson B., Fomproix N., Daneholt B. 2008. Close coupling between transcription and exit of mRNP from the cell nucleus. Exp. Cell Res. 314:1708–1720 10.1016/j.yexcr.2008.02.003 [DOI] [PubMed] [Google Scholar]
- Lemmers R.J.L.F., van der Vliet P.J., Klooster R., Sacconi S., Camaño P., Dauwerse J.G., Snider L., Straasheijm K.R., van Ommen G.J., Padberg G.W., et al. 2010. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 329:1650–1653 10.1126/science.1189044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O., et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 326:289–293 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L., Gassman K.L., Petell L.M., Wilson C.L., Bewersdorf J., Shopland L.S. 2009. The nuclear periphery of embryonic stem cells is a transcriptionally permissive and repressive compartment. J. Cell Sci. 122:3729–3737 10.1242/jcs.052555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra R., Kerr S.C., Harreman M.T., Apponi L.H., Fasken M.B., Ramineni S., Chaurasia S., Valentini S.R., Corbett A.H. 2007. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J. Biol. Chem. 282:3042–3049 10.1074/jbc.M608741200 [DOI] [PubMed] [Google Scholar]
- Markaki Y., Gunkel M., Schermelleh L., Beichmanis S., Neumann J., Heidemann M., Leonhardt H., Eick D., Cremer C., Cremer T. 2011. Functional nuclear organization of transcription and DNA replication: a topographical marriage between chromatin domains and the interchromatin compartment. Cold Spring Harbor Symp. Quant. Biol. In press [DOI] [PubMed] [Google Scholar]
- Meaburn K.J., Gudla P.R., Khan S., Lockett S.J., Misteli T. 2009. Disease-specific gene repositioning in breast cancer. J. Cell Biol. 187:801–812 10.1083/jcb.200909127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister P., Towbin B.D., Pike B.L., Ponti A., Gasser S.M. 2010. The spatial dynamics of tissue-specific promoters during C. elegans development. Genes Dev. 24:766–782 10.1101/gad.559610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. 2001. The concept of self-organization in cellular architecture. J. Cell Biol. 155:181–185 10.1083/jcb.200108110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. 2004. Spatial positioning; a new dimension in genome function. Cell. 119:153–156 10.1016/j.cell.2004.09.035 [DOI] [PubMed] [Google Scholar]
- Misteli T. 2007. Beyond the sequence: cellular organization of genome function. Cell. 128:787–800 10.1016/j.cell.2007.01.028 [DOI] [PubMed] [Google Scholar]
- Morey L., Helin K. 2010. Polycomb group protein-mediated repression of transcription. Trends Biochem. Sci. 35:323–332 10.1016/j.tibs.2010.02.009 [DOI] [PubMed] [Google Scholar]
- Naumova N., Dekker J. 2010. Integrating one-dimensional and three-dimensional maps of genomes. J. Cell Sci. 123:1979–1988 10.1242/jcs.051631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Németh A., Conesa A., Santoyo-Lopez J., Medina I., Montaner D., Péterfia B., Solovei I., Cremer T., Dopazo J., Längst G. 2010. Initial genomics of the human nucleolus. PLoS Genet. 6:e1000889 10.1371/journal.pgen.1000889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neusser M., Schubel V., Koch A., Cremer T., Müller S. 2007. Evolutionarily conserved, cell type and species-specific higher order chromatin arrangements in interphase nuclei of primates. Chromosoma. 116:307–320 10.1007/s00412-007-0099-3 [DOI] [PubMed] [Google Scholar]
- Newman M., Barabasi A.L., Watts D.J., 2006. The Structure and Dynamics of Networks. Princeton University Press, Princeton, NJ: 582 pp [Google Scholar]
- O’Carroll D., Erhardt S., Pagani M., Barton S.C., Surani M.A., Jenuwein T. 2001. The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 21:4330–4336 10.1128/MCB.21.13.4330-4336.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan J.M., Sontam D.M., Grierson R., Jones B. 2009. Repeated elements coordinate the spatial organization of the yeast genome. Yeast. 26:125–138 10.1002/yea.1657 [DOI] [PubMed] [Google Scholar]
- Papantonis A., Larkin J.D., Wada Y., Ohta Y., Ihara S., Kodama T., Cook P.R. 2010. Active RNA polymerases: mobile or immobile molecular machines? PLoS Biol. 8:e1000419 10.1371/journal.pbio.1000419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada L.A., McQueen P.G., Misteli T. 2004. Tissue-specific spatial organization of genomes. Genome Biol. 5:R44 10.1186/gb-2004-5-7-r44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D., Bracken A.P., Hansen J.B., Capillo M., Helin K. 2007. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 27:3769–3779 10.1128/MCB.01432-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D., Meuleman W., Pagie L., Bruggeman S.W.M., Solovei I., Brugman W., Gräf S., Flicek P., Kerkhoven R.M., van Lohuizen M., et al. 2010. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell. 38:603–613 10.1016/j.molcel.2010.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickersgill H., Kalverda B., de Wit E., Talhout W., Fornerod M., van Steensel B. 2006. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat. Genet. 38:1005–1014 10.1038/ng1852 [DOI] [PubMed] [Google Scholar]
- Pombo A., Jackson D.A., Hollinshead M., Wang Z., Roeder R.G., Cook P.R. 1999. Regional specialization in human nuclei: visualization of discrete sites of transcription by RNA polymerase III. EMBO J. 18:2241–2253 10.1093/emboj/18.8.2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragoczy T., Telling A., Sawado T., Groudine M., Kosak S.T. 2003. A genetic analysis of chromosome territory looping: diverse roles for distal regulatory elements. Chromosome Res. 11:513–525 10.1023/A:1024939130361 [DOI] [PubMed] [Google Scholar]
- Ragoczy T., Bender M.A., Telling A., Byron R., Groudine M. 2006. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 20:1447–1457 10.1101/gad.1419506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapakse I., Perlman M.D., Scalzo D., Kooperberg C., Groudine M., Kosak S.T. 2009. The emergence of lineage-specific chromosomal topologies from coordinate gene regulation. Proc. Natl. Acad. Sci. USA. 106:6679–6684 10.1073/pnas.0900986106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapakse I., Scalzo D., Tapscott S.J., Kosak S.T., Groudine M. 2010. Networking the nucleus. Mol. Syst. Biol. 6:395 10.1038/msb.2010.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy K.L., Zullo J.M., Bertolino E., Singh H. 2008. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 452:243–247 10.1038/nature06727 [DOI] [PubMed] [Google Scholar]
- Roix J.J., McQueen P.G., Munson P.J., Parada L.A., Misteli T. 2003. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat. Genet. 34:287–291 10.1038/ng1177 [DOI] [PubMed] [Google Scholar]
- Rouquette J., Genoud C., Vazquez-Nin G.H., Kraus B., Cremer T., Fakan S. 2009. Revealing the high-resolution three-dimensional network of chromatin and interchromatin space: a novel electron-microscopic approach to reconstructing nuclear architecture. Chromosome Res. 17:801–810 10.1007/s10577-009-9070-x [DOI] [PubMed] [Google Scholar]
- Rouquette J., Cremer C., Cremer T., Fakan S. 2010. Functional nuclear architecture studied by microscopy: present and future. Int. Rev. Cell Mol. Biol. 282:1–90 10.1016/S1937-6448(10)82001-5 [DOI] [PubMed] [Google Scholar]
- Ryba T., Hiratani I., Lu J., Itoh M., Kulik M., Zhang J., Schulz T.C., Robins A.J., Dalton S., Gilbert D.M. 2010. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 20:761–770 10.1101/gr.099655.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin A.J., Shiels C., Williamson J., Satijn D.P.E., Otte A.P., Sheer D., Freemont P.S. 1998. The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J. Cell Biol. 142:887–898 10.1083/jcb.142.4.887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Arib G., Laemmli C., Nishikawa J., Durussel T., Laemmli U.K. 2006. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol. Cell. 21:379–391 10.1016/j.molcel.2005.12.012 [DOI] [PubMed] [Google Scholar]
- Schoenfelder S., Sexton T., Chakalova L., Cope N.F., Horton A., Andrews S., Kurukuti S., Mitchell J.A., Umlauf D., Dimitrova D.S., et al. 2010. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat. Genet. 42:53–61 10.1038/ng.496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder-Reiter E., Pérez-Willard F., Zeile U., Wanner G. 2009. Focused ion beam (FIB) combined with high resolution scanning electron microscopy: a promising tool for 3D analysis of chromosome architecture. J. Struct. Biol. 165:97–106 10.1016/j.jsb.2008.10.002 [DOI] [PubMed] [Google Scholar]
- Sexton T., Umlauf D., Kurukuti S., Fraser P. 2007. The role of transcription factories in large-scale structure and dynamics of interphase chromatin. Semin. Cell Dev. Biol. 18:691–697 10.1016/j.semcdb.2007.08.008 [DOI] [PubMed] [Google Scholar]
- Shapiro J.A., von Sternberg R. 2005. Why repetitive DNA is essential to genome function. Biol. Rev. Camb. Philos. Soc. 80:227–250 10.1017/S1464793104006657 [DOI] [PubMed] [Google Scholar]
- Simonis M., Klous P., Splinter E., Moshkin Y., Willemsen R., de Wit E., van Steensel B., de Laat W. 2006. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet. 38:1348–1354 10.1038/ng1896 [DOI] [PubMed] [Google Scholar]
- Simonis M., Kooren J., de Laat W. 2007. An evaluation of 3C-based methods to capture DNA interactions. Nat. Methods. 4:895–901 10.1038/nmeth1114 [DOI] [PubMed] [Google Scholar]
- Skok J.A., Brown K.E., Azuara V., Caparros M.-L., Baxter J., Takacs K., Dillon N., Gray D., Perry R.P., Merkenschlager M., Fisher A.G. 2001. Nonequivalent nuclear location of immunoglobulin alleles in B lymphocytes. Nat. Immunol. 2:848–854 10.1038/ni0901-848 [DOI] [PubMed] [Google Scholar]
- Snow K.J., Wright S.M., Woo Y., Titus L.C., Mills K.D., Shopland L.S. 2011. Nuclear positioning, higher-order folding, and gene expression of Mmu15 sequences are refractory to chromosomal translocation. Chromosoma. 120:61–71 10.1007/s00412-010-0290-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei I., Kreysing M., Lanctôt C., Kösem S., Peichl L., Cremer T., Guck J., Joffe B. 2009. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 137:356–368 10.1016/j.cell.2009.01.052 [DOI] [PubMed] [Google Scholar]
- Somech R., Shaklai S., Geller O., Amariglio N., Simon A.J., Rechavi G., Gal-Yam E.N. 2005. The nuclear-envelope protein and transcriptional repressor LAP2beta interacts with HDAC3 at the nuclear periphery, and induces histone H4 deacetylation. J. Cell Sci. 118:4017–4025 10.1242/jcs.02521 [DOI] [PubMed] [Google Scholar]
- Soutoglou E., Misteli T. 2008. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 320:1507–1510 10.1126/science.1159051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack S.M., Brown D.B., Dewey W.C. 1977. Visualization of interphase chromosomes. J. Cell Sci. 26:281–299 [DOI] [PubMed] [Google Scholar]
- Strambio-De-Castillia C., Niepel M., Rout M.P. 2010. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat. Rev. Mol. Cell Biol. 11:490–501 10.1038/nrm2928 [DOI] [PubMed] [Google Scholar]
- Strickfaden H., Zunhammer A., van Koningsbruggen S., Köhler D., Cremer T. 2010. 4D chromatin dynamics in cycling cells: Theodor Boveri’s hypotheses revisited. Nucleus. 1:284–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strogatz S.H. 1994. Nonlinear Dynamics and Chaos: With Applications to Physics, Biology, Chemistry, and Engineering. Addison-Wesley Publishing Co., Inc., Reading, MA: 498 pp [Google Scholar]
- Strogatz S.H. 2001. Exploring complex networks. Nature. 410:268–276 10.1038/35065725 [DOI] [PubMed] [Google Scholar]
- Strogatz S.H. 2003. Sync: The Emerging Science of Spontaneous Order. Hyperion, New York: 338 pp [Google Scholar]
- Taddei A., Van Houwe G., Hediger F., Kalck V., Cubizolles F., Schober H., Gasser S.M. 2006. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 441:774–778 10.1038/nature04845 [DOI] [PubMed] [Google Scholar]
- Takizawa T., Gudla P.R., Guo L., Lockett S., Misteli T. 2008. Allele-specific nuclear positioning of the monoallelically expressed astrocyte marker GFAP. Genes Dev. 22:489–498 10.1101/gad.1634608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taslerová R., Kozubek S., Lukásová E., Jirsová P., Bártová E., Kozubek M. 2003. Arrangement of chromosome 11 and 22 territories, EWSR1 and FLI1 genes, and other genetic elements of these chromosomes in human lymphocytes and Ewing sarcoma cells. Hum. Genet. 112:143–155 [DOI] [PubMed] [Google Scholar]
- Tero A., Takagi S., Saigusa T., Ito K., Bebber D.P., Fricker M.D., Yumiki K., Kobayashi R., Nakagaki T. 2010. Rules for biologically inspired adaptive network design. Science. 327:439–442 10.1126/science.1177894 [DOI] [PubMed] [Google Scholar]
- Thompson M., Haeusler R.A., Good P.D., Engelke D.R. 2003. Nucleolar clustering of dispersed tRNA genes. Science. 302:1399–1401 10.1126/science.1089814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berkum N.L., Lieberman-Aiden E., Williams L., Imakaev M., Gnirke A., Mirny L.A., Dekker J., Lander E.S. 2010. Hi-C: a method to study the three-dimensional architecture of genomes. J. Vis. Exp. 39:1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B., Dekker J. 2010. Genomics tools for unraveling chromosome architecture. Nat. Biotechnol. 28:1089–1095 10.1038/nbt.1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez J., Müller M., Pirrotta V., Sedat J.W. 2006. The Mcp element mediates stable long-range chromosome-chromosome interactions in Drosophila. Mol. Biol. Cell. 17:2158–2165 10.1091/mbc.E06-01-0049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser A.E., Jaunin F., Fakan S., Aten J.A. 2000. High resolution analysis of interphase chromosome domains. J. Cell Sci. 113:2585–2593 [DOI] [PubMed] [Google Scholar]
- Volpi E.V., Chevret E., Jones T., Vatcheva R., Williamson J., Beck S., Campbell R.D., Goldsworthy M., Powis S.H., Ragoussis J., et al. 2000. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 113:1565–1576 [DOI] [PubMed] [Google Scholar]
- Wansink D.G., Schul W., van der Kraan I., van Steensel B., van Driel R., de Jong L. 1993. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J. Cell Biol. 122:283–293 10.1083/jcb.122.2.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R.R.E., Azuara V., Perry P., Sauer S., Dvorkina M., Jørgensen H., Roix J., McQueen P., Misteli T., Merkenschlager M., Fisher A.G. 2006. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J. Cell Sci. 119:132–140 10.1242/jcs.02727 [DOI] [PubMed] [Google Scholar]
- Zhao Z., Tavoosidana G., Sjölinder M., Göndör A., Mariano P., Wang S., Kanduri C., Lezcano M., Sandhu K.S., Singh U., et al. 2006. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 38:1341–1347 10.1038/ng1891 [DOI] [PubMed] [Google Scholar]