Abstract
The spatial organization of genes and chromosomes plays an important role in the regulation of several DNA processes. However, the principles and forces underlying this nonrandom organization are mostly unknown. Despite its small dimension, and thanks to new imaging and biochemical techniques, studies of the budding yeast nucleus have led to significant insights into chromosome arrangement and dynamics. The dynamic organization of the yeast genome during interphase argues for both the physical properties of the chromatin fiber and specific molecular interactions as drivers of nuclear order.
Introduction
Over the past decades, understanding how genomes are organized in the limited space of nuclei has become a major goal in cell biology, as it is becoming apparent that this organization affects genome expression, stability, and replication. It is striking that in all eukaryotic species analyzed so far, spatial genome arrangements are nonrandom: chromosomes or genomic loci occupy preferential positions with respect to each other and/or to nuclear landmarks (Parada and Misteli, 2002).
On a much smaller scale, the chromatin fiber, which results from the wrapping of the DNA double helix around nucleosomes, is usually described as a compact structure 30 nm in diameter. The 30-nm fiber is based on in vitro experiments, but its relevance in vivo is currently being challenged (Maeshima et al., 2010). How the chromatin fiber is organized at larger scales in order to fit into the limited volume of the nucleus is even less well known.
A small unicellular eukaryote, the budding yeast Saccharomyces cerevisiae, has played a major role in understanding chromosome organization in interphase. S. cerevisiae was the first eukaryote to have its entire genome sequenced (Goffeau et al., 1996). Each nucleus contains 16 relatively small chromosomes, comprising between 230 and 1,500 kb of DNA, plus ∼100–200 copies of ribosomal genes (rDNA) encompassing 1–2 Mb. Unlike most eukaryotes, budding yeast has several unique nuclear features: its nuclear envelope does not break down during mitosis, the nucleolus has a crescent shape abutting the nuclear envelope, the spindle pole body (SPB; the microtubule organizing center) is located opposite to the nucleolus and remains embedded in the nuclear envelope throughout the cell cycle, and the centromere has a fixed genomic location and is wrapped around a single nucleosome containing a specific histone variant not found in other nucleosomes.
An apparent limitation of S. cerevisiae for studies of chromatin organization is the small size of its nucleus, ∼1 µm radius in haploids, which is only a few times larger than the diffraction-limited resolution of conventional light microscopy. However, the recent progress in imaging techniques, including the development of novel computational analysis methods, have enabled mapping of yeast nuclear organization with subdiffraction resolution. Combined with new genome-wide biochemical techniques, S. cerevisiae is now providing important clues to fundamental questions regarding the structural and functional states of chromosomes.
Topology of 3D chromosomal architecture
Centromere clustering, telomere positioning, and the Rabl-like configuration.
Centromeres were initially shown to cluster in a rosette-like structure around the SPB by FISH (Guacci et al., 1997; Jin et al., 1998; Jin et al., 2000; Bystricky et al., 2004). Studies using the Cre/lox system (in which expression of bacterially derived Cre recombinase mediates the recombination between exogenously inserted loxP sites, thereby allowing the measurement of recombination frequency; Hoess and Abremski, 1984) indicated increased rates of recombination for loci close to the centromeres (Burgess and Kleckner, 1999). Centromere clustering was recently confirmed using chromosome conformation capture (3C) combined with massive sequencing performed on the yeast genome (Duan et al., 2010). The 3C technique relies on the capture by mild cross-linking of interacting chromatin segments in large populations of cells, followed by intramolecular ligation in diluted conditions. PCR or sequencing then identifies chimeric sequences, the abundance of which allows determination of the probabilities of interactions between pairs of chromatin loci. Initially restricted to the analysis of cis interactions on selected loci (Dekker et al., 2002), the technique has been extended to genome-wide interaction maps (Simonis et al., 2006; Lieberman-Aiden et al., 2009). This technique confirmed centromere clustering in a rosette-like structure, with the majority of interchromosomal contacts being concentrated in a 20-kb window around centromeres (Fig. 1; Rodley et al., 2009; Duan et al., 2010). Whether there is a preferential ordering of centromeres within the rosette remains to be determined.
Figure 1.
General configuration of yeast chromosomes in interphase yeast. (top) Subnuclear territories occupied by three loci and the SPB, obtained using the methodology and data described previously (Berger et al., 2008; Therizols et al., 2010). Thousands of nuclei were detected in 3D microscopy images and computationally oriented along a central axis (broken line in the bottom panel) defined by the nuclear center and the nucleolus center of mass (X, bottom), allowing the determination of both the radial distance of loci relative to the nuclear center and an elevation angle above the central axis. Each color represents a locus or the SPB; dark and light shades indicate high and low probabilities, respectively. Green, rDNA; blue, the SPB; red, subtelomere of the short (85 kb) chromosome arm 9R; cyan, subtelomere of the long (440 kb) chromosome arm 11L. Note how the rDNA, the site of nucleolar protein assembly, occupies a pole of the nucleus opposite the microtubule organization center (SPB). Bar, 1 µm. (bottom) Sketch of a Rabl-like chromosome configuration hypothesized on the basis of observed subnuclear positions (Therizols et al., 2010). Enlargements depict possible chromosome arrangements at smaller scales. The rightmost enlargement shows an array of nucleosomes in a loose chromatin fiber (Dekker, 2008), with a segment of nucleosome-free DNA looping out. The bottom panel is adapted from Therizols et al. (2010).
The mechanisms for centromere clustering have been well elucidated. Each centromere directs the assembly of the kinetochore, a protein complex of ∼70 subunits that binds the plus end of a single microtubule (Joglekar et al., 2009). During interphase, the 16 microtubules maintain the attachment between the centromeres and the SPB (Furuyama and Biggins, 2007). By fluorescent tagging of an SPB component and insertion of a specific bacterial operator sequence (recognized by a fluorescently tagged repressor) near the centromere, the distance between the SPB and a centromere has been measured with high resolution in living cells (Dorn et al., 2005). Although microtubules grow and shrink continuously, the SPB and centromere were never closer than ∼200 nm in G1 phase (Dorn et al., 2005). Importantly, disruption of the kinetochore–microtubule link leads to a perturbation of centromere clustering (Jin et al., 2000). One major driving force for chromosome organization in yeast thus appears to be the microtubule-dependent anchorage of centromeres to the SPB, and by extension the nuclear envelope, throughout the cell cycle.
An additional important feature of yeast nuclear organization is the positioning of chromosome extremities, i.e., telomeres and subtelomeres. In most eukaryotes, telomeres are stretches of repetitive DNA motifs. In S. cerevisiae, these repeats are roughly 250 bp long with repeating units of TG1–3. Subtelomeres are ∼30-kb regions of DNA upstream of telomeres, which include few nonessential genes. A highly conserved ∼500-bp core X sequence is shared by all subtelomeres, and 17 out of 32 subtelomeres contain Y′ elements, sequences with unknown function, which have variable lengths (4–8 kb) and numbers among chromosome arms and Saccharomyces species (Louis et al., 1994; Liti and Louis, 2005).
Telomeres and subtelomeres were initially observed by FISH to be located at the nuclear periphery. Furthermore, a relatively small number (3–8) of bright fluorescent spots was observed, which suggests that several chromosome ends are clustered in close proximity to each other (Gotta et al., 1996; Hediger et al., 2002). When subtelomeres were observed in a population of living cells, they were consistently found to be located preferentially near the nuclear periphery (Fig. 1; Hediger et al., 2002; Bystricky et al., 2005; Schober et al., 2008; Therizols et al., 2010). This holds true for 20 out of 32 yeast subtelomeres observed so far.
What keeps yeast chromosome ends in the vicinity of the nuclear edge? Several studies implicate proteins that interact directly or indirectly with telomeres and subtelomeres and are enriched at the nuclear periphery. For example, Sir4 (silent information regulator), which is part of the Sir2–Sir4 complex, is recruited to subtelomeres by the telomeric binding protein Rap1 (Hecht et al., 1995); Sir4 also interacts with Esc1, a protein located at the nuclear envelope (Andrulis et al., 2002; Taddei et al., 2004), and with Mps3, an integral SUN domain–containing protein of the nuclear membrane that is enriched at the SPB (Antoniacci et al., 2007; Bupp et al., 2007). Likewise, the protein Ku70, which together with Ku80 binds double-stranded telomeric DNA, interacts directly or indirectly with members of the nuclear envelope and/or the nuclear pore complex (NPC; Galy et al., 2000; Therizols et al., 2006). Ku80 can also bind telomerase subunits, which in turn require Mps3 to target a telomere at the envelope (Schober et al., 2009). These interactions, mediated by Sir4/Mps3 for subtelomeres and Ku70/80 for telomeres, have been proposed to be two partially redundant pathways that target chromosomal ends to the nuclear envelope, despite some variations depending on the chromosome arm and the phase of the cell cycle (Hediger et al., 2002; Taddei et al., 2004).
Centromere clustering near the SPB and telomere positioning close to the nuclear periphery imply a nonrandom and polarized arrangement of chromosomes in interphase yeast nuclei. Such a configuration was observed initially in epithelial salamander larvae cells by Carl Rabl and subsequently in rapidly dividing nuclei like in Drosophila melanogaster embryos and in many cereal species (Rabl, 1885; Cowan et al., 2001). The Rabl configuration observed in interphase is thought to result from the chromosome arrangement in anaphase, established in the preceding mitosis. In S. cerevisiae, centromere clustering does not necessarily require passage through anaphase, and telomeres are not strictly located at the opposite pole of the SPB; because of these differences, the chromosome configuration in interphase yeast has been named Rabl-like (Jin et al., 2000). Consistent with a Rabl configuration, subtelomeres of small arms (<∼300 kb) from distinct chromosomes are found in closer proximity than subtelomeres on arms of different sizes (Jin et al., 2000; Bystricky et al., 2005; Schober et al., 2008; Therizols et al., 2010). In addition, the observation that opposite ends of chromosomes are closer in space than ends of distinct chromosomes (at least for arms up to 435 kb) is consistent with a rosette configuration in which centromeres are at a finite distance from each other (Fig. 1; Therizols et al., 2010).
It is worth noting that although the Rabl configuration is not commonly found in human cells, other nuclear anchoring points exist. For example, the nucleolus interacts with many specific chromatin domains of human chromosomes (Németh et al., 2010; van Koningsbruggen et al., 2010). It has been hypothesized that these anchoring regions might play a similar role in establishing polarized chromosome configurations (van Koningsbruggen et al., 2010).
Chromosome configuration: between centromeres and telomeres.
In eukaryotes, DNA is wrapped around nucleosomes ∼10 nm in size. How and whether further compaction into the 30-nm chromatin fiber occurs in vivo is not well known. In yeast, some information on chromatin compaction has been derived from FISH experiments, in which spatial distances were measured between loci located at various genomic intervals along chromosomes (Guacci et al., 1994; Bystricky et al., 2004). By fitting a semiflexible polymer model to these distances, compaction and persistence lengths of 110–150 bp/nm (7–10 nucleosomes per 11 nm) and 170–220 nm have been determined, respectively (Bystricky et al., 2004). Although these compaction values are consistent with the canonical 30-nm fiber, a recent re-analysis of in vivo measurements in combination with 3C data for chromosome 3 supports a much looser structure, with ∼1.2–3.6 nucleosomes per 11 nm (Dekker, 2008). This variability might be explained by histone modifications and variations in the length of the linker DNA between consecutive nucleosomes, which are expected to modulate nucleosome occupancy and chromatin compaction (Routh et al., 2008). For instance, the number of molecules of Hho1, the histone H1 in yeast, is low and variable, and estimated as ∼1/37 nucleosomes to 1/4 nucleosomes (Freidkin and Katcoff, 2001; Downs et al., 2003). New insights on chromatin structure are likely to come through the combination of higher resolution microscopy methods and 3C approaches.
Beyond the fine scale arrangement of chromatin, what is the higher order structure of chromosomes? Chromosome painting studies in many metazoans systems showed that individual chromosomes occupy distinct, nonoverlapping subnuclear regions named chromosome territories (Cremer and Cremer, 2001; Branco and Pombo, 2006), but in yeast, the relative disposition and internal organization of potential chromosome territories is still largely unknown. In hybrids between two species of Saccharomyces—S. cerevisiae and S. paradoxus, whose genomes diverge by 8–20%—in situ hybridization with probes discriminating between chromosomes from either species revealed two nonoverlapping sets of chromosomes, which supports the existence of chromosome territories in yeast nuclei (Lorenz et al., 2002; Liti et al., 2009). On the contrary, experiments based on recombination after induction of a double-strand break (DSB) between sequences inserted at different positions in the genome showed a lack of territoriality (Haber and Leung, 1996). In this study, similar rates of recombination were obtained irrespective of the position of the break. In contrast, genome-wide 3C provided support for the existence of chromosome domains and chromosome territories in budding yeast (Duan et al., 2010). Imaging studies in which the positions of several loci were mapped with high resolution in a 2D coordinate system can be used to make additional predictions about chromosome organization (Berger et al., 2008). These maps showed a strong statistical confinement of most loci into “gene territories,” and revealed that loci on internal positions along different chromosomes were close to the SPB at small genomic distances from the centromere, and closer to the nucleolus at larger genomic distances (Berger et al., 2008). Further support for a close link between spatial positioning and genomic location came from the observation, with this method, of the angular position at the nuclear periphery of 12 subtelomeres. This angle, defined between the subtelomere and the axis joining the nuclear and nucleolar centers, increased as a function of chromosome arm length (Fig. 1; Therizols et al., 2010). Thus, the ends of short chromosome arms cannot explore the entire nuclear periphery, but are limited to a small region opposite the nucleolus, an observation consistent with the high frequency of interactions found between short chromosome arms (Duan et al., 2010; Therizols et al., 2010). Similarly, the observation that the end of long arms extends away from the SPB is compatible with the less frequent interactions observed between long chromosomes and any other chromosome (except the long chromosome arms 12R and 4R; Duan et al., 2010; Therizols et al., 2010). These data support a model in which the large-scale configuration of chromosomes is dictated by the length of the sequence, an arrangement that seems consistent with the generic behavior expected from a semiflexible polymer (Rosa and Everaers, 2008).
It may seem difficult to reconcile this apparent separation between short and long chromosome arms with the finding that five transfer RNA (tRNA) gene families (each containing 9–14 different members), scattered throughout the genome, were found clustered close to the nucleolus (Thompson et al., 2003). Such clustering would certainly imply significant constraints to the 3D organization of the chromosomes. However, hierarchical clustering analyses on interchromosomal contacts determined by 3C identified two clusters of colocalized tRNA genes: one close to the rDNA and one clustered with centromeres (Duan et al., 2010). It would be interesting to determine whether tRNAs identified at the nucleolus are preferentially close to the end of long chromosome arms, which would be consistent with the expectation that spatial proximity to the nucleolus requires large genomic distances from the centromere (Therizols et al., 2010). The fact that tRNA clustering in the nucleolus persists in the absence of microtubules further suggests that it is independent from centromeric clustering, which is in agreement with the observation that the relative positions of long arm subtelomeres are not affected by a defect in microtubule attachment (Haeusler et al., 2008; Therizols et al., 2010). Similarly, the recent identification in the promoters of many genes of “gene recruitment sequences” that are sufficient to confer physical interactions with the NPCs may impose other important constrains on chromosome configuration (Ahmed et al., 2010). It will be particularly interesting to precisely define gene recruitment sequence positions in nuclear space to understand how they may participate in certain chromosome configurations.
The special rDNA array on chromosome 12 and the nucleolus.
In S. cerevisiae, unlike in other yeasts and most other species, the tandem array of genes encoding ribosomal subunits (rDNA) is confined to a single genomic locus, on the right arm of chromosome 12 (12R). Under normal conditions, the S. cerevisiae nucleolus occupies roughly one third of the nuclear volume at one pole of the cell, opposite the SPB (Yang et al., 1989; Léger-Silvestre et al., 1999). Studies in many organisms, including yeast, have established that the nucleolus is assembled at the site of ribosomal RNA (rRNA) synthesis, which suggests that the nucleolus originates from rDNA by self-organization (for reviews see Misteli, 2001; Hernandez-Verdun, 2006). Chromatin immunoprecipitation assays with the inner nuclear membrane protein Heh1 (or Src1) have revealed an enrichment with rDNA repeated sequences and subtelomeres (Grund et al., 2008; Mekhail et al., 2008). The inferred association of the rDNA with the nuclear envelope has been proposed as a means to limit recombination between arrays by sequestering the rDNA repeats away from the recombination machinery (Mekhail et al., 2008). In fact, rDNA is transiently delocalized outside the nucleolus for repair when a DSB is induced in the arrays (Torres-Rosell et al., 2007). Imaging studies of ∼20 non-rDNA loci showed that all of these were excluded from the nucleolar volume, and that a reduction of this volume allowed subtelomeres on long chromosome arms to occupy a larger nucleoplasmic space (Berger et al., 2008; Therizols et al., 2010). In the case of arm 12R, the distal part, from the telomere to the rDNA, has been found to emerge from the inward face of the nucleolus (Fuchs and Loidl, 2004). Recent 3C-derived data have also revealed a dramatic dearth of interactions between DNA sequences located at opposite sides of the rDNA (Duan et al., 2010). The nucleolus thus appears to play a central role in the spatial organization of the genome by sequestering repeated arrays away from the rest of the genome.
Chromosomes in motion.
3C techniques, FISH, or static live cell imaging data provide a snapshot view of chromosomes that ignores the dynamic nature of chromatin. The first evidence for chromatin movements in interphase yeast nuclei came from pioneering studies using GFP-tagged loci tracked in vivo over time periods of 150–600 s (Robinett et al., 1996; Marshall et al., 1997; Heun et al., 2001). These studies used mean square displacement (MSD) analyses, a standard tool to characterize the nature and quantitative properties of stochastic motions from particle trajectories. For normal diffusion, such as the random (Brownian) motion of small particles in a liquid, the MSD is proportional to the time interval, and the proportionality coefficient provides the diffusion coefficient (within a scaling factor). Assuming normal diffusion, these initial studies used the measured MSD at small time intervals to estimate diffusion coefficients ranging from 0.5 to ∼3 × 10−3 µm2/s, depending on the locus, with subtelomeres and centromeres exhibiting slower diffusion than more internal loci. Because chromatin is restricted to the nuclear volume, the MSD cannot grow indefinitely as function of time, and must plateau at a value slightly smaller than the squared nuclear radius. Measured MSD curves indeed exhibited a plateau, though at smaller values, which suggests that chromatin loci are confined to regions smaller than the nucleus itself. Confinement radii estimated from these plateaus ranged from 0.3 to ∼0.7 µm, with subtelomeres and centromeres apparently more confined than internal loci (Marshall et al., 1997; Heun et al., 2001; Bystricky et al., 2005). Not surprisingly, centromere confinement can in part be explained by microtubule attachment because a centromere was less confined if microtubules were depolymerized (Marshall et al., 1997; Heun et al., 2001; Bystricky et al., 2005). In a more recent analysis, the dynamics of the GAL1 locus at smaller time scales (<60s) was characterized by an MSD curve proportional to the time interval at the power ∼0.4 (instead of 1 for Brownian motions), which instead suggests subdiffusive motion. This behavior was observed independently of the gene’s transcriptional status (Cabal et al., 2006). Interestingly, subdiffusion with a similar power law has been recently reported in bacterial loci (Weber et al., 2010), which suggests that the underlying mechanism is general. It seems likely that movements previously described as confined diffusion might in fact also obey subdiffusive behavior at similar time scales. Subdiffusion can arise as a result of several physical effects, including nuclear crowding, caging, viscoelasticity of the nucleoplasm, or polymer effects caused by the dynamic properties of the chromatin fiber. Recent theoretical studies have proposed that the observed subdiffusion arises from polymer physics rather than crowding or caging (Rosa and Everaers, 2008; Weber et al., 2010). Despite these average subdiffusive motions, occasional abrupt (≥0.5 µm) and rapid (≤10 s) jumps have also been observed (Heun et al., 2001). Because they were ATP dependent, these sudden motions appear to be powered by active mechanisms rather than by passive subdiffusion (Heun et al., 2001). Mechanisms for potential active motions are unknown in yeast, but in mammalian cells, nuclear actin, and myosin motors have been shown to play a role in movements that accompany transcriptional activation (Chuang et al., 2006; Dundr et al., 2007). In addition, it is possible that the mechanical coupling between the cytoskeleton and chromosomes recently evidenced in meiosis may also apply to the mitotic cycle and thus provide a force that originates outside the nucleus to move interphase chromosomes (King et al., 2008; Koszul et al., 2008). Further insights into the nature and mechanisms of chromatin dynamics are likely to result from the application of recent advances in high-speed and high-resolution microscopy combined with theoretical modeling (Manley et al., 2008; Hajjoul et al., 2009; Wombacher et al., 2010).
Functional relevance of chromosome organization
Chromosome organization and DNA metabolism are linked, as alterations of this organization are often associated with perturbations of replication, transcription, or DNA repair. Here, we will concentrate on the latter two functions. For a recent review on replication see Raghuraman and Brewer (2010).
Chromosome configuration and transcription.
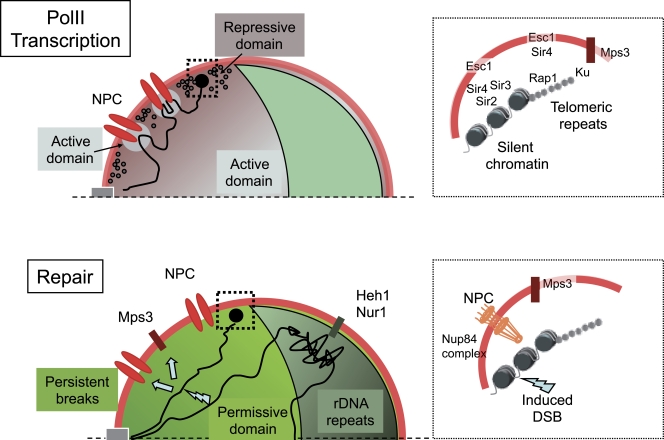
Repression of polII transcription often occurs at genes near the ends of chromosomes, which, as discussed in the section on telomere positioning, are preferentially found at the nuclear periphery. For example, the HML and HMR cassettes, which are required for S. cerevisiae mating type switch, are both subtelomeric and must be silent for cell-type identity to be maintained; polII-driven reporters close to telomeres, with or without some subtelomeric elements, generally display variegated expression (Gottschling et al., 1990; Renauld et al., 1993; Pryde and Louis, 1999; Bi, 2002; Halme et al., 2004). The view that the region close to the nuclear envelope is a key player in the repression of polII transcription is further based on the observation that artificial retention at the nuclear periphery of a locus deficient for silencing facilitates its transcriptional repression. This effect is related to the enrichment at the nuclear periphery of silencing Sir proteins, which are otherwise present in limiting amounts in the nucleus (Fig. 2; Maillet et al., 1996; Andrulis et al., 1998).
Figure 2.
Functional compartmentalization of the yeast nucleus. (top) PolII transcription domains. The nucleolus, where polI and polIII transcription occur, is shown in green. The nucleoplasm is partitioned between polII transcriptionally repressive domains (dark gray), where silencing proteins (black circles) are found, and domains permissive for transcription (light gray), in the nuclear interior and in the proximity of NPCs (red). Chromatin loops (black line) accompany the activation of inducible genes. Large black dot, chromosome end. The inset on the right shows molecular determinants of transcriptional silencing acting at telomeric repeats and the upstream subtelomere. (bottom) DNA repair domains. Permanent breaks induced either by HO or I-SceI endonucleases (lightning bolt symbol) are displaced (arrows) to the nuclear periphery through the action of the Nup84 subcomplex or the Mps3 protein (Therizols et al., 2006; Nagai et al., 2008; Oza et al., 2009). This displacement requires Mec1 and modification of the histone H2AZ. At the NPC, modification of repair proteins by SUMO ubiquitin ligases modifying enzymes Slx5 and Slx8 or the desumoylase Ulp1 might be important to ensure efficient repair (not depicted; Palancade et al., 2007; Nagai et al., 2008). In the nucleolus, recombination between rDNA repeats is prevented by their association with Heh1 and Nur1, proteins of the inner nuclear membrane (Mekhail et al., 2008). The inset on the right shows a permanent break induced at subtelomeres displaced toward the NPC or Mps3.
To understand whether silencing is restricted to the nuclear envelope, situations were investigated where peripheral targeting and expression can be uncoupled. When a DNA ring containing the HMR silent mating type cassette is excised from the chromosome and its position is tracked in the nuclei of ku70 or esc1 mutants, in which telomeric anchoring is defective, transcriptional repression of HMR can still occur in the nucleoplasm, away from the periphery, as long as silencing proteins are present (Gartenberg et al., 2004). Furthermore, some truncated subtelomeres can be found silenced in the nuclear interior, and some native subtelomeres, despite their peripheral location, can be transcriptionally active (Enomoto and Berman, 1998; Pryde and Louis, 1999; Tham et al., 2001; Mondoux et al., 2007). Thus, although constituting a generally repressive environment for polII transcription, the nuclear periphery, in some cases, appears to be permissive for expression. Likewise, in mammals, a position close to the nuclear edge can influence the expression of some but not all genes (Finlan et al., 2008; Guelen et al., 2008; Kumaran and Spector, 2008).
In fact, an environment favoring transcriptional activation is provided by a key constituent of the nuclear envelope, namely the NPC. Several inducible genes, including GAL1-10, GAL2, INO1, HSP104, and HKX1, are relocated from the nucleoplasm to the NPC when activated (Brickner and Walter, 2004; Cabal et al., 2006; Dieppois et al., 2006; Schmid et al., 2006; Taddei et al., 2006). In agreement with these observations, chromatin immunoprecipitation analyses showed an interaction between components of the NPC and a subset of highly transcribed genes (Casolari et al., 2004; Luthra et al., 2007). Again, transcription and positioning can be decoupled because mutations of the SAGA histone acetyltransferase complex abrogate perinuclear positioning without affecting transcription, and the rpb1-1 mutant of the largest RNA polII subunit still recruits GAL1 or INO1 genes to the NPC. These data further suggest that peripheral targeting is caused by the signal regulating transcriptional activation rather than transcription per se (Cabal et al., 2006; Dieppois et al., 2006; Schmid et al., 2006; Brickner et al., 2007).
The region close to the nuclear envelope thus emerges as a mosaic, with the vicinity of NPCs representing zones favorable to transcription, whereas the zones between NPCs are more repressive. Alternatively, one can speculate that in yeast, as recently reported for Drosophila cells, nucleoporins present in the nuclear interior could bind chromatin and regulate gene expression without requiring chromatin positioning at the periphery, or physical interactions with the NPCs (Capelson et al., 2010).
The observed relocation of inducible genes upon activation raises several questions. What are the mechanisms causing this repositioning? Do they involve molecular motors or do they result from a decondensation of the chromatin fiber? How are neighboring genes affected? Although the mechanism initiating these chromatin movements is still unknown, it is likely that changes in chromosomal conformation are stabilized and maintained through interaction with nuclear envelope proteins, such as NPC components. In fact, it has been shown for INO1 and GAL1 that the interaction of these genes with the NPC is regulated during the cell cycle; NPC interaction is lost during S phase because of Cdk1-dependent phosphorylation of Nup1 (Brickner and Brickner, 2010). Furthermore a link between nuclear positioning and changes in chromatin conformation is provided by the study of inducible genes such as GAL1. Upon activation, the 3′ and 5′ ends of this gene interact, thus forming a chromatin loop. This loop is required for rapid re-expression of the gene after a period of repression, thereby conferring a transcriptional memory that lasts for several generations (Brickner et al., 2007; Lainé et al., 2009; Tan-Wong et al., 2009; Light et al., 2010). A plausible explanation for this observation is that these loops may create a structure that favors recruitment or recycling of the transcription machinery at the NPC, even though polII has until now not been detected at the NPCs.
Chromosome configuration and DNA repair.
How does chromosome configuration impact DNA repair? DNA is constantly being damaged because of light-induced/oxidative stresses or as a result of replication fork stalling. The most deleterious form of DNA damage is a DSB. In haploid yeast cells, DSBs can be repaired by homologous recombination either with the sister chromatid once the genome has been replicated or with ectopic nonallelic homologous, and often repeated, regions. Another mechanism to repair DSBs is nonhomologous end joining, in which the broken DNA ends are religated. The latter process is used preferentially during G1, but is more prone to errors (Krogh and Symington, 2004).
Because DNA repair requires a physical contact between broken ends or homologous regions, one can expect a nonrandom organization of chromosomes in interphase to be reflected by variable repair efficiencies. Indeed, a lower efficiency of repair between spatially distant regions was observed in mammalian nuclei (Parada et al., 2002). A similar result is expected in yeast, based on recent work on yeast chromosome organization (Duan et al., 2010; Therizols et al., 2010). A connection between chromosome positioning at the nuclear periphery and genome stability has recently been revealed (Fig. 2; Therizols et al., 2006; Nagai et al., 2008; Kalocsay et al., 2009; Oza et al., 2009; Schober et al., 2009). It was first observed that a DSB induced by the I-SceI endonuclease on subtelomere 11L required interaction with the nucleoporin Nup84 subcomplex to be efficiently repaired (Therizols et al., 2006). Furthermore, a persistent DSB induced on the small chromosome arm 3R by the HO endonuclease (in the absence of HM homologous cassettes) migrates to and remains at the nuclear periphery by a process that requires the checkpoint protein Mec1 (ATR in humans) and the nuclear envelope protein Mps3 (Nagai et al., 2008; Kalocsay et al., 2009; Oza et al., 2009; Schober et al., 2009). Specific chromatin marks additionally come into play because the sumoylated histone variant H2AZ is required for persistent break relocation (Kalocsay et al., 2009).
The ends of chromosomes also exemplify the role played by chromosome architecture in recombination. Recombination can occur at subtelomeres but not at telomeres (Louis et al., 1994). Repression of homologous recombination at telomeres involves yKu and the core X sequence by a mechanism that does not require tethering at the nuclear periphery or silencing (Stavenhagen and Zakian, 1998; Marvin et al., 2009). It has been proposed that telomeres fold back on themselves, creating a loop structure that prevents recombination (Stavenhagen and Zakian, 1998; Marvin et al., 2009). This is in agreement with the observation that chromosome ends rarely associate with each other and that interactions between telomeres are, at most, transient (Therizols et al., 2010).
The repair of broken ends raises several questions. What forces underlie the observed peripheral repositioning? Can chromatin modifications such as H2AZ sumoylation or H2A phosphorylation initiate chromatin movements ending with a capture by nucleoporins or Mps3? Do broken ends remain in close proximity, as in mammalian cells in which a tagged DSB remains immobile for at least 24 h (Soutoglou et al., 2007)? Or do these ends freely diffuse in the nucleoplasm to search for potential homologous regions, as also observed in mammalian cells (Aten et al., 2004)? In the latter case, damaged chromosome loci must travel through the nucleus toward the repair center.
Nuclear foci versus chromatin interactions.
An important aspect of nuclear organization is the possible existence of preferred interactions between specific regions of chromatin or between chromatin-bound proteins. These features are functionally attractive, as they may offer a means to increase the local concentration of functional proteins and to prevent undesirable action of these proteins elsewhere in the nucleus. Visualization of chromatin regions by light microscopy does not allow detection of direct interactions, but, given the limited resolution of microscopy, only of their spatial proximity. Labeled regions located within the resolution limit lead to the appearance of aggregates, often named “nuclear foci.” An example of a nuclear focus in yeast is provided by the discovery that two, otherwise spatially distinct loci on different chromosomes colocalize to form a focus with proteins of the repair machinery upon induction of DSB at these two sites (Lisby et al., 2003). These repair foci may facilitate the rejoining of broken extremities and reduce aberrant or illegitimate rejoining. Similarly, concentration of yeast silencing proteins at the nuclear periphery in nuclear foci may prevent unwanted repression at other parts of the genome, thereby acting as a genetic control mechanism (Gartenberg et al., 2004).
A well-studied case of nuclear foci in yeast originated from the observation that Y′ subtelomeric sequences cluster into 3–8 foci at the nuclear envelope (Gotta et al., 1996; Enomoto et al., 1997; Galy et al., 2000; Feuerbach et al., 2002). In many mutants of subtelomeric silencing, including sir3, sir4, or rlf2, the transcriptional state of reporter genes close to telomeres is affected, and staining by telomeric Rap1 is more diffuse than in wild-type cells (Palladino et al., 1993; Gotta et al., 1996; Enomoto et al., 1997; Mondoux et al., 2007). However, the number of subtelomeric foci detected by Y′ probes remains almost unchanged in these mutants (Gotta et al., 1996; Enomoto et al., 1997; Mondoux et al., 2007). These observations suggest that subtelomeric chromatin might be affected without perturbation of Y′-labeled subtelomeric foci, and imply that factors required for Y′ associations are distinct from the silencing factors of telomere-adjacent genes. What, then, causes the Y′ sequence clusters seen in FISH data? Do these clusters reflect actual physical interactions? If so, proteins involved in nucleosome assembly are potential candidates for mediating interactions between subtelomeres (Miele et al., 2009). Alternatively, we asked if the FISH foci could arise in the absence of any interactions between telomeric sequences. Because yeast nuclei have a radius of only ∼1 µm and the resolution of conventional light microscopes is typically limited to ∼200 nm laterally and ∼500 nm or more axially, the fluorescence signals from two distinct Y′ sequences may frequently overlap and appear as a single spot or cluster even in the absence of actual interactions. To test this, we simulated wide-field microscopy images of 32 fluorescent telomeres located randomly and independently of each other in the nucleus. The telomeres were positioned at the nuclear periphery and excluded from the nucleolus, as previously observed (Therizols et al., 2010). The simulated images display a relatively small number of bright fluorescent spots reminiscent of Y′ FISH foci, which suggests that interactions may not be required to account for these observations (Fig. 3, left, top). If the 32 simulated telomeres are randomly located within the nuclear interior rather than positioned only at the nuclear periphery, the number of foci increases significantly, implying a smaller mean number of telomeres per focus (Fig. 3, left, bottom). This may agree with the observation that Y′ foci are more dispersed and numerous in mutants such as Δku70, where telomere tethering to the nuclear envelope is perturbed (Laroche et al., 1998). An increase in the number of foci is also predicted for telomeres in a larger nucleus (not depicted). Hence, the observation of a smaller number of nuclear foci than individual loci may arise as a consequence of limited imaging resolution and confinement to a nuclear territory, in this case the nuclear periphery. Physical interactions between such loci may nevertheless exist but need not necessarily be invoked to explain visualization of nuclear foci with conventional microscopy. Long-lasting interactions may also be necessary to explain functions like repair or silencing, which could be initiated by transient interactions.
Figure 3.
Nuclear foci can occur in absence of interactions. Simulated microscopy images of fluorescently tagged telomeres (green) in a yeast nucleus (left) and quantification of the distribution of simulated foci number (right). (left, top) Telomeres are confined to a portion of the nuclear periphery. (left, bottom) telomeres are randomly distributed in the nucleoplasm. In each simulation, 32 telomeres were randomly positioned independently of each other within a subvolume of a 1-µm-radius spherical nucleus. In the top row, this subvolume represents the “nuclear periphery” and is a spherical shell of inner and outer radii of 0.8 µm and 1 µm, respectively. In the bottom row, this subvolume consists of the entire nuclear sphere with the exception of a region representing the nucleolus. The 3D orientation of the nucleus was chosen randomly for each panel. Red, nucleoplasm. To account for limited resolution, the point spread function was defined according to typical microscopy parameters, with full width at half maximum of ∼0.2 µm and 0.6 µm along the lateral and axial directions, respectively. The simulated images were corrupted by a mixture of Poisson and additive Gaussian noise. Each panel shows maximum intensity projections along the axial direction of a 3D image stack. Histograms of the number of foci were obtained from 2,000 independent simulations each (counts are on the y axis). Two telomeres located at a distance <0.3 µm were considered to be part of the same focus. Some foci contained only one telomere. The number of foci increases when telomeres are allowed to explore most of the nuclear volume. The histograms indicate substantially more distinct foci than are visible in the images, partly because the projection images merge spatially separated foci and because the signal of single isolated telomeres is difficult to detect visually.
Conclusions
Budding yeast has been useful as a model system to understand many aspects of 3D chromosomal architecture, chromosome dynamics, and functional compartmentalization. Yet, many open questions remain, especially regarding the links between spatial chromosome organization and DNA-related processes. Given the implications for fundamental genome functions, a major objective is obviously to identify or better characterize the principles that drive nonrandom chromosome organization.
We would like to distinguish two types of organizing principles. Specific processes depend on the chemical identity of molecules involved in biochemical or genetic interactions, whereas generic principles do not depend on the exact compound, but on more general physical laws. For example, a specific mechanism is the tethering of chromosome ends to the nuclear envelope through the Ku/Sir/Mps3 protein pathways (Gotta et al., 1996; Laroche et al., 1998; Bupp et al., 2007) or the nucleolar clustering of tRNA genes (Thompson et al., 2003). These examples indicate sequence-specific interactions of loci with each other and a nuclear landmark, with potentially dramatic consequences on nuclear organization. Identifying such specific processes requires nailing down their molecular determinants in wild-type and mutant contexts. Generic processes include self-organization by macromolecular crowding, an attractive scenario to explain the existence of protein-based nuclear compartments, and polymer effects (Munkel and Langowski, 1998; Misteli, 2001; Rosa and Everaers, 2008). Polymer physics makes simple and robust predictions about chromosome configuration: for a given chromatin compaction and persistence length, the mean distance between the extremities of chromosomes containing a longer DNA sequence is predicted to be larger than for chromosomes with less DNA (Gehlen et al., 2006). In contrast, if chromosome configuration is mainly constrained by sequence- or nuclear landmark–specific interactions such as reported for tRNA, then such a simple relationship is expected to break down (Thompson et al., 2003). Our high-throughput subtelomere position analysis fits qualitatively with the simple prediction from polymer physics, as the ends of short arm chromosomes are closer to the SPB than the ends of longer chromosome arms (Therizols et al., 2010). The Rabl-like configuration can thus be understood as a result of the mitotic spindle forces and the basic properties of polymers. These simple effects can account for the nonrandom organization of chromosomes in the nucleus, reflected by differences in subnuclear territories of chromosome ends, despite stochastic motions of the chromatin. Such exclusively generic principles may give rise to features that do not necessarily require specific interactions, such as the apparent clustering of simulated independent telomeres. These considerations do not rule out specific interactions because fundamental DNA processes may be initiated by transient contacts that are the result of random dynamics. Specific interactions are also clearly involved in previously identified alterations of chromosome organization, e.g., in the repositioning of inducible genes upon transcriptional activation (Cabal et al., 2006; Berger et al., 2008; Brickner and Brickner, 2010).
Despite impressive recent advances in microscopy, genetics, and genomic technologies, further improvements in the resolution of imaging techniques and 3C-based maps of genomic contacts will likely be required to dissect the processes driving chromosome configuration at all scales. Although 3C methods can provide genome-wide interaction frequencies, these are averaged over large populations of fixed cells. Distinguishing between interactions that occur at high probability in a small fraction of cells from those occurring at a low probability in a large majority of cells, and determining which interactions occur in the same cells, is difficult, if not impossible. Imaging, in contrast, allows tracking locus positions in individual live cells and can be scaled up to analyze position distributions in populations of thousands of cells, but is limited to a small number of loci per experiment. Thus, these very different techniques should be used in combination to determine chromosome configuration and dynamics, and their variability within cell populations. In addition, further development of physics-based models and analysis techniques will be important to make full use of these data and to better understand the mechanisms underlying nuclear organization.
Understanding nuclear organization will be essential to address how nuclear processes are controlled and, potentially, how they evolved. In budding yeast, chromosome ends are spatially confined in a region that appears as a mosaic of repressive and permissive compartments for transcriptional regulation. Evolution may have taken advantage of this 3D organization by moving the genes that need to be switched on or off near chromosome ends. Consistent with this hypothesis, genes present in subtelomeric regions are often involved in adaptation of yeast to environmental changes (Halme et al., 2004; Fabre et al., 2005). More generally, it would be interesting to determine whether families of homologous genes found at genomic positions predicted to share a common nuclear space also share common transcriptional regulation.
The nucleus is a crowded environment of macromolecules and chromatin, and yet order exists. This is probably dictated in part by simple physical constraints, allowing nuclear functions to be harmoniously performed and conserved during evolution.
Acknowledgments
We thank K. Bystricky, F. Feuerbach, O. Gadal, R. Koszul, and P. Therizols for their critical reading of the manuscript.
C. Zimmer and E. Fabre are supported by Centre National de la Recherche Scientifique, Institut Pasteur, and Agence Nationale de la Recherche Programme Interdisciplinaire de Recherches sur les Systèmes Moléculaires et Cellulaires, et d’innovation Biomédicale (ANR-PIRIBIO) grant No. ANR-09-PIRI-0024.
Footnotes
Abbreviations used in this paper:
- DSB
- double-strand break
- MSD
- mean square displacement
- NPC
- nuclear pore complex
- SPB
- spindle pole body
- tRNA
- transfer RNA
References
- Ahmed S., Brickner D.G., Light W.H., Cajigas I., McDonough M., Froyshteter A.B., Volpe T., Brickner J.H. 2010. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat. Cell Biol. 12:111–118 10.1038/ncb2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis E.D., Neiman A.M., Zappulla D.C., Sternglanz R. 1998. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 394:592–595 10.1038/29100 [DOI] [PubMed] [Google Scholar]
- Andrulis E.D., Zappulla D.C., Ansari A., Perrod S., Laiosa C.V., Gartenberg M.R., Sternglanz R. 2002. Esc1, a nuclear periphery protein required for Sir4-based plasmid anchoring and partitioning. Mol. Cell. Biol. 22:8292–8301 10.1128/MCB.22.23.8292-8301.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniacci L.M., Kenna M.A., Skibbens R.V. 2007. The nuclear envelope and spindle pole body-associated Mps3 protein bind telomere regulators and function in telomere clustering. Cell Cycle. 6:75–79 10.4161/cc.6.1.3647 [DOI] [PubMed] [Google Scholar]
- Aten J.A., Stap J., Krawczyk P.M., van Oven C.H., Hoebe R.A., Essers J., Kanaar R. 2004. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 303:92–95 10.1126/science.1088845 [DOI] [PubMed] [Google Scholar]
- Berger A.B., Cabal G.G., Fabre E., Duong T., Buc H., Nehrbass U., Olivo-Marin J.C., Gadal O., Zimmer C. 2008. High-resolution statistical mapping reveals gene territories in live yeast. Nat. Methods. 5:1031–1037 10.1038/nmeth.1266 [DOI] [PubMed] [Google Scholar]
- Bi X. 2002. Domains of gene silencing near the left end of chromosome III in Saccharomyces cerevisiae. Genetics. 160:1401–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco M.R., Pombo A. 2006. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 4:e138 10.1371/journal.pbio.0040138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner D.G., Brickner J.H. 2010. Cdk phosphorylation of a nucleoporin controls localization of active genes through the cell cycle. Mol. Biol. Cell. 21:3421–3432 10.1091/mbc.E10-01-0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner D.G., Cajigas I., Fondufe-Mittendorf Y., Ahmed S., Lee P.C., Widom J., Brickner J.H. 2007. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 5:e81 10.1371/journal.pbio.0050081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner J.H., Walter P. 2004. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2:e342 10.1371/journal.pbio.0020342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bupp J.M., Martin A.E., Stensrud E.S., Jaspersen S.L. 2007. Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J. Cell Biol. 179:845–854 10.1083/jcb.200706040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S.M., Kleckner N. 1999. Collisions between yeast chromosomal loci in vivo are governed by three layers of organization. Genes Dev. 13:1871–1883 10.1101/gad.13.14.1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystricky K., Heun P., Gehlen L., Langowski J., Gasser S.M. 2004. Long-range compaction and flexibility of interphase chromatin in budding yeast analyzed by high-resolution imaging techniques. Proc. Natl. Acad. Sci. USA. 101:16495–16500 10.1073/pnas.0402766101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystricky K., Laroche T., van Houwe G., Blaszczyk M., Gasser S.M. 2005. Chromosome looping in yeast: telomere pairing and coordinated movement reflect anchoring efficiency and territorial organization. J. Cell Biol. 168:375–387 10.1083/jcb.200409091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabal G.G., Genovesio A., Rodriguez-Navarro S., Zimmer C., Gadal O., Lesne A., Buc H., Feuerbach-Fournier F., Olivo-Marin J.C., Hurt E.C., Nehrbass U. 2006. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 441:770–773 10.1038/nature04752 [DOI] [PubMed] [Google Scholar]
- Capelson M., Liang Y., Schulte R., Mair W., Wagner U., Hetzer M.W. 2010. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 140:372–383 10.1016/j.cell.2009.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari J.M., Brown C.R., Komili S., West J., Hieronymus H., Silver P.A. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 117:427–439 10.1016/S0092-8674(04)00448-9 [DOI] [PubMed] [Google Scholar]
- Chuang C.H., Carpenter A.E., Fuchsova B., Johnson T., de Lanerolle P., Belmont A.S. 2006. Long-range directional movement of an interphase chromosome site. Curr. Biol. 16:825–831 10.1016/j.cub.2006.03.059 [DOI] [PubMed] [Google Scholar]
- Cowan C.R., Carlton P.M., Cande W.Z. 2001. The polar arrangement of telomeres in interphase and meiosis. Rabl organization and the bouquet. Plant Physiol. 125:532–538 10.1104/pp.125.2.532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T., Cremer C. 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2:292–301 10.1038/35066075 [DOI] [PubMed] [Google Scholar]
- Dekker J. 2008. Mapping in vivo chromatin interactions in yeast suggests an extended chromatin fiber with regional variation in compaction. J. Biol. Chem. 283:34532–34540 10.1074/jbc.M806479200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J., Rippe K., Dekker M., Kleckner N. 2002. Capturing chromosome conformation. Science. 295:1306–1311 10.1126/science.1067799 [DOI] [PubMed] [Google Scholar]
- Dieppois G., Iglesias N., Stutz F. 2006. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol. Cell. Biol. 26:7858–7870 10.1128/MCB.00870-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn J.F., Jaqaman K., Rines D.R., Jelson G.S., Sorger P.K., Danuser G. 2005. Yeast kinetochore microtubule dynamics analyzed by high-resolution three-dimensional microscopy. Biophys. J. 89:2835–2854 10.1529/biophysj.104.058461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs J.A., Kosmidou E., Morgan A., Jackson S.P. 2003. Suppression of homologous recombination by the Saccharomyces cerevisiae linker histone. Mol. Cell. 11:1685–1692 10.1016/S1097-2765(03)00197-7 [DOI] [PubMed] [Google Scholar]
- Duan Z., Andronescu M., Schutz K., McIlwain S., Kim Y.J., Lee C., Shendure J., Fields S., Blau C.A., Noble W.S. 2010. A three-dimensional model of the yeast genome. Nature. 465:363–367 10.1038/nature08973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M., Ospina J.K., Sung M.H., John S., Upender M., Ried T., Hager G.L., Matera A.G. 2007. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J. Cell Biol. 179:1095–1103 10.1083/jcb.200710058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto S., Berman J. 1998. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 12:219–232 10.1101/gad.12.2.219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto S., McCune-Zierath P.D., Gerami-Nejad M., Sanders M.A., Berman J. 1997. RLF2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev. 11:358–370 10.1101/gad.11.3.358 [DOI] [PubMed] [Google Scholar]
- Fabre E., Muller H., Therizols P., Lafontaine I., Dujon B., Fairhead C. 2005. Comparative genomics in hemiascomycete yeasts: evolution of sex, silencing, and subtelomeres. Mol. Biol. Evol. 22:856–873 10.1093/molbev/msi070 [DOI] [PubMed] [Google Scholar]
- Feuerbach F., Galy V., Trelles-Sticken E., Fromont-Racine M., Jacquier A., Gilson E., Olivo-Marin J.C., Scherthan H., Nehrbass U. 2002. Nuclear architecture and spatial positioning help establish transcriptional states of telomeres in yeast. Nat. Cell Biol. 4:214–221 10.1038/ncb756 [DOI] [PubMed] [Google Scholar]
- Finlan L.E., Sproul D., Thomson I., Boyle S., Kerr E., Perry P., Ylstra B., Chubb J.R., Bickmore W.A. 2008. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 4:e1000039 10.1371/journal.pgen.1000039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freidkin I., Katcoff D.J. 2001. Specific distribution of the Saccharomyces cerevisiae linker histone homolog HHO1p in the chromatin. Nucleic Acids Res. 29:4043–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs J., Loidl J. 2004. Behaviour of nucleolus organizing regions (NORs) and nucleoli during mitotic and meiotic divisions in budding yeast. Chromosome Res. 12:427–438 10.1023/B:CHRO.0000034726.05374.db [DOI] [PubMed] [Google Scholar]
- Furuyama S., Biggins S. 2007. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl. Acad. Sci. USA. 104:14706–14711 10.1073/pnas.0706985104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V., Olivo-Marin J.C., Scherthan H., Doye V., Rascalou N., Nehrbass U. 2000. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature. 403:108–112 10.1038/47528 [DOI] [PubMed] [Google Scholar]
- Gartenberg M.R., Neumann F.R., Laroche T., Blaszczyk M., Gasser S.M. 2004. Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell. 119:955–967 10.1016/j.cell.2004.11.008 [DOI] [PubMed] [Google Scholar]
- Gehlen L.R., Rosa A., Klenin K., Langowski J., Gasser S.M., Bystricky K. 2006. Spatially confined polymer chains: implications of chromatin fibre flexibility and peripheral anchoring on telomere–telomere interaction. J. Phys. Condens. Matter. 18:S245–S252 10.1088/0953-8984/18/14/S09 [DOI] [Google Scholar]
- Goffeau A., Barrell B.G., Bussey H., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., et al. 1996. Life with 6000 genes. Science. 274:546–567: 563–567 10.1126/science.274.5287.546 [DOI] [PubMed] [Google Scholar]
- Gotta M., Laroche T., Formenton A., Maillet L., Scherthan H., Gasser S.M. 1996. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 134:1349–1363 10.1083/jcb.134.6.1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling D.E., Aparicio O.M., Billington B.L., Zakian V.A. 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 63:751–762 10.1016/0092-8674(90)90141-Z [DOI] [PubMed] [Google Scholar]
- Grund S.E., Fischer T., Cabal G.G., Antúnez O., Pérez-Ortín J.E., Hurt E. 2008. The inner nuclear membrane protein Src1 associates with subtelomeric genes and alters their regulated gene expression. J. Cell Biol. 182:897–910 10.1083/jcb.200803098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V., Hogan E., Koshland D. 1994. Chromosome condensation and sister chromatid pairing in budding yeast. J. Cell Biol. 125:517–530 10.1083/jcb.125.3.517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V., Hogan E., Koshland D. 1997. Centromere position in budding yeast: evidence for anaphase A. Mol. Biol. Cell. 8:957–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L., Pagie L., Brasset E., Meuleman W., Faza M.B., Talhout W., Eussen B.H., de Klein A., Wessels L., de Laat W., van Steensel B. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 453:948–951 10.1038/nature06947 [DOI] [PubMed] [Google Scholar]
- Haber J.E., Leung W.Y. 1996. Lack of chromosome territoriality in yeast: promiscuous rejoining of broken chromosome ends. Proc. Natl. Acad. Sci. USA. 93:13949–13954 10.1073/pnas.93.24.13949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler R.A., Pratt-Hyatt M., Good P.D., Gipson T.A., Engelke D.R. 2008. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 22:2204–2214 10.1101/gad.1675908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjoul H., Kocanova S., Lassadi I., Bystricky K., Bancaud A. 2009. Lab-on-Chip for fast 3D particle tracking in living cells. Lab Chip. 9:3054–3058 10.1039/b909016a [DOI] [PubMed] [Google Scholar]
- Halme A., Bumgarner S., Styles C., Fink G.R. 2004. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell. 116:405–415 10.1016/S0092-8674(04)00118-7 [DOI] [PubMed] [Google Scholar]
- Hecht A., Laroche T., Strahl-Bolsinger S., Gasser S.M., Grunstein M. 1995. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 80:583–592 10.1016/0092-8674(95)90512-X [DOI] [PubMed] [Google Scholar]
- Hediger F., Neumann F.R., Van Houwe G., Dubrana K., Gasser S.M. 2002. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr. Biol. 12:2076–2089 10.1016/S0960-9822(02)01338-6 [DOI] [PubMed] [Google Scholar]
- Hernandez-Verdun D. 2006. Nucleolus: from structure to dynamics. Histochem. Cell Biol. 125:127–137 10.1007/s00418-005-0046-4 [DOI] [PubMed] [Google Scholar]
- Heun P., Laroche T., Shimada K., Furrer P., Gasser S.M. 2001. Chromosome dynamics in the yeast interphase nucleus. Science. 294:2181–2186 10.1126/science.1065366 [DOI] [PubMed] [Google Scholar]
- Hoess R.H., Abremski K. 1984. Interaction of the bacteriophage P1 recombinase Cre with the recombining site loxP. Proc. Natl. Acad. Sci. USA. 81:1026–1029 10.1073/pnas.81.4.1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q., Trelles-Sticken E., Scherthan H., Loidl J. 1998. Yeast nuclei display prominent centromere clustering that is reduced in nondividing cells and in meiotic prophase. J. Cell Biol. 141:21–29 10.1083/jcb.141.1.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q.W., Fuchs J., Loidl J. 2000. Centromere clustering is a major determinant of yeast interphase nuclear organization. J. Cell Sci. 113:1903–1912 [DOI] [PubMed] [Google Scholar]
- Joglekar A.P., Bloom K., Salmon E.D. 2009. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr. Biol. 19:694–699 10.1016/j.cub.2009.02.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalocsay M., Hiller N.J., Jentsch S. 2009. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol. Cell. 33:335–343 10.1016/j.molcel.2009.01.016 [DOI] [PubMed] [Google Scholar]
- King M.C., Drivas T.G., Blobel G. 2008. A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell. 134:427–438 10.1016/j.cell.2008.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R., Kim K.P., Prentiss M., Kleckner N., Kameoka S. 2008. Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell. 133:1188–1201 10.1016/j.cell.2008.04.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh B.O., Symington L.S. 2004. Recombination proteins in yeast. Annu. Rev. Genet. 38:233–271 10.1146/annurev.genet.38.072902.091500 [DOI] [PubMed] [Google Scholar]
- Kumaran R.I., Spector D.L. 2008. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J. Cell Biol. 180:51–65 10.1083/jcb.200706060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lainé J.P., Singh B.N., Krishnamurthy S., Hampsey M. 2009. A physiological role for gene loops in yeast. Genes Dev. 23:2604–2609 10.1101/gad.1823609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche T., Martin S.G., Gotta M., Gorham H.C., Pryde F.E., Louis E.J., Gasser S.M. 1998. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr. Biol. 8:653–656 10.1016/S0960-9822(98)70252-0 [DOI] [PubMed] [Google Scholar]
- Léger-Silvestre I., Trumtel S., Noaillac-Depeyre J., Gas N. 1999. Functional compartmentalization of the nucleus in the budding yeast Saccharomyces cerevisiae. Chromosoma. 108:103–113 10.1007/s004120050357 [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O., et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 326:289–293 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light W.H., Brickner D.G., Brand V.R., Brickner J.H. 2010. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol. Cell. 40:112–125 10.1016/j.molcel.2010.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M., Mortensen U.H., Rothstein R. 2003. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat. Cell Biol. 5:572–577 10.1038/ncb997 [DOI] [PubMed] [Google Scholar]
- Liti G., Louis E.J. 2005. Yeast evolution and comparative genomics. Annu. Rev. Microbiol. 59:135–153 10.1146/annurev.micro.59.030804.121400 [DOI] [PubMed] [Google Scholar]
- Liti G., Carter D.M., Moses A.M., Warringer J., Parts L., James S.A., Davey R.P., Roberts I.N., Burt A., Koufopanou V., et al. 2009. Population genomics of domestic and wild yeasts. Nature. 458:337–341 10.1038/nature07743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz A., Fuchs J., Trelles-Sticken E., Scherthan H., Loidl J. 2002. Spatial organisation and behaviour of the parental chromosome sets in the nuclei of Saccharomyces cerevisiae x S. paradoxus hybrids. J. Cell Sci. 115:3829–3835 10.1242/jcs.00066 [DOI] [PubMed] [Google Scholar]
- Louis E.J., Naumova E.S., Lee A., Naumov G., Haber J.E. 1994. The chromosome end in yeast: its mosaic nature and influence on recombinational dynamics. Genetics. 136:789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra R., Kerr S.C., Harreman M.T., Apponi L.H., Fasken M.B., Ramineni S., Chaurasia S., Valentini S.R., Corbett A.H. 2007. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J. Biol. Chem. 282:3042–3049 10.1074/jbc.M608741200 [DOI] [PubMed] [Google Scholar]
- Maeshima K., Hihara S., Eltsov M. 2010. Chromatin structure: does the 30-nm fibre exist in vivo? Curr. Opin. Cell Biol. 22:291–297 10.1016/j.ceb.2010.03.001 [DOI] [PubMed] [Google Scholar]
- Maillet L., Boscheron C., Gotta M., Marcand S., Gilson E., Gasser S.M. 1996. Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev. 10:1796–1811 10.1101/gad.10.14.1796 [DOI] [PubMed] [Google Scholar]
- Manley S., Gillette J.M., Patterson G.H., Shroff H., Hess H.F., Betzig E., Lippincott-Schwartz J. 2008. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat. Methods. 5:155–157 10.1038/nmeth.1176 [DOI] [PubMed] [Google Scholar]
- Marshall W.F., Straight A., Marko J.F., Swedlow J., Dernburg A., Belmont A., Murray A.W., Agard D.A., Sedat J.W. 1997. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr. Biol. 7:930–939 10.1016/S0960-9822(06)00412-X [DOI] [PubMed] [Google Scholar]
- Marvin M.E., Griffin C.D., Eyre D.E., Barton D.B., Louis E.J. 2009. In Saccharomyces cerevisiae, yKu and subtelomeric core X sequences repress homologous recombination near telomeres as part of the same pathway. Genetics. 183:441–451 10.1534/genetics.109.106674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhail K., Seebacher J., Gygi S.P., Moazed D. 2008. Role for perinuclear chromosome tethering in maintenance of genome stability. Nature. 456:667–670 10.1038/nature07460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele A., Bystricky K., Dekker J. 2009. Yeast silent mating type loci form heterochromatic clusters through silencer protein-dependent long-range interactions. PLoS Genet. 5:e1000478 10.1371/journal.pgen.1000478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. 2001. The concept of self-organization in cellular architecture. J. Cell Biol. 155:181–185 10.1083/jcb.200108110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondoux M.A., Scaife J.G., Zakian V.A. 2007. Differential nuclear localization does not determine the silencing status of Saccharomyces cerevisiae telomeres. Genetics. 177:2019–2029 10.1534/genetics.107.079848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkel C., Langowski J. 1998. Chromosome structure predicted by a polymer model. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 57:5888–5896 10.1103/PhysRevE.57.5888 [DOI] [Google Scholar]
- Nagai S., Dubrana K., Tsai-Pflugfelder M., Davidson M.B., Roberts T.M., Brown G.W., Varela E., Hediger F., Gasser S.M., Krogan N.J. 2008. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 322:597–602 10.1126/science.1162790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Németh A., Conesa A., Santoyo-Lopez J., Medina I., Montaner D., Péterfia B., Solovei I., Cremer T., Dopazo J., Längst G. 2010. Initial genomics of the human nucleolus. PLoS Genet. 6:e1000889 10.1371/journal.pgen.1000889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oza P., Jaspersen S.L., Miele A., Dekker J., Peterson C.L. 2009. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 23:912–927 10.1101/gad.1782209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palancade B., Liu X., Garcia-Rubio M., Aguilera A., Zhao X., Doye V. 2007. Nucleoporins prevent DNA damage accumulation by modulating Ulp1-dependent sumoylation processes. Mol. Biol. Cell. 18:2912–2923 10.1091/mbc.E07-02-0123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino F., Laroche T., Gilson E., Axelrod A., Pillus L., Gasser S.M. 1993. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell. 75:543–555 10.1016/0092-8674(93)90388-7 [DOI] [PubMed] [Google Scholar]
- Parada L., Misteli T. 2002. Chromosome positioning in the interphase nucleus. Trends Cell Biol. 12:425–432 10.1016/S0962-8924(02)02351-6 [DOI] [PubMed] [Google Scholar]
- Parada L.A., McQueen P.G., Munson P.J., Misteli T. 2002. Conservation of relative chromosome positioning in normal and cancer cells. Curr. Biol. 12:1692–1697 10.1016/S0960-9822(02)01166-1 [DOI] [PubMed] [Google Scholar]
- Pryde F.E., Louis E.J. 1999. Limitations of silencing at native yeast telomeres. EMBO J. 18:2538–2550 10.1093/emboj/18.9.2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl C. 1885. Uber Zellteilung. Morphologishes Jarbuch. 10:214-330 [Google Scholar]
- Raghuraman M.K., Brewer B.J. 2010. Molecular analysis of the replication program in unicellular model organisms. Chromosome Res. 18:19–34 10.1007/s10577-009-9099-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renauld H., Aparicio O.M., Zierath P.D., Billington B.L., Chhablani S.K., Gottschling D.E. 1993. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 7(7A):1133–1145 10.1101/gad.7.7a.1133 [DOI] [PubMed] [Google Scholar]
- Robinett C.C., Straight A., Li G., Willhelm C., Sudlow G., Murray A., Belmont A.S. 1996. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 135:1685–1700 10.1083/jcb.135.6.1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodley C.D., Bertels F., Jones B., O’Sullivan J.M. 2009. Global identification of yeast chromosome interactions using Genome conformation capture. Fungal Genet. Biol. 46:879–886 10.1016/j.fgb.2009.07.006 [DOI] [PubMed] [Google Scholar]
- Rosa A., Everaers R. 2008. Structure and dynamics of interphase chromosomes. PLOS Comput. Biol. 4:e1000153 10.1371/journal.pcbi.1000153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routh A., Sandin S., Rhodes D. 2008. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc. Natl. Acad. Sci. USA. 105:8872–8877 10.1073/pnas.0802336105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Arib G., Laemmli C., Nishikawa J., Durussel T., Laemmli U.K. 2006. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol. Cell. 21:379–391 10.1016/j.molcel.2005.12.012 [DOI] [PubMed] [Google Scholar]
- Schober H., Kalck V., Vega-Palas M.A., Van Houwe G., Sage D., Unser M., Gartenberg M.R., Gasser S.M. 2008. Controlled exchange of chromosomal arms reveals principles driving telomere interactions in yeast. Genome Res. 18:261–271 10.1101/gr.6687808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober H., Ferreira H., Kalck V., Gehlen L.R., Gasser S.M. 2009. Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev. 23:928–938 10.1101/gad.1787509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M., Klous P., Splinter E., Moshkin Y., Willemsen R., de Wit E., van Steensel B., de Laat W. 2006. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet. 38:1348–1354 10.1038/ng1896 [DOI] [PubMed] [Google Scholar]
- Soutoglou E., Dorn J.F., Sengupta K., Jasin M., Nussenzweig A., Ried T., Danuser G., Misteli T. 2007. Positional stability of single double-strand breaks in mammalian cells. Nat. Cell Biol. 9:675–682 10.1038/ncb1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavenhagen J.B., Zakian V.A. 1998. Yeast telomeres exert a position effect on recombination between internal tracts of yeast telomeric DNA. Genes Dev. 12:3044–3058 10.1101/gad.12.19.3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A., Hediger F., Neumann F.R., Bauer C., Gasser S.M. 2004. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J. 23:1301–1312 10.1038/sj.emboj.7600144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A., Van Houwe G., Hediger F., Kalck V., Cubizolles F., Schober H., Gasser S.M. 2006. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 441:774–778 10.1038/nature04845 [DOI] [PubMed] [Google Scholar]
- Tan-Wong S.M., Wijayatilake H.D., Proudfoot N.J. 2009. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 23:2610–2624 10.1101/gad.1823209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tham W.H., Wyithe J.S., Ko Ferrigno P., Silver P.A., Zakian V.A. 2001. Localization of yeast telomeres to the nuclear periphery is separable from transcriptional repression and telomere stability functions. Mol. Cell. 8:189–199 10.1016/S1097-2765(01)00287-8 [DOI] [PubMed] [Google Scholar]
- Therizols P., Duong T., Dujon B., Zimmer C., Fabre E. 2010. Chromosome arm length and nuclear constraints determine the dynamic relationship of yeast subtelomeres. Proc. Natl. Acad. Sci. USA. 107:2025–2030 10.1073/pnas.0914187107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therizols P., Fairhead C., Cabal G.G., Genovesio A., Olivo-Marin J.C., Dujon B., Fabre E. 2006. Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J. Cell Biol. 172:189–199 10.1083/jcb.200505159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M., Haeusler R.A., Good P.D., Engelke D.R. 2003. Nucleolar clustering of dispersed tRNA genes. Science. 302:1399–1401 10.1126/science.1089814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Rosell J., Sunjevaric I., De Piccoli G., Sacher M., Eckert-Boulet N., Reid R., Jentsch S., Rothstein R., Aragón L., Lisby M. 2007. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat. Cell Biol. 9:923–931 10.1038/ncb1619 [DOI] [PubMed] [Google Scholar]
- van Koningsbruggen S., Gierlinski M., Schofield P., Martin D., Barton G.J., Ariyurek Y., den Dunnen J.T., Lamond A.I. 2010. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol. Biol. Cell. 21:3735–3748 10.1091/mbc.E10-06-0508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S.C., Theriot J.A., Spakowitz A.J. 2010. Subdiffusive motion of a polymer composed of subdiffusive monomers. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 82:011913 10.1103/PhysRevE.82.011913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wombacher R., Heidbreder M., van de Linde S., Sheetz M.P., Heilemann M., Cornish V.W., Sauer M. 2010. Live-cell super-resolution imaging with trimethoprim conjugates. Nat. Methods. 7:717–719 10.1038/nmeth.1489 [DOI] [PubMed] [Google Scholar]
- Yang C.H., Lambie E.J., Hardin J., Craft J., Snyder M. 1989. Higher order structure is present in the yeast nucleus: autoantibody probes demonstrate that the nucleolus lies opposite the spindle pole body. Chromosoma. 98:123–128 10.1007/BF00291048 [DOI] [PubMed] [Google Scholar]