The GAP activity of OATL1, which is recruited to autophagosomes by Atg8, regulates autophagosome–lysosome fusion.
Abstract
Macroautophagy is a bulk degradation system conserved in all eukaryotic cells. A ubiquitin-like protein, Atg8, and its homologues are essential for autophagosome formation and act as a landmark for selective autophagy of aggregated proteins and damaged organelles. In this study, we report evidence demonstrating that OATL1, a putative Rab guanosine triphosphatase–activating protein (GAP), is a novel binding partner of Atg8 homologues in mammalian cells. OATL1 is recruited to isolation membranes and autophagosomes through direct interaction with Atg8 homologues and is involved in the fusion between autophagosomes and lysosomes through its GAP activity. We further provide evidence that Rab33B, an Atg16L1-binding protein, is a target substrate of OATL1 and is involved in the fusion between autophagosomes and lysosomes, the same as OATL1. Because both its GAP activity and its Atg8 homologue–binding activity are required for OATL1 to function, we propose a model that OATL1 uses Atg8 homologues as a scaffold to exert its GAP activity and to regulate autophagosomal maturation.
Introduction
Macroautophagy (referred to as autophagy hereafter) is a conserved mechanism for degrading cytosolic proteins and organelles in eukaryotic cells. Autophagy in mammalian cells not only supplies nutrients under starved conditions but also protects against human diseases by degrading aggregated proteins and damaged organelles (Mizushima et al., 2008). The degradation of cytoplasmic components by autophagy is achieved as follows. Isolation membranes (also called phagophores) emerge in the cytoplasm and elongate to envelop cytoplasmic components. The resulting spherical structures, called autophagosomes, fuse with endosomes and lysosomes, and their intermediate organelles, called autolysosomes, are ultimately transformed into lysosomes. These membrane dynamics involved in autophagosome formation are well recognized, but the mechanism that regulates membrane trafficking during autophagy remains largely unknown (Yoshimori, 2004; Mizushima, 2007).
A set of genes essential for autophagy (ATG genes) was originally identified by genetic analysis of the budding yeast Saccharomyces cerevisiae (Klionsky et al., 2003; Nakatogawa et al., 2009), and their presence was subsequently demonstrated in a variety of eukaryotic cells, including mammalian cells. One of their gene products, Atg8, and its homologues (e.g., LC3, GABARAP, and GATE-16 in mammals) are ubiquitin-like proteins conjugated to phosphatidylethanolamine (PE), and they have been found to be localized at elongating isolation membranes and autophagosomes but not at autolysosomes or lysosomes (Ichimura et al., 2000; Kabeya et al., 2000, 2004). In vitro analysis has shown that Atg8-PE forms an oligomer and induces liposome clustering and hemifusion (Nakatogawa et al., 2007), indicating that membrane remodeling is a function of Atg8 homologues. Actually, isolation membranes devoid of Atg8 homologues elongate but do not form mature autophagosomes in mammalian cells (Fujita et al., 2008b; Sou et al., 2008; Weidberg et al., 2010). Another function of Atg8 homologues is as an adapter for selective autophagy because, for example, p62/SQSTM1 (referred to as p62 hereafter) and NBR1 directly interact with both Atg8 homologues and polyubiquitin and thereby facilitate the clearance of polyubiquitinated proteins by autophagy in mammalian cells (Komatsu et al., 2007; Pankiv et al., 2007; Ichimura et al., 2008; Kirkin et al., 2009). Atg8 homologues seem to have additional functions because they interact with a variety of proteins, including GABA receptor γ2, clathrin heavy chain, and calreticulin (Mohrlüder et al., 2009), but the physiological functions of their interactions have yet to be determined.
Rab-type small GTPases are evolutionarily conserved membrane trafficking proteins (Pfeffer, 2001; Zerial and McBride, 2001; Stenmark, 2009), and it has been suggested that some members of the Rab family regulate autophagy. Rab7 (or Ypt7 in budding yeasts), which is responsible for the function of lysosomes (or vacuoles), is important for the fusion between autophagosomes and lysosomes and the subsequent degradation of autophagosomal contents (Kirisako et al., 1999; Gutierrez et al., 2004; Jäger et al., 2004). Rab11 regulates fusion between multivesicular bodies and autophagosomes in mammalian cells (Fader et al., 2008), and ER-resident Rab24 and Rab32 are involved in autophagosome formation, although the precise molecular mechanisms of their involvement are largely unknown (Munafó and Colombo, 2002; Hirota and Tanaka, 2009). We have previously reported finding that Golgi-resident Rab33B interacts with Atg16L1, an essential factor for isolation membrane elongation (Mizushima et al., 2003; Cadwell et al., 2008; Saitoh et al., 2008) and that it affects LC3 lipidation (Itoh et al., 2008). However, the regulatory mechanism of these Rab proteins in the autophagy process is totally unknown because their regulatory factors have never been identified.
In general, Rab proteins are thought to be activated and inactivated by guanine nucleotide exchange factor and GTPase-activating protein (GAP), respectively. A TBC (Tre-2/Bub2/Cdc16) domain was the first Rab-GAP domain reported in the budding yeasts (Strom et al., 1993), and ∼40 TBC domain–containing proteins (referred to as TBC proteins hereafter) are found in humans and mice (Bernards, 2003; Fukuda, 2011). Although the target Rabs of mammalian TBC proteins are being identified step by step (Fukuda, 2011), no TBC protein has yet been reported to be directly involved in autophagy.
In this study, we screened for mammalian TBC proteins involved in the autophagy process by expressing 41 different TBC proteins in mouse embryonic fibroblast (MEF) cells and monitoring them for colocalization with endogenous LC3. We succeeded in identifying OATL1/TBC1D25 (referred to as OATL1 hereafter) and TBC1D2B as autophagosome-resident TBC proteins and characterized the function of OATL1 in autophagy. The results of the present study show that OATL1 is recruited to autophagosomes through direct interaction with Atg8 homologues and that its overexpression inhibits conversion from autophagosomes to autolysosomes (i.e., autophagosomal maturation). They also reveal that Rab33B, an Atg16L1-binding Rab protein (Itoh et al., 2008), is a target of OATL1. Possible mechanisms of OATL1-mediated regulation of autophagosomal maturation are discussed based on these findings.
Results
Screening for TBC proteins involved in autophagy
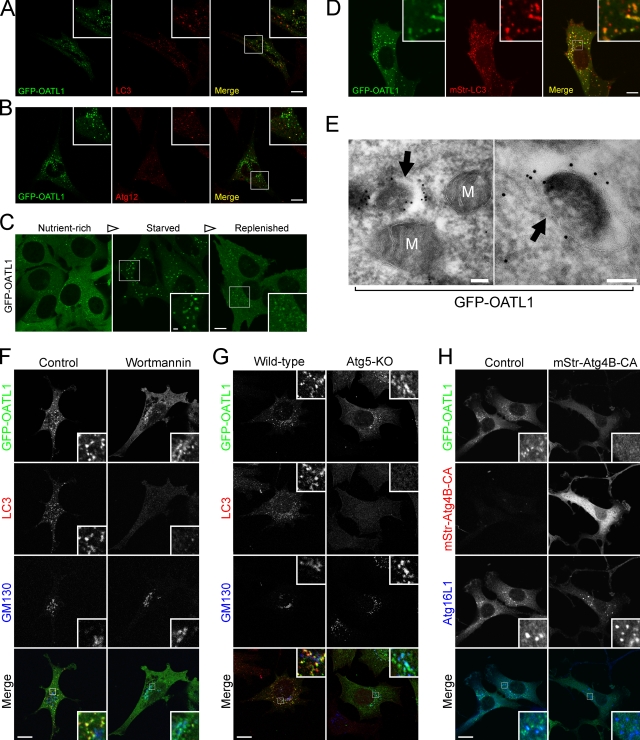
To search for TBC proteins showing autophagosomal localization and/or that affect autophagosomal biogenesis, we transiently expressed EGFP-tagged human or mouse TBC proteins in MEF cells and compared their localization with endogenous LC3 (i.e., isolation membranes and autophagosomes) under starved conditions. Two of the 41 TBC proteins we tested, OATL1 and TBC1D2B (recently reported as an LC3-binding protein; Behrends et al., 2010), were clearly colocalized with LC3 (Fig. 1 A and Fig. S1), and TBC1D11 showed partial colocalization with LC3 (Fig. S1). Some portions of GFP-OATL1–positive dot structures were also overlapped with Atg12, an isolation membrane marker protein (Fig. 1 B), suggesting that GFP-OATL1 was localized at both isolation membranes and autophagosomes. To confirm the autophagosomal localization of OATL1, we investigated the localization of GFP-OATL1 in living cells under various nutrient conditions. Under nutrient-rich conditions, GFP-OATL1 was dispersed in the cytoplasm and formed small punctate structures (Fig. 1 C, left), whereas after amino acid and serum starvation, GFP-OATL1 formed ring-shaped structures (Fig. 1 C, center). These ring structures likely corresponded to autophagosomes because merged images showed that they almost perfectly coincided with monomeric strawberry (mStr)–tagged LC3 (mStr-LC3; Fig. 1 D) and had diminished in number after replenishment with nutrient-rich medium (Fig. 1 C, right). Immuno-EM revealed that GFP-OATL1 was clearly localized at autophagosome-like structures (Fig. 1 E, arrows). These findings indicate that GFP-OATL1 is localized at isolation membranes and autophagosomes under starved conditions.
Figure 1.
OATL1 was localized at LC3-positive isolation membranes and autophagosomes. (A and B) GFP-OATL1 was colocalized with autophagic marker proteins. NIH3T3 cells transiently expressing GFP-OATL1 under starved conditions were fixed and stained with anti-LC3 antibody (A) or anti-Atg12 antibody (B). Merged images are shown at the right. (C) GFP-OATL1 was localized at ring-shaped organelles only under starved conditions. MEF cells stably expressing GFP-OATL1 were cultured under nutrient-rich, starved, and replenished conditions. Images of living cells are shown. (D) GFP-OATL1 was clearly colocalized with mStr-LC3 in living cells. MEF cells stably expressing GFP-OATL1 and mStr-LC3 cultured under starved conditions were observed. A merged image is shown at the right. (E) Immuno-EM revealed colloidal gold particles, indicating the presence of GFP-OATL1, on the isolation membranes of autophagosome-like structures (arrows) under starved conditions. M, mitochondria. (F–H) GFP-OATL1 was localized at the Golgi apparatus in autophagy-deficient cells. (F) MEF cells transiently expressing GFP-OATL1 were cultured under starved conditions in the presence (wortmannin) or absence (control) of 100 nM wortmannin, then fixed, and stained with anti-LC3 antibody and anti-GM130 antibody. (G) Wild-type and Atg5-knockout (Atg5-KO) MEF cells transiently expressing GFP-OATL1 were cultured under starved conditions, fixed, and stained with anti-LC3 antibody and anti-GM130 antibody. (H) NIH3T3 cells stably expressing mStr-Atg4B-CA or not expressing mStr-Atg4B-CA (control) were transiently transfected with pEGFP-C1-OATL1 (GFP-OATL1). The cells were cultured under starved conditions, fixed, and stained with anti-Atg16L1 antibody. In F–H, the signal color in the merged images is indicated by the color of the typeface. The insets show magnified views of the boxed areas. Bars: (A–D and F–H) 10 µm; (C, inset) 2 µm; (E) 100 nm.
Subcellular localization of GFP-OATL1 in autophagy-deficient cells
To determine whether the localization of OATL1 depends on autophagosome formation, we investigated the subcellular localization of GFP-OATL1 by using the three types of autophagy-deficient cells. First, we investigated the localization of GFP-OATL1 in cells treated with wortmannin, a phosphatidylinositol 3-kinase inhibitor that inhibits isolation membrane formation (Blommaart et al., 1997; Mizushima et al., 2001). GFP-OATL1 was dispersed in the cytoplasm and partially colocalized with GM130, a cis-Golgi marker protein in the wortmannin-treated cells, whereas in the control cells, GFP-OATL1 was colocalized with endogenous LC3-positive structures (Fig. 1 F). The same change in the distribution of GFP-OATL1 was observed in the second type of autophagy-deficient cells, Atg5-knockout (KO) MEF cells, in which the formation of isolation membranes is completely abolished (Fig. 1 G; Mizushima et al., 2001). The third type of autophagy-deficient cells we used were NIH3T3 cells expressing an Atg4B mutant protein with an alanine substitution for the 74th catalytic cysteine residue (Atg4B-CA cells; Fujita et al., 2008b). In contrast to the wortmannin-treated cells and Atg5-KO cells, the Atg4B-CA–expressing cells possess isolation membranes, but such isolation membranes are devoid of Atg8 homologues, including LC3 (Fujita et al., 2008b). GFP-OATL1 did not form any punctate structures in the Atg4B-CA–expressing cells, but the isolation membrane marker protein Atg16L1 clearly formed punctate structures, the same as in the control cells (Fig. 1 H). These findings collectively suggest that OATL1 is targeted to Atg8 homologue–decorated isolation membranes and autophagosomes.
OATL1 directly binds Atg8 homologues
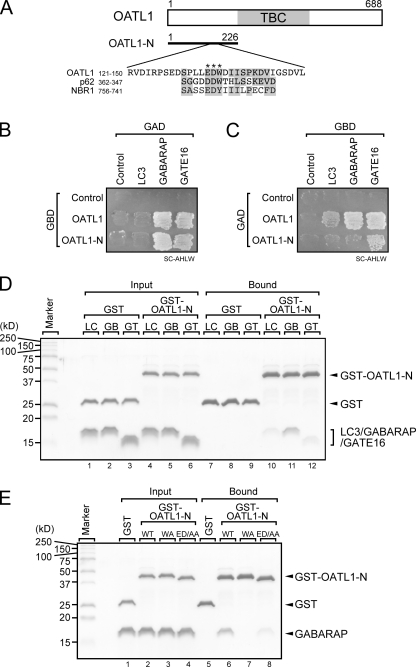
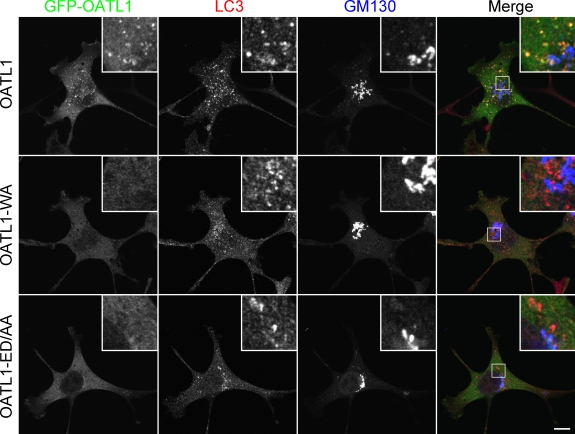
The specific localization of OATL1 on Atg8 homologue–resident isolation membranes and autophagosomes led us to hypothesize that OATL1 physically interacts with Atg8 homologues. To verify our hypothesis, we attempted to analyze the interaction between OATL1 and Atg8 homologues by yeast two-hybrid (Y2H) assays. We tested the interaction of OATL1 with LC3-β (referred to as LC3 hereafter), GABARAP, and GATE-16 as representatives of the three subfamilies classified by their primary sequences (Fig. S2). The results showed that OATL1 interacted with all three Atg8 homologues, although the Y2H interactions depend on the combination of bait and prey (Fig. 2, B and C). To determine which region of OATL1 is responsible for the interaction with Atg8 homologues, we constructed three truncated mutants of OATL1 (Fig. 2 A and Fig. S3 A). The results showed that the GFP-tagged N-terminal region of OATL1 (GFP–OATL1-N) alone was sufficient to colocalize with LC3 and that the GFP-tagged TBC domain of OATL1 (GFP–OATL1-TBC) and C-terminal region of OATL1 (GFP–OATL1-C) were not (Fig. S3, B–D). Consistent with the results of the immunofluorescence analysis, OATL1-N alone interacted with Atg8 homologues in the Y2H assay (Fig. 2, B and C), indicating that the N-terminal domain of OATL1 is sufficient for the interaction between OATL1 and Atg8 homologues. Furthermore, direct interactions between OATL1 and the three Atg8 homologues were observed when purified components were used (Fig. 2 D, lanes 10–12), although GABARAP seemed to show the highest affinity for OATL1-N among the three Atg8 homologues tested (Fig. 2 D, compare lane 11 with lanes 10 and 12). Moreover, we discovered a unique sequence in the N-terminal region of OATL1 that is similar to the LC3 recognition sequence (LRS) of p62 and NBR1 (Fig. 2 A; Ichimura et al., 2008; Kirkin et al., 2009). We especially noted the presence of the 136th tryptophan and diacidic residues (i.e., 134th glutamic acid and 135th aspartic acid) of OATL1 because mutations of the corresponding residues in p62 and NBR1 abrogated LC3-binding ability (Ichimura et al., 2008; Kirkin et al., 2009). As expected, mutant proteins in which these residues were replaced by alanines, i.e., OATL1-N-WA and OATL1-N-ED/AA, completely abolished and dramatically reduced, respectively, the interaction with GABARAP (Fig. 2 E, compare lane 6 and lanes 7 or 8). Consistent with the results of the in vitro binding assays, neither GFP-OATL1-WA nor GFP-OATL1-ED/AA was localized at LC3-positive structures (Fig. 3). We therefore concluded that OATL1 is localized at autophagosomes through a direct interaction with Atg8 homologues.
Figure 2.
OATL1 directly interacted with mammalian Atg8 homologues. (A) Schematic representation of the OATL1 protein. The solid bar indicates a truncated mutant of OATL1 (OATL1-N) that includes a putative LRS. The sequence alignment of the LRS in OATL1, p62, and NBR1 is shown. Residues in the sequences that are conserved and similar are shown against a shaded background. The asterisks indicate the positions of amino acid residues that were substituted in the OATL1 mutants in this study (see E). TBC, the Tre-2/Bub2/Cdc16 domain. (B and C) Yeast cells (pJ69-4A) possessing the indicated genes on pGAD-C1 (prey) or pGBD-C1 (bait) vectors were patched on synthetic complete medium lacking adenine, histidine, leucine, and uracil (SC-AHLW; selection medium). The baits and preys in B are reversed in C. (D) Purified LC3 (LC), GABARAP (GB), or GATE-16 (GT) proteins were mixed with purified GST or GST–OATL1-N proteins and then incubated with glutathione–Sepharose beads. Proteins bound to the beads were analyzed by 14% SDS-PAGE followed by Coomassie Brilliant Blue R-250 staining. (E) Purified GABARAP was mixed with GST, GST–OATL1-N (WT), GST–OATL1-N-W136A (WA), or GST–OATL1-N-E134A/D135A (ED/AA) and then incubated with glutathione–Sepharose beads. Proteins bound to the beads were analyzed by 14% SDS-PAGE followed by Coomassie Brilliant Blue R-250 staining. All proteins used in D and E were produced in bacteria and affinity purified by standard protocols.
Figure 3.
Atg8 homologue–binding-deficient mutants of OATL1 were localized at the Golgi apparatus instead of at autophagosomes. Wild-type MEF cells transiently expressing GFP-OATL1 (top), GFP–OATL1-WA (middle), or GFP–OATL1-ED/AA (bottom) under starved conditions were fixed and stained with anti-LC3 antibody and anti-GM130 antibody. Merged images between GFP-OATL1, LC3, and GM130 are shown on the right. The insets show magnified views of the boxed areas. Bar, 10 µm.
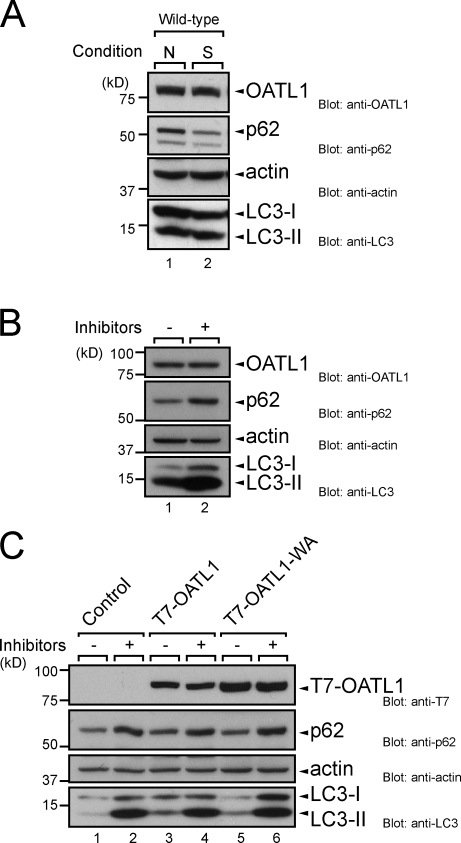
OATL1 is not susceptible to lysosomal degradation through autophagy
Because two Atg8 homologue–binding proteins, p62 and NBR1, have been shown to be degraded in the process of selective autophagy (Komatsu et al., 2007; Pankiv et al., 2007; Ichimura et al., 2008; Kirkin et al., 2009), it seemed highly likely that OATL1 is a substrate of autophagy. To investigate this possibility, we compared the amount of endogenous OATL1 protein in MEF cells under nutrient-rich and starved conditions. To our surprise, however, the amount of OATL1 in MEF cells was unaltered, even when autophagy was activated under starved conditions (Fig. 4 A, compare lanes 1 and 2, top), whereas the amount of p62 was clearly decreased under the same conditions (Fig. 4 A, compare lanes 1 and 2, second panel). Moreover, the amount of OATL1 was not increased, even by treatment with lysosomal protease inhibitors (E64-d and pepstatin A; Fig. 4 B, top), whereas the amounts of p62 and LC3-II were dramatically increased (Fig. 4 B, second and bottom panel, respectively). To rule out the possibility of transcriptional regulation of OATL1, we also tested MEF cells expressing T7-OATL1 under a retrovirus promoter. The same as the endogenous OATL1 level, the T7-OATL1 protein level was unaltered by treatment with the lysosomal protease inhibitors (Fig. 4 C, lanes 3 and 4). Although a point mutation in p62 or NBR1 that impairs their interaction with Atg8 homologues has been shown to abrogate efficient degradation of p62 or NBR1 (Ichimura et al., 2008; Kirkin et al., 2009), the WA mutation in OATL1 did not affect the amount of OATL1 at all (Fig. 4 C, lanes 5 and 6). We therefore concluded that OATL1 is not an autophagic substrate and that it is not susceptible to lysosomal degradation through autophagy.
Figure 4.
OATL1 was not a substrate of autophagy. (A) OATL1 protein level was unaltered by autophagic activity. Cell lysates from MEF cells under nutrient-rich (N) and starved (S) conditions were analyzed by immunoblotting with anti-OATL1 antibody (top), anti-p62 antibody (second panel), antiactin antibody (third panel), and anti-LC3 antibody (bottom). (B) Exposure to lysosomal protease inhibitors did not result in an increase in OATL1 protein level. Cell lysates from MEF cells cultured under nutrient-rich conditions with (+) or without (–) 100 nM E64-d and 100 µg/ml pepstatin A for 24 h were analyzed by immunoblotting with anti-OATL1 antibody (top), anti-p62 antibody (second panel), antiactin antibody (third panel), and anti-LC3 antibody (bottom). (C) Binding of OATL1 with Atg8 homologues did not affect the OATL1 protein level. Cell lysates from MEF cells stably expressing the proteins indicated (top) and cultured under nutrient-rich conditions with (+) or without (–) 100 nM E64-d and 100 µg/ml pepstatin A for 24 h were analyzed by immunoblotting with anti-T7 tag antibody (top), anti-p62 antibody (second panel), antiactin antibody (third panel), and anti-LC3 antibody (bottom).
Overexpression of OATL1 causes a delay in autophagosomal maturation
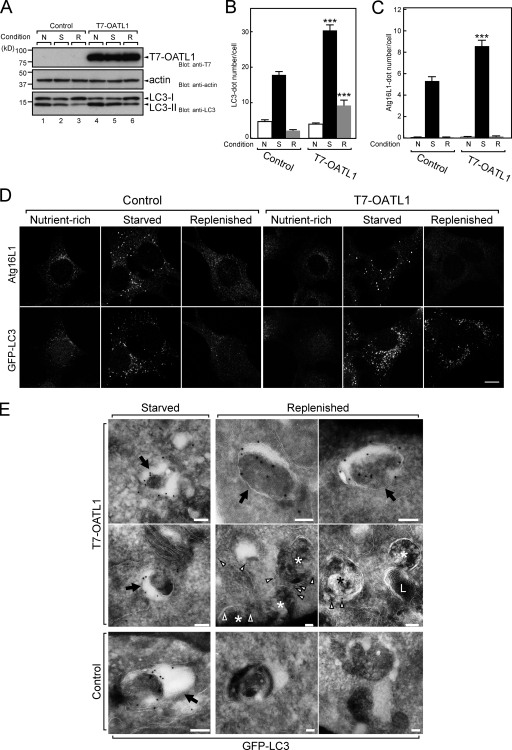
We next investigated the effect of overexpression of OATL1 on autophagy under various nutrient conditions by monitoring the profile of LC3 lipidation and the numbers of isolation membranes and autophagosomes in the cytoplasm. MEF cells stably expressing T7-tagged OATL1 (named OATL1 cells) contained a slightly increased amount of LC3-II under starved and replenished conditions in comparison with the control cells (Fig. 5 A, compare lanes 2 and 5 and lanes 3 and 6). Consistent with the results of the immunoblot analysis, the number of LC3-positive dots detected by the anti-LC3 antibody in OATL1 cells under both starved and replenished conditions was significantly higher than in the control cells (Fig. 5 B). This increased number of LC3 dots in the OATL1 cells might have been attributable to either an activation of autophagosome formation or an inactivation of autophagosomal maturation. To determine which of these two possibilities was actually responsible for the increase, we counted the number of Atg16L1-positive dots (corresponding to isolation membranes) under the same conditions because if OATL1 had accelerated autophagosome formation, the number of isolation membranes would have increased. Quantitative analysis revealed that the number of isolation membranes in the OATL1 cells was significantly higher than in the control cells under starved conditions but comparable with the number in the control cells under replenished conditions (Fig. 5 C). Furthermore, simultaneous observation of endogenous Atg16L1 (isolation membranes) and GFP-LC3 (isolation membranes and autophagosomes) in cells expressing GFP-LC3 confirmed that OATL1 overexpression resulted in an increase in GFP-LC3 dots alone under replenished conditions (Fig. 5 D). It should be noted that prolonged incubation under replenished conditions resulted in a gradual decrease in the number of LC3 dots to the control level (unpublished data). Immuno-EM observations revealed that, under starved conditions, GFP-LC3 was localized at typical autophagosomes in both control cells and OATL1 cells (Fig. 5 E, left, arrows). Under replenished conditions, the GFP-LC3 in OATL1 cells was mainly localized at autophagosome-like structures (Fig. 5 E, right, arrows) and occasionally at amphisome-like structures that contained multiple vesicles (Fig. 5 E, asterisks). Because we did not find any LC3-positive structures having a similar appearance in the control cells (Fig. 5 E, bottom right two images), these structures at the EM level most likely correspond to the residual LC3-positive organelles observed by immunofluorescence microscopy (Fig. 5 D). These findings allowed us to conclude that overexpression of OATL1 delays a process that occurs after the formation of autophagosomes and that would adequately explain the increased number of isolation membranes under starved conditions in OATL1 cells. The retarded maturation of the autophagosomes in OATL1 cells may reduce the negative feedback by nutrients from lysosomes and also contribute to the increase in the number of isolation membranes.
Figure 5.
Overexpression of OATL1 delayed autophagosomal maturation. (A) Overexpression of OATL1 resulted in an increase in the amount of LC3-II. MEF cells stably expressing T7-OATL1 or not expressing T7-OATL1 (control) were cultured under nutrient-rich (N), starved (S), or replenished (R) conditions, and their lysates were analyzed by immunoblotting with anti-T7 tag antibody (top), antiactin antibody (middle), and anti-LC3 antibody (bottom). (B and C) Wild-type MEF cells under the same conditions as in A were fixed and stained with anti-LC3 antibody or anti-Atg16L1 antibody. The mean numbers of LC3-positive (B) or Atg16L1-positive (C) dots per cell are shown. Error bars represent the means ± SEM of representative data (n ≥ 100) from two independent experiments. ***, P < 0.001; Student’s unpaired t test (compared with the control under the same conditions). (D) Overexpression of OATL1 increased the number of residual autophagosomes under replenished conditions. MEF cells stably expressing GFP-LC3 alone (control) or GFP-LC3 and T7-OATL1 (T7-OATL1) under the same conditions as in A were fixed and stained with anti-Atg16L1 antibody. GFP-LC3 and Atg16L1 are shown in the top and bottom, respectively. Bar, 10 µm. (E) EM analysis of the residual GFP-LC3–positive structures. MEF cells stably expressing GFP-LC3 and T7-OATL1 (T7-OATL1) or GFP-LC3 alone (control) were cultured under starved or replenished conditions and then fixed, and ultrathin cryosections were examined by immuno-EM. Under starved conditions, colloidal gold particles, indicating the presence of GFP-LC3, were detected in typical autophagosome-like structures (arrows) in both control cells and T7-OATL1–expressing cells. In T7-OATL1–expressing cells, numerous gold particles (some indicated by arrowheads) were detected in typical autophagosome-like structures (arrows) and occasionally on amphisome-like structures (asterisks), which contained multiple vesicles under replenished conditions. L, multilamellar lysosomal structure. Bars, 100 nm.
OATL1 delays autophagosomal maturation by inhibiting the encounter between autophagosomes and lysosomes
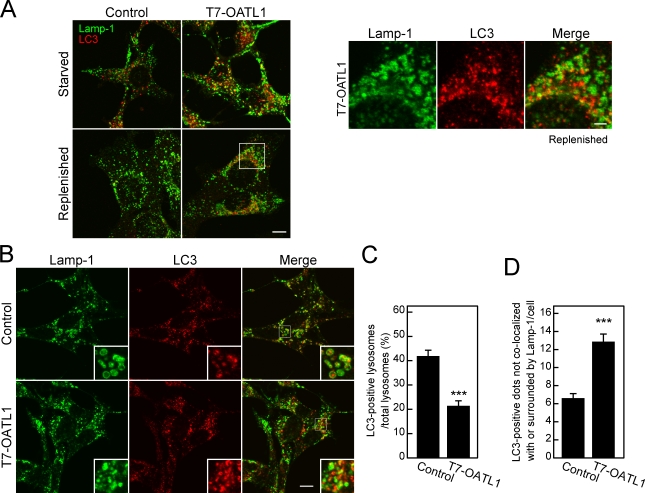
Because autophagosomal maturation is achieved by the fusion of autophagosomes with endosomes and lysosomes (Yoshimori, 2004), we attempted to determine whether the residual LC3-positive structures possess the properties of lysosomes. To do so, we monitored Lamp-1 as a lysosomal marker in OATL1 cells. However, colocalization between Lamp-1 and LC3 was hardly detected in OATL1 cells under replenished conditions (Fig. 6 A), suggesting a possibility that residual autophagosomes in OATL1 cells do not encounter with lysosomes. To investigate this possibility, we treated OATL1 cells with an inhibitor of vacuolar-type ATPase, bafilomycin A1, which decreases lysosomal protease activity and LC3 degradation in lysosomes. In the presence of bafilomycin A1, the LC3 in control cells was often localized in lysosomes (i.e., surrounded by Lamp-1; Fig. 6 B, top). However, overexpression of OATL1 under such conditions decreased the ratio of LC3-positive lysosomes and increased the number of Lamp-1–negative LC3 dots (i.e., autophagosomes; Fig. 6, B [bottom], C, and D). These results suggest that overexpression of OATL1 delayed a process that fuses autophagosomes and lysosomes.
Figure 6.
Overexpression of OATL1 inhibited the encounter between autophagosomes and lysosomes. (A) The residual LC3-positive structures did not contain Lamp-1. MEF cells stably expressing T7-OATL1 or not expressing T7-OATL1 (control) were cultured under starved conditions and replenished conditions. The cells cultured under each condition were fixed and stained with anti-LC3 antibody and anti–Lamp-1 antibody. Merged images are shown. Higher magnification views of the boxed area are shown on the right. (B–D) Overexpression of OATL1 caused a reduction in the ratio of LC3-positive lysosomes. The cells treated with bafilomycin A1 under starved conditions were fixed and stained with anti-LC3 antibody (red) and anti–Lamp-1 antibody (green). Merged images are shown on the right. Higher magnification views of the boxed areas are shown as insets. The ratios of LC3-positive lysosomes and the numbers of Lamp-1–negative LC3 dots per cell are shown in C and D, respectively. Error bars represent the means ± SEM of representative data (n ≥ 100) from two independent experiments. ***, P < 0.001; Student’s unpaired t test (compared with the control under the same conditions). Bars: (A [left] and B) 10 µm; (A, right) 2 µm.
Atg8 homologue–binding activity and GAP activity of OATL1 are required for the delayed autophagosomal maturation
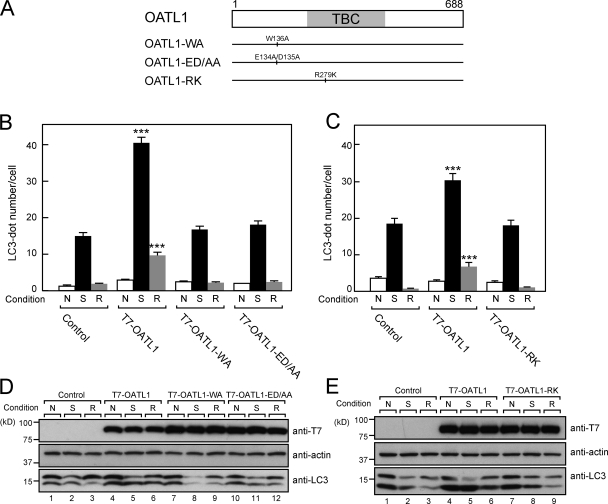
Because OATL1 contains both an LRS-like motif and a TBC Rab-GAP domain, we also investigated the requirement of these domains for the inhibitory activity of OATL1 in autophagosomal maturation. To do so, we established three additional MEF cell lines stably expressing T7-tagged OATL1-WA, OATL1-ED/AA, and OATL1-RK (in which the 279th catalytic arginine residue in OATL1 is replaced by lysine; Fig. 7 A). The results showed no effect of any of the three mutants on either the profile of LC3 lipidation (Fig. 7, D and E) or the number of autophagosomes (Fig. 7, B and C), although the OATL1-RK mutant retained the ability to be recruited to autophagosomes and to bind LC3 (Fig. S4, A and B). These findings indicated that both the Atg8 homologue–binding activity and the GAP activity of OATL1 are essential for its effects on autophagosome maturation.
Figure 7.
Both Atg8 homologue–binding activity and GAP activity of OATL1 were required for the delayed autophagosomal maturation. (A) Schematic representation of the OATL1 mutants used in this study. (B) MEF cells stably expressing T7-OATL1, T7–OATL1-WA, T7–OATL1-ED/AA, or none of these (control) were cultured under nutrient-rich (N), starved (S), or replenished (R) conditions. The cells cultured under each condition were fixed and stained with anti-LC3 antibody. The mean numbers of LC3-positive dots per cell are shown. Error bars represent the means ± SEM of representative data (n ≥ 100) from two independent experiments. ***, P < 0.001; one-way analysis of variance and Tukey posthoc test (compared with the control under the same conditions). (C) The same as in B, except that T7–OATL1-RK was used instead of T7–OATL1-WA or T7–OATL1-ED/AA. (D) Cell lysates from control, T7-OATL1–, T7–OATL1-WA–, and T7–OATL1-ED/AA–expressing cells cultured under the same conditions as in B were analyzed by immunoblotting with anti-T7 tag antibody (top), antiactin antibody (middle), and anti-LC3 antibody (bottom). (E) The same as in D, except that T7–OATL1-RK was used instead of T7–OATL1-WA or T7–OATL1-ED/AA.
Because OATL1 is endogenously expressed in MEF cells (Fig. 4 A), we also evaluated the effect of OATL1 knockdown on autophagy by using specific siRNA. However, although the amount of p62 was slightly decreased in the OATL1 knockdown cells (∼70% of the amount in the control cells), OATL1 knockdown had no effect on the profile of LC3, autophagic flux, or the number of LC3 dots (Fig. S5, A, C, and E). We speculate that OATL1 and p62 compete with each other for binding to Atg8 homologues in vivo and that down-regulation of OATL1 enables more frequent access of p62 to Atg8 homologues, thereby resulting in the decrease in p62.
Rab33B is the target of OATL1
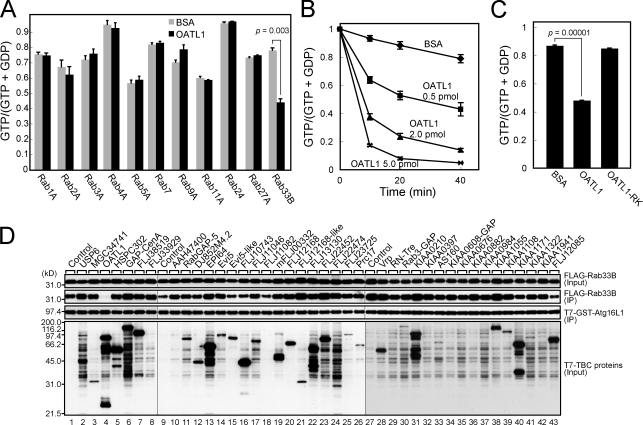
Because the GAP activity of OATL1 is important for its function, we next searched for its substrate among the Rab family members. We purified GST-tagged OATL1 proteins from COS-7 cells and assessed their GAP activity toward eleven Rabs purified from bacteria. The results showed that OATL1 significantly promoted the GTP hydrolysis activity of Rab33B in a dose-dependent manner (Fig. 8 B), whereas it did not exhibit any significant GAP activity toward other Rabs (Fig. 8 A). Although we previously reported that OATL1 has GAP activity toward Rab2A (Itoh et al., 2006), a higher dose of OATL1 is required for clear Rab2A-GAP activity than for Rab33B-GAP activity (2 pmol for Rab2A vs. 0.5 pmol for Rab33B), indicating that OATL1 prefers Rab33B as a substrate. Because the RK mutation completely abolished Rab33B-GAP activity (Fig. 8 C), OATL1 should exert its GAP activity by the conserved mechanism (Pan et al., 2006). The Rab33B-GAP activity of OATL1 was also confirmed by a GTP-Rab pull-down assay (Itoh and Fukuda, 2006) in which the amount of active Rab33B was estimated by using beads coupled with Atg16L1, an effector of Rab33B (Itoh et al., 2008). The amount of active Rab33B was dramatically reduced only when Rab33B was coexpressed with OATL1 (Fig. 8 D, second panel, compare lanes 1 and 4). It should be noted that no other TBC proteins possessed as strong Rab33B-GAP activity as OATL1 (Fig. 8 D), indicating that the relationship between OATL1 and Rab33B is highly specific.
Figure 8.
OATL1 functions as a GAP specific for Rab33B. (A) Rab33B is a specific substrate of OATL1. The Rab-GAP activity of OATL1 toward indicated Rab proteins is summarized. The GTPase activity of each Rab protein in the presence of BSA or OATL1 is shown. A significant p-value is shown (Student’s unpaired t test). (B) Dose dependency of the GAP activity of OATL1 is shown. GTP hydrolysis by Rab33B in the presence of BSA or the concentrations of OATL1 indicated was measured. (C) A conserved catalytic arginine residue in the TBC domain of OATL1 is essential for its GAP activity. GTP hydrolysis by Rab33B in the presence of BSA, OATL1, or OATL1-RK was measured. A significant p-value is shown (Student’s unpaired t test). (B and C) The results are expressed as the amount of the GTP-bound form of Rab33B after the reaction as a percentage of the amount before the reaction. Error bars represent the means ± SEM of data from three determinations. (D) OATL1 is the only TBC protein that possesses strong Rab33B-GAP activity. Total lysates of COS-7 cells expressing FLAG-Rab33B alone (control; lanes 1, 9, and 27) or FLAG-Rab33B and the T7-tagged TBC proteins indicated (lanes 2–8, 10–26, and 28–43) were incubated with glutathione–Sepharose beads that had been coupled with purified T7–GST-Atg16L1 used as a trapper of active Rab33B. Total lysates (input; top and bottom) and proteins trapped by the Atg16L1 beads (IP, immunoprecipitation; middle two panels) were analyzed by immunoblotting with anti-FLAG tag antibody (top two panels) and anti-T7 tag antibody (bottom two panels). Input means 1/60 volume of the reaction mixture. Black lines indicate that intervening lanes have been spliced out.
Effect of Rab33B-QL overexpression on autophagosomal maturation
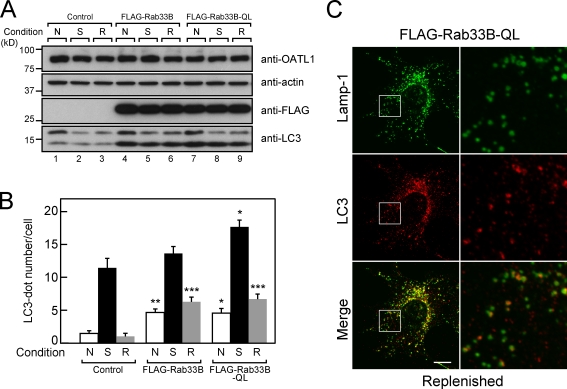
If Rab33B is a target of OATL1 during autophagosomal maturation in vivo, Rab33B should also affect autophagosomal maturation. As expected, expression of FLAG-tagged Rab33B (FLAG-Rab33B) or Rab33B-QL (i.e., a GTPase-deficient mutant; FLAG–Rab33B-QL) increased both the LC3-II level (Fig. 9 A) and the number of LC3-positive dots in the cytoplasm (Fig. 9 B) even under replenished conditions. Because monomeric RFP–Rab33B-TN (a constitutively negative form mutant) did not increase LC3-II under nutrient-rich or starved conditions (Itoh et al., 2008), Rab33B-TN is unlikely to affect autophagosomal maturation. In contrast to the OATL1-expressing cells (Fig. 6 A), most of the LC3-positive dots in the cytoplasm of FLAG–Rab33-QL–expressing cells appeared to be colocalized with Lamp-1, especially when viewed under low magnification (Fig. 9 C, left). However, close inspection of the immunostaining under high magnification revealed that most of the LC3-positive dots did not completely overlap with the Lamp-1–positive dots (Fig. 9 C, right), in contrast to their appearance in the bafilomycin A1–treated cells (Fig. 6 B). Based on these findings, together with the fact that Rab33B is a target of OATL1, we speculate that the LC3-positive autophagosomes in Rab33B-QL–expressing cells are not efficiently fused with lysosomes.
Figure 9.
Overexpression of the wild-type and active form Rab33B inhibited fusion between autophagosomes and lysosomes. (A) Wild-type Rab33B and the active form Rab33B mutant (Rab33B-QL) increased the amount of LC3-II. Cell lysates from MEF cells stably expressing FLAG-Rab33B, FLAG–Rab33B-QL, or neither (control) were cultured under nutrient-rich (N), starved (S), or replenished (R) conditions, and their lysates were analyzed by immunoblotting with anti-OATL1 antibody (top), antiactin antibody (second panel), anti-FLAG tag antibody (third panel), and anti-LC3 antibody (bottom). (B) Expression of the wild-type and active form Rab33B increased the number of LC3-positive dots. MEF cells expressing FLAG-Rab33B, FLAG–Rab33B-QL, or neither (control) were cultured under the same conditions as in A. The cells cultured under each condition were fixed and stained with anti-LC3 antibody, and the numbers of LC3-positive dots per cell were counted (n ≥ 100). Error bars represent the means ± SEM of representative data (n ≥ 100) from two independent experiments. ***, P < 0.001; **, P < 0.01; *, P < 0.05; one-way analysis of variance and Tukey posthoc test (compared with the control under the same conditions). (C) Overexpression of Rab33B-QL induced LC3-positive dots that were closely associated with Lamp-1–positive dots. MEF cells stably expressing FLAG–Rab33B-QL were cultured under replenished conditions, then fixed, and stained with anti-LC3 antibody (red) and anti–Lamp-1 antibody (green). Merged images are shown on the bottom. Higher magnification views of the boxed areas are shown at the right. Bar, 10 µm.
Discussion
Although many genes that are essential for autophagy have been identified during the past decade, little is known about the mechanism that regulates membrane trafficking in the autophagy process. In particular, involvement of a Rab-GAP in autophagy had never been reported, even though several Rab proteins have been shown to be involved in autophagy (Fukuda and Itoh, 2008). In the present study, we, for the first time, screened for Rab-GAPs associated with autophagy based on the localization of TBC proteins, and we succeeded in identifying OATL1 as a prime candidate. We also identified two additional TBC proteins, TBC1D2B and TBC1D11, which showed clear colocalization and partial colocalization, respectively, with LC3 in MEF cells (Fig. S1). These results are consistent with a study in a recent paper that some TBC proteins (TBC1D2B, TBC1D5, TBC1D11, and TBC1D15) form a complex with Atg8 homologues (Behrends et al., 2010). Although we do not know exactly why TBC1D5 and TBC1D15 did not show clear colocalization with LC3 despite having the ability to bind Atg8 homologues, cell type–specific expression of Atg8 homologues and/or of a cofactor that facilitates autophagosomal localization may be responsible for this discrepancy.
OATL1 was found to be localized at LC3-positive isolation membranes and autophagosomes (Fig. 1) and to directly interact with Atg8 homologues (Fig. 2). We found that an LRS-like sequence in OATL1 is responsible for its binding to Atg8 homologues, the same as in other Atg8 homologue–binding proteins, p62 and NBR1 (Komatsu et al., 2007; Pankiv et al., 2007; Ichimura et al., 2008; Kirkin et al., 2009). In contrast to p62 and NBR1, however, neither endogenous nor exogenous OATL1 showed any evidence of autophagic substrates (Fig. 4). Furthermore, WA and ED/AA mutations of OATL1, which abrogated the interaction between OATL1 and Atg8 homologues, completely abolished membranous localization of the mutant proteins without altering their protein expression level (Fig. 3). We therefore concluded that the LRS in OATL1 is not required for degradation of OATL1 but is required for targeting OATL1 on the organelle surface. One important remaining question is why OATL1 is not degraded during autophagy because Atg8 homologues, landmarks of OATL1, are localized at both the inner and outer surface of autophagosomes (Kabeya et al., 2004). Further extended investigations will be needed to answer this question.
Another important finding is that OATL1 inactivates Rab33B, a medial Golgi-resident Rab involved in retrograde transport (Zheng et al., 1998; Valsdottir et al., 2001; Jiang and Storrie, 2005; Starr et al., 2010), both in vitro and in cultured cells (Fig. 8). We especially noted the fact that Rab33B directly interacts with Atg16L1 (Itoh et al., 2008). The Atg12–5–16L1 complex interacts with Atg3, an E2 enzyme that catalyzes LC3 lipidation and facilitates the encounter between LC3 and PE on the membrane, like an E3 enzyme in the ubiquitin conjugation system (Hanada et al., 2007; Fujita et al., 2008a). Overexpression of Rab33B had been found to induce excess lipidation of LC3, suggesting that Rab33B accelerates the E3 activity of the Atg12–5–16L1 complex (Itoh et al., 2008), but the physiological significance of LC3 lipidation by Rab33B had remained unclear. Because the results of the present study indicated that these four factors, i.e., Rab33B, the Atg12–5–16L1 complex, Atg8 homologues, and OATL1, physically and/or enzymatically interact, it is tempting to speculate that they form a feedback loop in which (a) activated Rab33B recruits the Atg12–5–16L1 complex near the membrane through direct interaction with Atg16L1, (b) Atg8 homologues are conjugated with PE by an E3 activity of Atg12–5–16L1, (c) OATL1 recognizes the Atg8 homologues anchored with the membrane near Rab33B, and (d) OATL1 inactivates Rab33B through its GAP activity (Fig. S5 F).
The aforementioned molecular mechanism is inferred to be involved in autophagosomal maturation because overexpression of either OATL1 or Rab33B induced a delay in autophagosomal maturation (Fig. 5 and Fig. 9). Overexpression of OATL1 inhibited the process before the encounter between autophagosomes and lysosomes (Fig. 6), whereas overexpression of Rab33B or Rab33B-QL, a GTPase-deficient mutant of Rab33B, appears to have inhibited the fusion between autophagosomes and lysosomes (Fig. 9). These data suggest a model in which Rab33B plays a role in the tethering/docking step and/or fusion step between autophagosomes and lysosomes. Therefore, inactivation of Rab33B by OATL1 overexpression reduces encounters between autophagosomes and lysosomes, whereas expression of the active form–fixed Rab33B reduces the rate of fusion between autophagosomes and lysosomes.
However, inconsistent with this model, knockdown of OATL1 (or its substrate Rab33B) did not cause any significant phenotype in terms of autophagy (Fig. S5, A–E). The following four possibilities could explain the lack of a knockdown effect on autophagy. (1) The first possibility is a technical problem. Almost complete knockdown has been shown to be required for inhibition of autophagy by RNA interference of ATG genes (Hosokawa et al., 2006; Yoshimura et al., 2006), and thus, the down-regulation of OATL1 (or Rab33B) in our study may not have been sufficient to induce obvious phenotypes. (2) The second possibility is the existence of redundant pathways. It has previously been shown that impairment of Rab7 function induces autolysosome accumulation (Gutierretz et al., 2004; Jäger et al., 2004; Kimura et al., 2007). Because OATL1 was unable to accelerate the GTPase activity of Rab7 (Fig. 8 A), MEF cells are likely to possess two independent pathways for autophagosomal maturation, a Rab33B–OATL1 pathway and a Rab7 pathway. If the Rab7 pathway were dominant in MEF cells, knockdown of OATL1 or Rab33B might not result in any clear phenotype. (3) The third possibility is that the Rab33B–OATL1 pathway is essential for autophagy in specific types of cells but not for starvation-induced autophagy. Because the cytoplasmic components should be quickly degraded under starved conditions, regulation of fusion between autophagosomes and lysosomes may be less important in canonical autophagy. In contrast, in antigen-presenting cells (e.g., dendritic cells), for example, autophagosomes fuse with the major histocompatibility complex class II compartment, a kind of lysosome-related organelle (Schmid et al., 2007). In this particular case, there may be a distinct regulatory mechanism of autophagosomal maturation, and the Rab33B–OATL1 pathway may be involved in this mechanism. (4) The forth possibility is that the function of the Rab33B–OATL1 pathway is involved in classical membrane trafficking. Actually, GABARAP was originally identified as a binding protein of GABAA receptors (Wang et al., 1999), and a recent study has shown that GABARAP is required for ER-Golgi transport of cadherin (Nakamura et al., 2008). GATE-16 was first identified as a positive regulator of intra-Golgi transport (Legesse-Miller et al., 2000) and reported to interact with NSF and GOS-28 (Sagiv et al., 2000). Thus, the Atg8 homologues are also likely to be involved in classical membrane trafficking (e.g., ER-Golgi transport). Because Rab33B has been suggested to be involved in retrograde transport from the Golgi (Zheng et al., 1998; Valsdottir et al., 2001; Jiang and Storrie, 2005; Starr et al., 2010), the pivotal function of the Rab33B–OATL1 pathway may be in regulation of the classical membrane trafficking pathway. Further extensive research will be necessary to investigate all of these possibilities and determine the exact function of the Rab33B–OATL1 pathway.
In summary, we have identified OATL1, a previously uncharacterized Rab-GAP, as a novel Atg8 homologue–binding protein and discovered that OATL1 inactivates Rab33B, an Atg16L1-binding protein. We have also demonstrated that overexpression of OATL1 or Rab33B delays the maturation of autophagosomes by modulating the fusion between autophagosomes and lysosomes. We suggest that OATL1 is recruited to autophagosomes through direct interaction with Atg8 homologues and then inactivates Rab33B. Although the significance of the Rab33B–OATL1 pathway remains to be fully elucidated, based on the results of the present study, we propose a novel concept of the function of Atg8 homologues, i.e., as a scaffold for signaling molecules that regulates autophagosomal maturation.
Materials and methods
Materials
Anti-OATL1 and anti-Atg16L1 rabbit polyclonal antibodies were produced by using GST–OATL1-N and GST–Atg16L1-M (middle region of Atg16L1), respectively, as an antigen, and they were affinity purified as described previously (Fukuda and Mikoshiba, 1999). The anti-OATL1 antibody used in this study recognizes endogenous OATL1 when used for immunoblotting (Fig. 4 A), but it was incapable of doing so in an immunofluorescence analysis. Anti-LC3 rabbit polyclonal antibody was prepared as described previously (Itoh et al., 2008). HRP-conjugated anti-FLAG tag (M2) mouse monoclonal antibody was obtained from Sigma-Aldrich. HRP-conjugated anti-T7 tag mouse monoclonal antibody and anti-T7 tag antibody-conjugated agarose were purchased from Merck Biosciences Novagen. Alexa Fluor 488–, Alexa Fluor 594–, and Alexa Fluor 633–conjugated secondary antibodies were obtained from Invitrogen. Anti-GM130 mouse monoclonal antibody (BD), antiactin goat polyclonal antibody (Santa Cruz Biotechnology, Inc.), anti-Atg12 (or Apg12) rabbit polyclonal antibody (Invitrogen), anti-p62 rabbit polyclonal antibody (Enzo Life Sciences, Inc.), and anti–Lamp-1 rat monoclonal antibody (1D4b; BD) were obtained commercially. α-[32P]GTP and glutathione–Sepharose 4B were purchased from GE Healthcare. GTP-γS and GDP were obtained from Roche. E64-d and pepstatin A were purchased from Peptide Laboratory. Bafilomycin A1 and wortmannin were purchased from EMD and Merck KGaA, respectively.
Plasmid construction
The cDNA encoding the human OATL1 was cloned, and the OATL1-RK mutant cDNA was constructed as described previously (Itoh et al., 2006). pEF-T7-GST-OATL1, pEF-T7-OATL1, and pEGFP-C1-OATL1 were prepared as described previously (Itoh and Fukuda, 2006; Itoh et al., 2006). pEF-T7-GST-Atg16L1, pEF-FLAG-Rab33B, pMRX-puro-FLAG-Rab33B, and pMRX-puro-FLAG-Rab33B-QL were also prepared as described previously (Itoh et al., 2008). The cDNAs and plasmids of the other TBC proteins have been previously described elsewhere (Itoh et al., 2006; Ishibashi et al., 2009). The cDNA encoding the mouse LC3-β, GABARAP, or GATE-16 was cloned from mouse brain cDNAs by using conventional PCR techniques and the following pairs of oligonucleotides with a BamHI or BglII linker (underlined) or a stop codon (boldface): 5′-GGATCCATGCCGTCCGAGAAGACCTT-3′ and 5′-TTACACAGCCATTGCTGTCC-3′ for LC3-β, 5′-GGATCCATGAAGTTCGTGTACAAA-3′ and 5′-TCACAGACCATAGACGCT-3′ for GABARAP, and 5′-AGATCTATGAAGTGGATGTTTAAG-3′ and 5′-TCAGAAGCCAAAAGTGTT-3′ for GATE-16. To obtain a mutant cDNA fragment encoding OATL1-WA and a mutant cDNA containing OATL1-ED/AA, KOD plus ver. 2 and the pEGFP-C1-OATL1 plasmid were used as the polymerase for PCR (Toyobo Co., Ltd.) and as the template, respectively. The following primers were used for amplification: 5′-CATTGCTAGAAGACGCGGACATAATCAGCC-3′ and 5′-GGCTGATTATGTCCGCGTCTTCTAGCAATG-3′ for OATL1-W136A (OATL1-WA) and 5′-AGCCCATTGCTAGCAGCCTGGGACATAATC-3′ and 5′-GATTATGTCCCAGGCTGCTAGCAATGGGCT-3′ for OATL1-E134A/D135A (OATL1-ED/AA). The linear DNA fragments obtained were circularized with an In-Fusion recombination enzyme (BD). Deletion mutants of OATL1 (see Fig. S3 A for details) were similarly constructed by using conventional PCR techniques and the following pairs of oligonucleotides with a BglII linker (underlined) or a stop codon (boldface): 5′-AGATCTATGGCAACAGCCTCCGGG-3′ and 5′-TCAATGATAGATCCGCAGGCG-3′ for OATL1-N, OATL1-N-WA, or OATL1-N-ED/AA; 5′-AGATCTATAATCAGCCCCAAAGAT-3′ and 5′-TCACCTCCTGGAGCCCATGTT-3′ for OATL1-TBC; and 5′-AGATCTGGTGGCCACAGGGGGTGG-3′ and 5′-TCAAGATGCGGCTGTGGC-3′ for OATL1-C. These mutant OATL1 cDNA fragments of OATL1 were transferred to the pEF-T7 tag mammalian expression vector (modified from pEF-BOS; Fukuda et al., 1994, 1999), pEGFP-C1 vector, pGBD-C1 vector (James et al., 1996), pGAD-C1 vector (James et al., 1996), pGEX-4T3 vector (GE Healthcare), pMRX-IRES-puro-EGFP vector, or pMRX-IRES-puro-T7 vector by standard techniques. pMRX-IRES-puro/bsr-EGFP/T7/FLAG vectors are variants of pMRX-IRES-puro/bsr (donated by S. Yamaoka, Tokyo Medical and Dental University, Tokyo, Japan; Saitoh et al., 2003). We used DNA sequencing to confirm that no unexpected mutations had occurred in the open reading frame of these cDNAs.
Y2H assay
Y2H assays were performed by using the strain (pJ69-4A), vectors (pGBD-C1 and pGAD-C1), and the selection medium (synthetic complete medium lacking adenine, histidine, leucine, and tryptophan) as described previously (James et al., 1996).
Cell culture, transfection, and infection
Wild-type and Atg5-KO MEF cells were a gift from N. Mizushima (Tokyo Medical and Dental University, Tokyo, Japan). NIH3T3 cells stably expressing the Atg4B-CA mutant and control NIH3T3 cells were a gift from T. Yoshimori (Osaka University, Osaka, Japan). Plat-E cells were donated by T. Kitamura (University of Tokyo, Tokyo, Japan). COS-7, NIH3T3, and MEF cells were maintained in DME (Sigma-Aldrich or Wako Chemicals USA, Inc.) containing 10% fetal bovine serum and antibiotics under 5% CO2 at 37°C. Starved conditions were achieved by washing the cells once with HBSS (Sigma-Aldrich) and transferring them to HBSS for 1.5 h. Replenished conditions were achieved by additional incubation in DME containing serum for 1 h after starvation. Transfection of plasmids into COS-7 cells (for immunoprecipitation) and NIH3T3 and MEF cells (for immunofluorescence) was performed by using Lipofectamine Plus and Lipofectamine 2000 (Invitrogen), respectively, each according to the manufacturer’s instructions. Plat-E cell culture and retrovirus infection were performed essentially as described previously (Morita et al., 2000). Lysosomal proteases were inhibited by exposing cells to 100 nM E64-d and 100 µg/ml pepstatin A for 24 h.
Immunofluorescence and image analyses
Immunostaining was performed essentially as described previously (Itoh and Fukuda, 2006). In brief, cultured cells were fixed with 4% paraformaldehyde or ice-cold methanol and stained with specific antibodies. The stained cells were examined for fluorescence with a confocal fluorescence microscope (IX81 and Fluoview 1000; Olympus) through an objective lens (100× magnification, NA 1.45; Olympus) and with Fluoview software (version 2.1c; Olympus). Live-cell images were acquired at room temperature briefly. The images acquired were processed with GNU image manipulation program software (version 2.2 and 2.6). For quantitative analysis, images of the cells were captured at random with the confocal microscope, and the number of the fluorescent dots (e.g., LC3 dots and Atg16L1 dots) was counted with MetaMorph software (version 6.3r3; MDS Analytical Technologies) and ImageJ software (version 1.43u; National Institutes of Health).
GAP assay
GTP-Rab33B pull-down assay was performed by using T7–GST-Atg16L1 as a trapper of active Rab33B (Itoh and Fukuda, 2006). Bacterially produced Rab proteins and T7–GST-OATL1 protein produced by COS-7 cells were purified (Fukuda and Kanno, 2005) and used for the in vitro GAP assay as described previously (Itoh et al., 2006). To elucidate the Rab-GAP specificity of OATL1, an in vitro GAP assay was performed using 0.5 pmol OATL1 protein. Because the intrinsic GTPase activity of the Rab family members differs, the reaction time was varied according to the Rab isoform tested (Rab1A for 40 min, Rab2A for 40 min, Rab3A for 20 min, Rab4A for 20 min, Rab5A for 20 min, Rab7 for 40 min, Rab9A for 20 min, Rab11A for 40 min, Rab24 for 40 min, Rab27A for 40 min, and Rab33B for 40 min).
RNA interference
Double-strand RNAs were purchased from Eurogentec. The siRNA target sequence in OATL1 is 5′-GGAGCCTTCACTTCGTTTT-3′. For Rab33B knockdown, we used stealth RNA against mouse Rab33B purchased from Invitrogen (Rab33b-MSS208364 and Rab33b-MSS208365). Transfection of siRNA was performed by using RNAiMAX (Invitrogen) according to the manufacturer’s instructions.
EM analysis
Cells expressing each GFP fusion protein were fixed with 4% paraformaldehyde and 0.1% glutaraldehyde in phosphate buffer, pH 7.4, and ultrathin cryosections were examined by immuno-EM. The procedures used for cell freezing, sectioning, and the immunoreactions have been described previously (Waguri and Komatsu, 2009). Rabbit polyclonal antibody against GFP (ab6556; Abcam) was used as the first antibody. The sections were viewed with an electron microscope (JEM1200; JEOL).
Online supplemental material
Fig. S1 shows a summary of the Rab-GAP screening. Fig. S2 shows a sequence alignment and phylogenic tree of animal and yeast Atg8 homologues. Fig. S3 shows the localization of truncated OATL1 mutant proteins. Fig. S4 shows the localization of OATL1-RK mutant proteins and their binding ability with LC3. Fig. S5 shows the results of OATL1 knockdown and Rab33B knockdown experiments and a schematic model of the inactivation of Rab33B by OATL1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201008107/DC1.
Acknowledgments
We thank Dr. Noboru Mizushima for donating wild-type and Atg5-KO MEF cells, Dr. Toshio Kitamura for donating Plat-E cells and retroviral vectors, Dr. Shoji Yamaoka for pMRX-IRES-puro/bsr vectors, Dr. Tamotsu Yoshimori for NIH3T3 cells stably expressing Atg4B-CA mutant and control cells, Atsuko Yabashi and Katsuyuki Kanno for help in EM, and Dr. Shunsuke Kimura, Dr. Naonobu Fujita, and members of the Fukuda Laboratory for valuable discussions.
This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, and Technology (MEXT) of Japan (to M. Fukuda and T. Itoh), by a grant from the Global Center of Excellence Program (Basic and Translational Research Center for Global Brain Science) of the MEXT of Japan (to M. Fukuda), and by a grant from the Uehara Memorial Foundation (to T. Itoh).
Footnotes
Abbreviations used in this paper:
- GAP
- GTPase-activating protein
- KO
- knockout
- LRS
- LC3 recognition sequence
- MEF
- mouse embryonic fibroblast
- mStr
- monomeric strawberry
- PE
- phosphatidylethanolamine
- Y2H
- yeast two-hybrid
References
- Behrends C., Sowa M.E., Gygi S.P., Harper J.W. 2010. Network organization of the human autophagy system. Nature. 466:68–76 10.1038/nature09204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards A. 2003. GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim. Biophys. Acta. 1603:47–82 [DOI] [PubMed] [Google Scholar]
- Blommaart E.F.C., Krause U., Schellens J.P.M., Vreeling-Sindelárová H., Meijer A.J. 1997. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur. J. Biochem. 243:240–246 10.1111/j.1432-1033.1997.0240a.x [DOI] [PubMed] [Google Scholar]
- Cadwell K., Liu J.Y., Brown S.L., Miyoshi H., Loh J., Lennerz J.K., Kishi C., Kc W., Carrero J.A., Hunt S., et al. 2008. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 456:259–263 10.1038/nature07416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fader C.M., Sánchez D., Furlán M., Colombo M.I. 2008. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 9:230–250 10.1111/j.1600-0854.2007.00677.x [DOI] [PubMed] [Google Scholar]
- Fujita N., Itoh T., Omori H., Fukuda M., Noda T., Yoshimori T. 2008a. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell. 19:2092–2100 10.1091/mbc.E07-12-1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N., Hayashi-Nishino M., Fukumoto H., Omori H., Yamamoto A., Noda T., Yoshimori T. 2008b. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol. Biol. Cell. 19:4651–4659 10.1091/mbc.E08-03-0312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M. 2011. TBC proteins: GAPs for mammalian small GTPase Rab? Biosci. Rep. 31:159–168 [DOI] [PubMed] [Google Scholar]
- Fukuda M., Itoh T. 2008. Direct link between Atg protein and small GTPase Rab: Atg16L functions as a potential Rab33 effector in mammals. Autophagy. 4:824–826 [DOI] [PubMed] [Google Scholar]
- Fukuda M., Kanno E. 2005. Analysis of the role of Rab27 effector Slp4-a/Granuphilin-a in dense-core vesicle exocytosis. Methods Enzymol. 403:445–457 10.1016/S0076-6879(05)03039-9 [DOI] [PubMed] [Google Scholar]
- Fukuda M., Mikoshiba K. 1999. A novel alternatively spliced variant of synaptotagmin VI lacking a transmembrane domain. Implications for distinct functions of the two isoforms. J. Biol. Chem. 274:31428–31434 10.1074/jbc.274.44.31428 [DOI] [PubMed] [Google Scholar]
- Fukuda M., Aruga J., Niinobe M., Aimoto S., Mikoshiba K. 1994. Inositol-1,3,4,5-tetrakisphosphate binding to C2B domain of IP4BP/synaptotagmin II. J. Biol. Chem. 269:29206–29211 [PubMed] [Google Scholar]
- Fukuda M., Kanno E., Mikoshiba K. 1999. Conserved N-terminal cysteine motif is essential for homo- and heterodimer formation of synaptotagmins III, V, VI, and X. J. Biol. Chem. 274:31421–31427 10.1074/jbc.274.44.31421 [DOI] [PubMed] [Google Scholar]
- Gutierrez M.G., Munafó D.B., Berón W., Colombo M.I. 2004. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J. Cell Sci. 117:2687–2697 10.1242/jcs.01114 [DOI] [PubMed] [Google Scholar]
- Hanada T., Noda N.N., Satomi Y., Ichimura Y., Fujioka Y., Takao T., Inagaki F., Ohsumi Y. 2007. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 282:37298–37302 10.1074/jbc.C700195200 [DOI] [PubMed] [Google Scholar]
- Hirota Y., Tanaka Y. 2009. A small GTPase, human Rab32, is required for the formation of autophagic vacuoles under basal conditions. Cell. Mol. Life Sci. 66:2913–2932 10.1007/s00018-009-0080-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Hara Y., Mizushima N. 2006. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 580:2623–2629 10.1016/j.febslet.2006.04.008 [DOI] [PubMed] [Google Scholar]
- Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., et al. 2000. A ubiquitin-like system mediates protein lipidation. Nature. 408:488–492 10.1038/35044114 [DOI] [PubMed] [Google Scholar]
- Ichimura Y., Kumanomidou T., Sou Y.-S., Mizushima T., Ezaki J., Ueno T., Kominami E., Yamane T., Tanaka K., Komatsu M. 2008. Structural basis for sorting mechanism of p62 in selective autophagy. J. Biol. Chem. 283:22847–22857 10.1074/jbc.M802182200 [DOI] [PubMed] [Google Scholar]
- Ishibashi K., Kanno E., Itoh T., Fukuda M. 2009. Identification and characterization of a novel Tre-2/Bub2/Cdc16 (TBC) protein that possesses Rab3A-GAP activity. Genes Cells. 14:41–52 10.1111/j.1365-2443.2008.01251.x [DOI] [PubMed] [Google Scholar]
- Itoh T., Fukuda M. 2006. Identification of EPI64 as a GTPase-activating protein specific for Rab27A. J. Biol. Chem. 281:31823–31831 10.1074/jbc.M603808200 [DOI] [PubMed] [Google Scholar]
- Itoh T., Satoh M., Kanno E., Fukuda M. 2006. Screening for target Rabs of TBC (Tre-2/Bub2/Cdc16) domain-containing proteins based on their Rab-binding activity. Genes Cells. 11:1023–1037 10.1111/j.1365-2443.2006.00997.x [DOI] [PubMed] [Google Scholar]
- Itoh T., Fujita N., Kanno E., Yamamoto A., Yoshimori T., Fukuda M. 2008. Golgi-resident small GTPase Rab33B interacts with Atg16L and modulates autophagosome formation. Mol. Biol. Cell. 19:2916–2925 10.1091/mbc.E07-12-1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger S., Bucci C., Tanida I., Ueno T., Kominami E., Saftig P., Eskelinen E.L. 2004. Role for Rab7 in maturation of late autophagic vacuoles. J. Cell Sci. 117:4837–4848 10.1242/jcs.01370 [DOI] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E.A. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 144:1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Storrie B. 2005. Cisternal rab proteins regulate Golgi apparatus redistribution in response to hypotonic stress. Mol. Biol. Cell. 16:2586–2596 10.1091/mbc.E04-10-0861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720–5728 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y., Yoshimori T. 2004. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 117:2805–2812 10.1242/jcs.01131 [DOI] [PubMed] [Google Scholar]
- Kimura S., Noda T., Yoshimori T. 2007. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 3:452–460 [DOI] [PubMed] [Google Scholar]
- Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. 1999. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147:435–446 10.1083/jcb.147.2.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V., Lamark T., Sou Y.-S., Bjørkøy G., Nunn J.L., Bruun J.A., Shvets E., McEwan D.G., Clausen T.H., Wild P., et al. 2009. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 33:505–516 10.1016/j.molcel.2009.01.020 [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., Cregg J.M., Dunn W.A., Jr, Emr S.D., Sakai Y., Sandoval I.V., Sibirny A., Subramani S., Thumm M., Veenhuis M., Ohsumi Y. 2003. A unified nomenclature for yeast autophagy-related genes. Dev. Cell. 5:539–545 10.1016/S1534-5807(03)00296-X [DOI] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Koike M., Sou Y.-S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., et al. 2007. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 131:1149–1163 10.1016/j.cell.2007.10.035 [DOI] [PubMed] [Google Scholar]
- Legesse-Miller A., Sagiv Y., Glozman R., Elazar Z. 2000. Aut7p, a soluble autophagic factor, participates in multiple membrane trafficking processes. J. Biol. Chem. 275:32966–32973 10.1074/jbc.M000917200 [DOI] [PubMed] [Google Scholar]
- Mizushima N. 2007. Autophagy: process and function. Genes Dev. 21:2861–2873 10.1101/gad.1599207 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T. 2001. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152:657–668 10.1083/jcb.152.4.657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Kuma A., Kobayashi Y., Yamamoto A., Matsubae M., Takao T., Natsume T., Ohsumi Y., Yoshimori T. 2003. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J. Cell Sci. 116:1679–1688 10.1242/jcs.00381 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. 2008. Autophagy fights disease through cellular self-digestion. Nature. 451:1069–1075 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrlüder J., Schwarten M., Willbold D. 2009. Structure and potential function of gamma-aminobutyrate type A receptor-associated protein. FEBS J. 276:4989–5005 10.1111/j.1742-4658.2009.07207.x [DOI] [PubMed] [Google Scholar]
- Morita S., Kojima T., Kitamura T. 2000. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7:1063–1066 10.1038/sj.gt.3301206 [DOI] [PubMed] [Google Scholar]
- Munafó D.B., Colombo M.I. 2002. Induction of autophagy causes dramatic changes in the subcellular distribution of GFP-Rab24. Traffic. 3:472–482 10.1034/j.1600-0854.2002.30704.x [DOI] [PubMed] [Google Scholar]
- Nakamura T., Hayashi T., Nasu-Nishimura Y., Sakaue F., Morishita Y., Okabe T., Ohwada S., Matsuura K., Akiyama T. 2008. PX-RICS mediates ER-to-Golgi transport of the N-cadherin/β-catenin complex. Genes Dev. 22:1244–1256 10.1101/gad.1632308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H., Ichimura Y., Ohsumi Y. 2007. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 130:165–178 10.1016/j.cell.2007.05.021 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. 2009. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10:458–467 10.1038/nrm2708 [DOI] [PubMed] [Google Scholar]
- Pan X., Eathiraj S., Munson M., Lambright D.G. 2006. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature. 442:303–306 10.1038/nature04847 [DOI] [PubMed] [Google Scholar]
- Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- Pfeffer S.R. 2001. Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol. 11:487–491 10.1016/S0962-8924(01)02147-X [DOI] [PubMed] [Google Scholar]
- Sagiv Y., Legesse-Miller A., Porat A., Elazar Z. 2000. GATE-16, a membrane transport modulator, interacts with NSF and the Golgi v-SNARE GOS-28. EMBO J. 19:1494–1504 10.1093/emboj/19.7.1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T., Nakayama M., Nakano H., Yagita H., Yamamoto N., Yamaoka S. 2003. TWEAK induces NF-kappaB2 p100 processing and long lasting NF-kappaB activation. J. Biol. Chem. 278:36005–36012 10.1074/jbc.M304266200 [DOI] [PubMed] [Google Scholar]
- Saitoh T., Fujita N., Jang M.H., Uematsu S., Yang B.G., Satoh T., Omori H., Noda T., Yamamoto N., Komatsu M., et al. 2008. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature. 456:264–268 10.1038/nature07383 [DOI] [PubMed] [Google Scholar]
- Schmid D., Pypaert M., Münz C. 2007. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 26:79–92 10.1016/j.immuni.2006.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sou Y.S., Waguri S., Iwata J., Ueno T., Fujimura T., Hara T., Sawada N., Yamada A., Mizushima N., Uchiyama Y., et al. 2008. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol. Biol. Cell. 19:4762–4775 10.1091/mbc.E08-03-0309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T., Sun Y., Wilkins N., Storrie B. 2010. Rab33b and Rab6 are functionally overlapping regulators of Golgi homeostasis and trafficking. Traffic. 11:626–636 10.1111/j.1600-0854.2010.01051.x [DOI] [PubMed] [Google Scholar]
- Stenmark H. 2009. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10:513–525 10.1038/nrm2728 [DOI] [PubMed] [Google Scholar]
- Strom M., Vollmer P., Tan T.J., Gallwitz D. 1993. A yeast GTPase-activating protein that interacts specifically with a member of the Ypt/Rab family. Nature. 361:736–739 10.1038/361736a0 [DOI] [PubMed] [Google Scholar]
- Valsdottir R., Hashimoto H., Ashman K., Koda T., Storrie B., Nilsson T. 2001. Identification of rabaptin-5, rabex-5, and GM130 as putative effectors of rab33b, a regulator of retrograde traffic between the Golgi apparatus and ER. FEBS Lett. 508:201–209 10.1016/S0014-5793(01)02993-3 [DOI] [PubMed] [Google Scholar]
- Waguri S., Komatsu M. 2009. Biochemical and morphological detection of inclusion bodies in autophagy-deficient mice. Methods Enzymol. 453:181–196 10.1016/S0076-6879(08)04009-3 [DOI] [PubMed] [Google Scholar]
- Wang H., Bedford F.K., Brandon N.J., Moss S.J., Olsen R.W. 1999. GABA(A)-receptor-associated protein links GABA(A) receptors and the cytoskeleton. Nature. 397:69–72 10.1038/16264 [DOI] [PubMed] [Google Scholar]
- Weidberg H., Shvets E., Shpilka T., Shimron F., Shinder V., Elazar Z. 2010. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 29:1792–1802 10.1038/emboj.2010.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimori T. 2004. Autophagy: a regulated bulk degradation process inside cells. Biochem. Biophys. Res. Commun. 313:453–458 10.1016/j.bbrc.2003.07.023 [DOI] [PubMed] [Google Scholar]
- Yoshimura K., Shibata M., Koike M., Gotoh K., Fukaya M., Watanabe M., Uchiyama Y. 2006. Effects of RNA interference of Atg4B on the limited proteolysis of LC3 in PC12 cells and expression of Atg4B in various rat tissues. Autophagy. 2:200–208 [DOI] [PubMed] [Google Scholar]
- Zerial M., McBride H. 2001. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2:107–117 10.1038/35052055 [DOI] [PubMed] [Google Scholar]
- Zheng J.Y., Koda T., Fujiwara T., Kishi M., Ikehara Y., Kakinuma M. 1998. A novel Rab GTPase, Rab33B, is ubiquitously expressed and localized to the medial Golgi cisternae. J. Cell Sci. 111:1061–1069 [DOI] [PubMed] [Google Scholar]