Autocatalytic processing of the Hedgehog ligand from its precursor protein relies on protein disulfide isomerase and ER-associated degradation.
Abstract
The Hedgehog (Hh) signaling pathway has important functions during metazoan development. The Hh ligand is generated from a precursor by self-cleavage, which requires a free cysteine in the C-terminal part of the protein and results in the production of the cholesterol-modified ligand and a C-terminal fragment. In this paper, we demonstrate that these reactions occur in the endoplasmic reticulum (ER). The catalytic cysteine needs to form a disulfide bridge with a conserved cysteine, which is subsequently reduced by protein disulfide isomerase. Generation of the C-terminal fragment is followed by its ER-associated degradation (ERAD), providing the first example of an endogenous luminal ERAD substrate that is constitutively degraded. This process requires the ubiquitin ligase Hrd1, its partner Sel1, the cytosolic adenosine triphosphatase p97, and degradation by the proteasome. Processing-defective mutants of Hh are degraded by the same ERAD components. Thus, processing of the Hh precursor competes with its rapid degradation, explaining the impaired Hh signaling of processing-defective mutants, such as those causing human holoprosencephaly.
Introduction
The Hedgehog (Hh) signaling pathway is initiated by the binding of the secreted Hh ligand to its cell surface receptor, Patched (Marigo et al., 1996; Stone et al., 1996). This binding event inactivates Patched, resulting ultimately in the activation of a specific transcriptional program, which is important in embryonic development, adult stem cell maintenance, and carcinogenesis (Lum and Beachy, 2004; Ogden et al., 2004; Kalderon, 2005). The secreted Hh ligand is generated through a unique process. Hh is synthesized as a precursor that is translocated into the ER. The precursor undergoes cholesterol-dependent self-cleavage, resulting in N- and C-terminal fragments (Fig. S1 A; Lee et al., 1994; Porter et al., 1995, 1996a,b). This process is driven by the intein-like activity of the C-terminal fragment in two steps (Hall et al., 1997). In the first step, a conserved catalytic cysteine in the C terminus attacks the polypeptide backbone and forms a thioester intermediate. In the second step, the 3β-hydroxyl group of a cholesterol molecule displaces the C-terminal fragment, generating an ester linkage with the carboxyl group of the N-terminal fragment. Hh processing and cholesterol modification are critical for normal Hh signaling, and mutations in human Sonic Hh (Shh [HShh]) that impair processing cause holoprosencephaly, one of the most common congenital malformations of the brain (Traiffort et al., 2004; Maity et al., 2005; Roessler et al., 2009). The cholesterol-modified N-terminal fragment, further modified by palmitylation at its N terminus (Chamoun et al., 2001), is ultimately released from cells and is responsible for all the signaling effects of the Hh pathway. It is currently unknown where in the secretory pathway the processing of the Hh precursor occurs. In addition, the fate of the C-terminal fragment generated during the processing of the precursor is unclear.
Here, we demonstrate that the self-cleavage of the Hh precursor occurs in the ER, requiring the reduction of a disulfide bond between the catalytic cysteine and another conserved cysteine in the C-terminal fragment by protein disulfide isomerase (PDI). After cleavage, the C-terminal fragment is degraded by the ER-associated degradation (ERAD) pathway (Hirsch et al., 2009; Xie and Ng, 2010), providing the first example of an endogenous luminal ERAD substrate that is constitutively degraded. Degradation requires key ERAD components previously implicated in the degradation of misfolded ER proteins, including the ubiquitin ligase Hrd1p (Bordallo et al., 1998; Bays et al., 2001a), its interaction partner Sel1 (Gardner et al., 2000; Mueller et al., 2006, 2008), and the p97 ATPase (Bays et al., 2001b; Ye et al., 2001; Jarosch et al., 2002; Rabinovich et al., 2002). Our results indicate that the generation of the N-terminal signaling domain of Hh in the ER is accompanied by the disposal of the C-terminal fragment by ERAD. We also show that processing-defective mutants of Hh, such as those causing human holoprosencephaly, are quickly degraded by the same ERAD pathway. Our results suggest that ERAD plays a critical role in birth defects caused by Hh precursor mutations.
Results
Purified Hh precursor processing requires a conserved noncatalytic cysteine
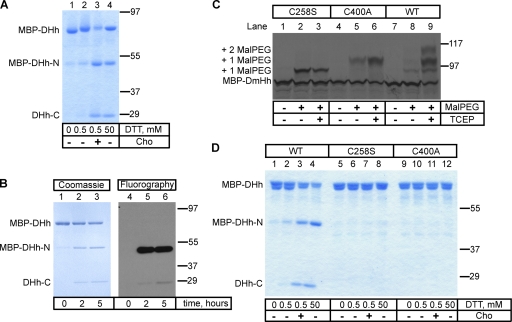
We first investigated the in vitro processing of the purified Drosophila melanogaster Hh (DHh) precursor (Lee et al., 1994; Porter et al., 1996a). A fusion protein was generated that contains maltose-binding protein (MBP), the last 15 amino acids of the N-terminal fragment, and the entire C-terminal fragment of DHh (MBP-DHh). The protein was expressed in Escherichia coli and purified as a soluble protein on an amylose affinity column. When incubated with high concentrations of DTT or with low concentrations of DTT and cholesterol, MBP-DHh underwent cleavage, generating an N-terminal fragment (MBP-DHh–N) and a C-terminal fragment (DHh-C; Fig. 1 A) as previously described (Porter et al., 1996b). The N-terminal fragment was modified with cholesterol as shown by the change in its electrophoretic mobility compared with the unmodified fragment (Fig. 1 A, lane 4 vs. lane 3) and by the incorporation of 3H-labeled cholesterol (Fig. 1 B).
Figure 1.
Processing of the purified Hh precursor. (A) A fusion was generated between maltose-binding protein (MBP), the last 15 amino acids of the N-terminal fragment of Drosophila Hh (DHh), and the entire C-terminal fragment of DHh (MBP-DHh). The purified protein was incubated for 5 h at room temperature with different concentrations of DTT in the absence or presence of cholesterol (Cho). The samples were analyzed by nonreducing SDS-PAGE and Coomassie staining. MBP-DHh–N and DHh-C are the N- and C-terminal fragments generated by Hh processing. (B) As in A, but the reaction contained 3H-labeled cholesterol. The samples were analyzed by reducing SDS-PAGE followed by either Coomassie staining or fluorography. (C) In vitro translated 35S-labeled wild-type (WT) MBP-DHh or the indicated cysteine mutants were incubated with 5-kD maleimide-polyethylene glycol (Mal-PEG) as indicated, in the presence or absence of the reducing agent tris(2-carboxyethyl)phosphine (TCEP). The samples were analyzed by reducing SDS-PAGE and autoradiography. The positions of singly and doubly Mal-PEG–modified species are indicated. The singly modified species have a different mobility, depending on which cysteine is modified. (D) As in A, but comparing wild-type MBP-DHh with the two cysteine mutants. Molecular masses are given in kilodaltons.
We noticed that the precursor migrated slower on nonreducing SDS-PAGE gels when treated with even low concentrations of DTT (Fig. 1 A, lane 1 vs. lane 2). Because MBP-DHh contains only two cysteines, this suggests that the catalytically active cysteine (C258) is disulfide bonded with C400. Both cysteines are absolutely conserved among all Hh proteins across phyla. In the crystal structure of the C-terminal fragment of DHh (Hall et al., 1997), C258 is in close proximity to C400, suggesting the possibility of such a disulfide bond. Given that C258 needs to be reduced to act as a nucleophile in Hh processing, this suggests that the disulfide-bonded species is an inactive precursor; the known requirement of a reducing agent for Hh processing in vitro (Porter et al., 1996b) could be explained by the need to reduce this conserved disulfide bond.
To test whether the two cysteines indeed form a disulfide bridge in MBP-DHh, we expressed the same protein by in vitro translation in reticulocyte lysate and treated it with maleimide-polyethylene glycol (Mal-PEG), a reagent that adds ∼5 kD for each modified free cysteine. Whereas nonreduced wild-type MBP-DHh showed little modification (Fig. 1 C, lane 8), prior disulfide bond reduction with tris(2-carboxyethyl)phosphine resulted in the appearance of significant levels of singly and doubly Mal-PEG–modified species (Fig. 1 C, lane 9). When either of the two cysteines was mutated, only a singly modified protein was detected, even without reduction (Fig. 1 C, lanes 2 and 3 and lanes 5 and 6). These data indicate the formation of a disulfide bond between C258 and C400. As expected, mutation of the catalytic C258 completely blocked cleavage of MBP-DHh (Fig. 1 D, lanes 5–8). Interestingly, mutation of the noncatalytic C400 also abolished cleavage, both in the presence of high concentrations of DTT and low concentrations of DTT and cholesterol (Fig. 1 C, lanes 9–12). Thus, despite the presence of a reduced C258, the mutant is inactive, perhaps because a disulfide bond between C258 and C400 is required for the folding of the C terminus into a catalytically active conformation. In all Hh proteins, the noncatalytic cysteine is part of a conserved SCY sequence, and the mutation of the other two residues also abolishes cleavage of DHh (unpublished data). Collectively, these experiments suggest that a disulfide bridge needs to form between the conserved cysteines, which subsequently would have to be reduced for generating the thiol group required for intein catalysis.
Hh processing in extracts and intact cells also requires the conserved noncatalytic cysteine
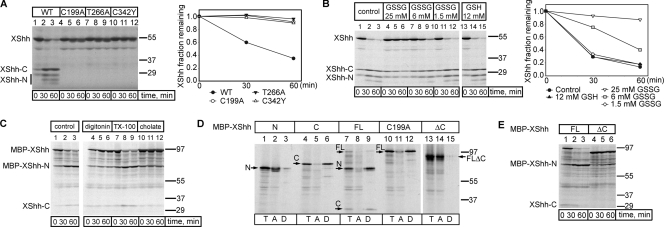
To test whether the conserved noncatalytic cysteine is essential for the processing of a full-length vertebrate Hh precursor, we established a novel cell-free assay based on Xenopus laevis egg extracts. Radioactively labeled Xenopus Shh (XShh; Lai et al., 1995) precursor, generated by in vitro translation in reticulocyte lysate, was efficiently processed when incubated with Xenopus egg extracts (Fig. 2 A, lanes 1–3). As in the purified system, both the catalytic cysteine (C199) and the noncatalytic cysteine (C342) were required for cleavage (Fig. 2 A, lanes 4–6 and 10–12). Mutation of another conserved residue shown to be important for cleavage in DHh (T266) also abolished processing (Fig. 2 A, lanes 7–9). Identical results were obtained with the corresponding DHh constructs, generated by in vitro translation, and incubated with Xenopus egg extracts (unpublished data). Consistent with our assumption that reduction of the disulfide bridge in the Hh precursor is required for processing, addition of increasing concentrations of oxidized glutathione (GSSG) inhibited XShh precursor cleavage (Fig. 2 B, lanes 1–12).
Figure 2.
Hh processing in Xenopus egg extracts. (A) The in vitro translated 35S-labeled Xenopus Sonic Hh (XShh) wild-type (WT) precursor was incubated at room temperature with Xenopus egg extracts for the indicated times. Parallel experiments were performed with three point mutants. The samples were analyzed by SDS-PAGE and autoradiography. The graph shows the quantification of the Hh precursor. (B) As in A, but the extract was supplemented with the indicated concentrations of oxidized or reduced glutathione (GSSG or GSH). (A and B) n = 3 time points. (C) In vitro translated 35S-labeled XShh fused at its N terminus to the maltose-binding protein (MBP-XShh) was incubated at room temperature with Xenopus egg extracts for the indicated times, in the absence or presence of 0.5% of either the cholesterol-sequestering detergents digitonin or cholate, or the control detergent Triton X-100 (TX-100). The samples were analyzed by SDS-PAGE and autoradiography. (D) In vitro translation was used to generate 35S-labeled fusions of MBP and either the N-terminal fragment of XShh (N), the C-terminal fragment of XShh (C), full-length XShh (FL), full-length XShh with a cysteine mutation in the active site (C199A), or XShh lacking the last 93 amino acids (ΔC). The fusions were incubated for 1 h with Xenopus egg extracts and subjected to Triton X-114 partitioning. Aliquots of the input (T), of the aqueous phase (A), or of the detergent phase (D) were analyzed by SDS-PAGE and autoradiography. (E) As in A, but with an MBP fusion of either wild-type XShh (FL) or a mutant lacking the last 93 amino acids (ΔC). Molecular masses are given in kilodaltons.
We next tested whether the N-terminal fragment generated in Xenopus extracts is cholesterol modified. Indeed, the cleavage reaction was blocked by cholesterol-sequestering detergents (Fig. 2 C, lanes 4–6 and 10–12 vs. lanes 1–3 and 7–9). Furthermore, the N-terminal fragment generated in extracts partitioned into the Triton X-114 phase (Fig. 2 D, lanes 7–9) in contrast to the N-terminal fragment generated directly by in vitro translation (Fig. 2 D, lanes 1–3). The C-terminal fragment is also hydrophobic, as it partitioned into the detergent phase, whether alone or contained in the full-length protein (Fig. 2 D, lanes 4–6 and 7–12). Deletion of the last 93 amino acids from full-length XShh (XShhΔC) rendered the protein hydrophilic (Fig. 2 D, lanes 13–15). The deleted region indeed contains several hydrophobic amino acids and might interact with cholesterol during the intein reaction (Hall et al., 1997). This deletion greatly delayed, but did not completely abolish, processing of the precursor in Xenopus extracts (Fig. 2 E). Finally, when the purified MBP-DHh precursor was added to Xenopus extracts, mass spectrometry identified cholesterol attached to the N-terminal fragment (unpublished data). These data demonstrate that cholesterol is properly attached to Hh proteins in Xenopus egg extracts.
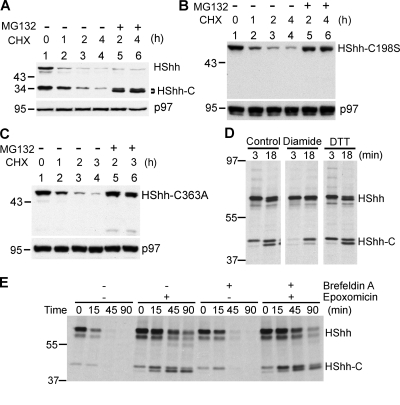
To test whether the conserved noncatalytic cysteine is also essential for the processing of the Hh precursor in intact cells, we stably expressed wild type or cysteine mutants of HShh C-terminally tagged with an HA epitope (HShh-HA) in 293T cells. After inhibiting protein synthesis with cycloheximide, the wild-type Hh precursor and the processed C-terminal fragment (HShh-C) were observed at early time points (Fig. 3 A). As in the in vitro system, no processing was observed when either the catalytic cysteine (C198) or the noncatalytic cysteine (C363) was mutated (Fig. 3, B and C, lanes 1). For both cysteine mutants, the block in processing correlated with a complete absence of active Hh ligand from 293T cell supernatants as assayed using Hh-responsive NIH-3T3 cells; in contrast, wild-type HShh-HA expressed in 293T cells resulted in a robust secretion of active Hh ligand (unpublished data). Consistent with the postulated role of the two conserved cysteines, a reagent that makes the ER more oxidizing (diamide) significantly slows down Hh precursor processing in mammalian cells, whereas DTT had a small accelerating effect as shown by pulse-chase experiments (Fig. 3 D and not depicted). These data support the idea that Hh processing requires the formation and subsequent reduction of a disulfide bridge between the conserved cysteines.
Figure 3.
Processing of HShh is dependent on disulfide bridge formation and reduction. (A) HShh-HA was stably expressed in 293T cells. Protein synthesis was inhibited with cycloheximide (CHX), and the fate of the protein was followed by SDS-PAGE and immunoblotting with anti-HA antibodies. Immunoblotting for p97 was used as a loading control. (B) As in A, but with HShh-HA containing a mutation in the catalytic cysteine (C198S). (C) As in A, but with HShh-HA containing a mutation in the conserved noncatalytic cysteine (C363A). (D) HA-tagged HShh was stably expressed in 293T cells. The cells were pulsed with [35S]methionine for 3 min and chase incubated with unlabeled methionine for the indicated times. 200 µM diamide or 0.5 mM DTT was added 10 min before the pulse and was present during the pulse and chase. The proteasome inhibitor epoxomicin (1 µM) was present, beginning at 1 h before the pulse. The samples were analyzed by immunoprecipitation with HA antibodies followed by reducing SDS-PAGE and fluorography. An equal number of cells were processed for each condition. (E) As in D, except that, where indicated, 10 µM brefeldin A and 1 µM epoxomicin were present, which were added 1 h before the pulse. The samples were analyzed as in D. Molecular masses are given in kilodaltons.
We noted that, after the addition of cycloheximide, both the Hh precursor and HShh-C disappeared in a time-dependent manner (Fig. 3, A–C). When the cycloheximide chase of wild-type HShh-HA was performed in the presence of the proteasome inhibitor MG132, HShh-C accumulated, indicating that the precursor was efficiently processed and that HShh-C was normally degraded by the cytosolic proteasome (Fig. 3 A, lanes 5 and 6). Similar experiments with the processing-defective cysteine mutants showed that the Hh precursor is also degraded by the proteasome (Fig. 3, B and C, lanes 5 and 6). When HShh-C was expressed by itself, it was also degraded but at a slower rate (Fig. S1, B and C), indicating that its degradation is most efficient when generated during normal processing.
Hh processing does not require vesicular transport out of the ER
We reasoned that the remodeling of the conserved disulfide bridge in Hh occurs in the ER, the site of all known disulfide bond formation and reduction in the secretory pathway. To test this assumption, we performed pulse-chase experiments after blocking vesicular transport from the ER to the Golgi by brefeldin A (Fig. 3 E). No effect on Hh processing was observed at concentrations that caused the complete disappearance of the Golgi (Fig. S2 A) and regardless of whether or not a proteasome inhibitor was present (Fig. 3 E). These results strongly argue that both the processing of the Hh precursor and the degradation of HShh-C occur in the ER.
The Hh protein is a substrate for PDI
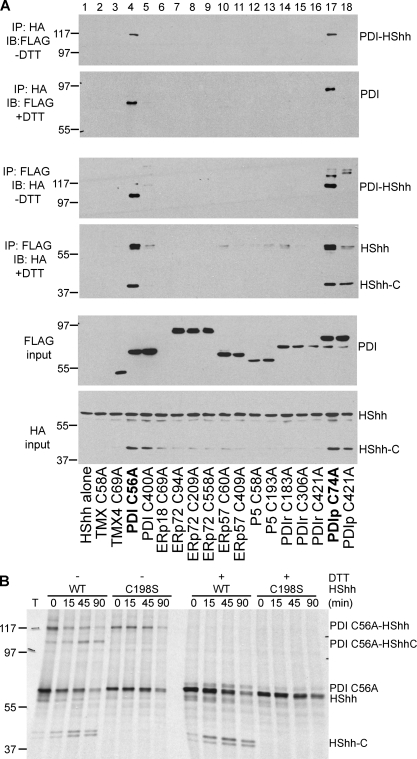
It seemed likely that remodeling of the conserved disulfide bridge in Hh is catalyzed by a member of the ER-localized thioredoxin (Trx)-like oxidoreductases, a class of enzymes generally responsible for such reactions. These enzymes contain at least one CXXC motif, the first cysteine of which forms a transient mixed disulfide bridge with the substrate; this mixed disulfide intermediate can be trapped by mutating the second cysteine in the CXXC motif (CXXA mutants). We therefore screened CXXA mutants of ER-localized human Trx-like proteins for the formation of a mixed disulfide bridge with Hh. We coexpressed in 293T cells FLAG-tagged CXXA mutants of nine different Trx-like ER proteins together with HShh-HA. Because some of the proteins contain more than one CXXC motif, we tested a total of 17 different constructs (Schulman et al., 2010). In each case, the formation of a mixed disulfide bridge was assayed by immunoprecipitation with HA or FLAG antibodies followed by nonreducing SDS-PAGE and immunoblotting. The strongest interactions were observed for PDI and the closely related PDIp protein (Fig. 4 A, first and third panels). As expected, both mixed disulfide adducts were sensitive to DTT treatment (Fig. 4 A, second and fourth panels), and no adducts were seen when one of the two components was omitted (Fig. S2 B). PDI and PDIp contain two CXXC motifs, but the reaction with Hh occurred overwhelmingly with the N-terminal motif. This data implies that the first CXXC motif of PDI is dedicated to substrate interaction, whereas the second CXXC motif interacts with the oxidase Ero1p (Tsai and Rapoport, 2002).
Figure 4.
PDI and PDIp are involved in the remodeling of the conserved disulfide bridge in HShh. (A) HShh-HA and FLAG-tagged Trx-like ER proteins, in which one of their CXXC motifs was changed to CXXA, were coexpressed in 293T cells. Cell extracts were subjected to immunoprecipitation (IP) with HA or FLAG antibodies followed by SDS-PAGE and immunoblotting (IB) with FLAG and HA antibodies. Where indicated, the immunoprecipitated samples were reduced with DTT before electrophoresis. (B) Wild-type (WT) HShh-HA or the processing-defective C198A mutant was coexpressed with a FLAG-tagged CXXA mutant of PDI (C56A) in 293T cells. The cells were pulse labeled with [35S]methionine for 3 min and chase incubated for different time periods. The proteasome inhibitor epoxomicin (1 µM) was added 1 h before the pulse. All samples were subjected to immunoprecipitation with HA antibodies followed by SDS-PAGE and fluorography. Where indicated, the samples were reduced with DTT before electrophoresis. Molecular masses are given in kilodaltons.
PDI and PDIp reacted with both the Hh precursor and HShh-C (Fig. 4 A, fourth panel) as expected from the fact that both contain the two conserved cysteines. Pulse-chase experiments in the presence of a proteasome inhibitor demonstrated that the formation of the mixed disulfide between the Hh precursor and the CXXA mutant of PDI occurs rapidly and precedes Hh processing, including the appearance of the mixed disulfide adduct between HShh-C and the PDI mutant (Fig. 4 B). Interestingly, the mixed disulfide species can undergo the intein reaction with about the same kinetics as the Hh precursor itself. This also suggests that the noncatalytic cysteine of Hh is linked to PDI. Furthermore, when Hh processing was blocked by mutation of the catalytic cysteine, the Hh precursor formed a mixed disulfide bond with the PDI mutant.
These data are consistent with a model in which PDI function is linked and required for Hh processing. Finally, it should be noted that overexpression of the PDI and PDIp mutants caused the accumulation of HShh-C (Fig. 4 A, bottom), likely because a mixed disulfide adduct with PDI is not susceptible to degradation.
HShh-C is degraded in the ER
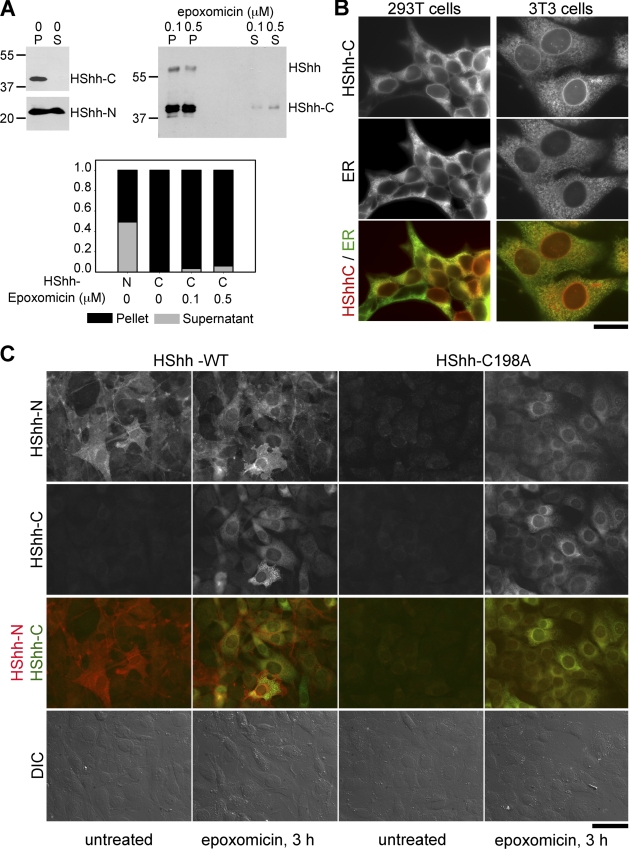
To study the fate of HShh-C, we first considered the possibility that it might be secreted together with the N-terminal fragment (HShh-N). HShh-HA was stably expressed in 293T cells, and HShh-N and HShh-C were analyzed by immunoblotting with Hh and HA antibodies, respectively, both in cells and in equivalent amounts of culture medium. Whereas HShh-N partitioned equally between cells and medium, HShh-C was present only in cells (Fig. 5 A, left). Even when HShh-C accumulated in cells after treatment with a proteasome inhibitor, only very small amounts of HShh-C were found in the medium (Fig. 5 A, right). These results show that HShh-C is not secreted, in contrast to HShh-N, the Hh ligand. Rather, the instability of HShh-C and its stabilization by proteasome inhibitors, even under conditions where vesicular transport out of the ER is blocked, suggest that HShh-C is degraded in the ER. Previous experiments on the secretion of HShh-C can be explained by its massive overexpression and by the lack of quantification (Bumcrot et al., 1995).
Figure 5.
HShh-C is not secreted and is degraded in the ER. (A) HShh-HA was stably expressed in 293T cells. The cells were washed and incubated for 12 h with DME containing 0.5% fetal bovine serum. The cell pellets and equivalent amounts of culture medium were analyzed for the presence of HShh-N and HShh-C by immunoblotting with HShh-N antibodies and HA antibodies. Where indicated, the proteasome inhibitor epoxomicin was present during the last 3 h of incubation. The graph shows the distribution of HShh-N and HShh-C between cells and medium. P, pellet. S, supernatant. Molecular masses are given in kilodaltons. (B) HShh was tagged with mCherry at its C terminus and stably expressed in 293T or in NIH-3T3 cells. Its localization was determined by fluorescence microscopy. The ER was revealed by immunostaining with rabbit antibodies against calnexin. The bottom row shows merged images. Bar, 20 µm. (C) Wild-type HShh-HA or the processing-defective mutant HShh-C198A-HA was stably expressed in NIH-3T3 cells. Cells were immunostained with rat HA and rabbit HShh-N antibodies followed by goat anti–rat Alexa Fluor 488 (green) and goat anti–rabbit Alexa Fluor 594 (red) secondary antibodies. The cells were incubated for 3 h with or without the proteasome inhibitor epoxomicin (1 µM). The third row shows merged images of the green and red channels. The bottom row shows differential interference contrast (DIC) images. Bar, 50 µm.
To confirm that HShh-C is degraded in the ER, we tagged HShh with mCherry at its C terminus and visualized the protein by fluorescence microscopy. The protein showed the typical ER staining, colocalizing with calnexin (Fig. 5 B), in both 293T and in NIH-3T3 cells stably expressing HShh-mCherry. When NIH-3T3 cells stably expressing HShh-HA were treated with the proteasome inhibitor epoxomicin, the intensity of the staining increased significantly, which is consistent with HShh-C being degraded in the ER (Fig. 5 C, second row; and Fig. S2 C). Identical results were obtained with the proteasome inhibitors MG132 and bortezomib (not depicted) and in 293T cells (Fig. S2 C). The immunofluorescent staining observed under these conditions corresponds mostly to HShh-C (Fig. 5 A, immunoblots). In contrast to HShh-C, HShh-N was not degraded, as demonstrated by staining with antibodies directed against the N terminus of HShh (Fig. 5 C, first row). The subcellular localization of HShh-N was also different from that of HShh-C, with much of HShh-N localizing to the plasma membrane (Fig. 5 C, overlay in third row). The processing-defective HShh precursor mutant (HShh-C198A) was as unstable as HShh-C when analyzed by antibodies against either the N or C terminus, and it localized to the ER when stabilized by proteasome inhibitors (Fig. 5 C, third and fourth panels). These data demonstrate that failure of processing results in the rapid degradation of the full-length HShh precursor in the ER.
Hh is degraded by ERAD
Next, we examined whether HShh-C generated in the ER lumen is degraded by the ERAD pathway. Luminal glycosylated ERAD substrates are generally processed by glycosidases in their carbohydrate moiety, which is subsequently recognized by lectins. The substrates are then translocated into the cytosol, polyubiquitinated by the ubiquitin ligase Hrd1, moved into the cytosol by the p97 ATPase, and finally degraded by the proteasome. Proteolysis is often preceded by deglycosylation (Wiertz et al., 1996). We tested which aspects of the ERAD pathway apply to the degradation of HShh-C.
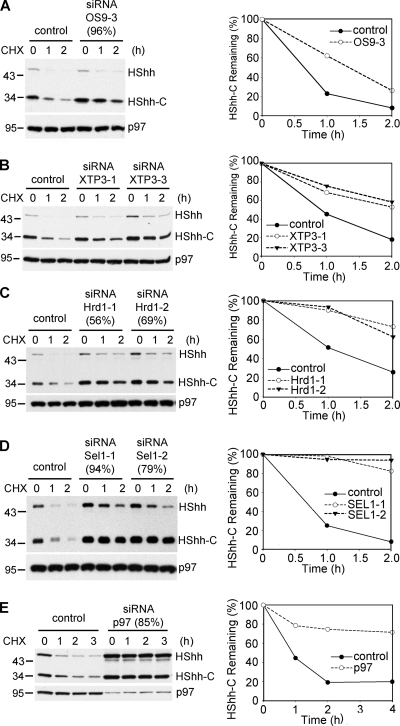
We used RNAi to identify ERAD components required for the degradation of HShh-C. Depletion of the lectins implicated in recognizing glycosylated ERAD substrates, OS9 or XTP3 (Christianson et al., 2008; Hosokawa et al., 2008, 2009; Bernasconi et al., 2010), caused a significant inhibition of degradation (Fig. 6, A and B, quantification on the right). No further inhibition was seen when both lectins were depleted at the same time (Fig. S3). Depletion of the ubiquitin ligase Hrd1 (Bays et al., 2001a) and of its interacting partner Sel1 strongly stabilized HShh-C (Fig. 6, C and D). Finally, depletion of the ATPase p97 (Ye et al., 2001) also had a drastic inhibitory effect (Fig. 6 E); both the HShh precursor and HShh-C accumulated, indicating that they both undergo ERAD. The depletion by RNAi of other ERAD components, such as the Ring finger ubiquitin ligases gp78 (Fang et al., 2001), TRC8 (Stagg et al., 2009), and TEB4 (Hassink et al., 2005), as well as Derlin-1 (Lilley and Ploegh, 2004; Ye et al., 2004), Herp (Kokame et al., 2000; Schulze et al., 2005; Carvalho et al., 2006), BiP (Denic et al., 2006), and ERdj5 (Ushioda et al., 2008), had no effect on HShh-C degradation (Fig. S3). Addition of kifunensine or 1-deoxymannojirimycin (Elbein, 1991) had no significant effect on the degradation of HShh-C (Fig. S3), indicating that mannosidase I is not required for processing of the glycan on HShh-C.
Figure 6.
ERAD components required for the degradation of HShh-C. (A) Cells were depleted of the ER luminal lectin OS9 by siRNA, and the fate of stably expressed HShh-HA was followed after cycloheximide (CHX) addition. The extent of OS9 depletion (in parentheses) was determined by quantitative RT-PCR. Controls were treated with an unrelated siRNA. All samples were analyzed by SDS-PAGE and immunoblotting with HA antibodies. The right graph shows quantification of HShh-C in the experiment. All samples were also analyzed by immunoblotting for p97 (loading control). (B) As in A, but with depletion of the ER luminal lectin XTP3 by two different siRNAs. (C) As in A, but with depletion of the ubiquitin ligase Hrd1 by two different siRNAs. (D) As in A, but with depletion of the Hrd1-interacting protein Sel1 by two different siRNAs. (A–D) n = 3 time points. (E) As in A, but with depletion of the ATPase p97 by siRNA. n = 4 time points. Molecular masses are given in kilodaltons.
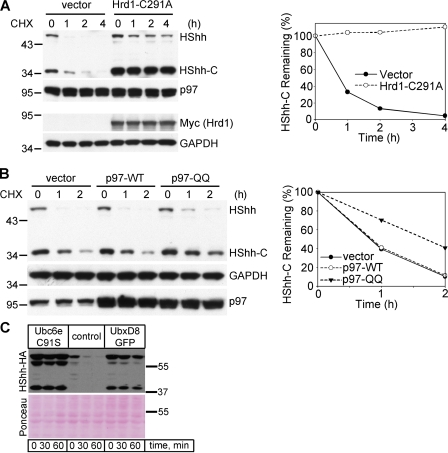
The role of various ERAD components in the degradation of HShh was also tested by the expression of dominant-negative constructs. A catalytically inactive mutant of the ubiquitin ligase Hrd1, in which a cysteine in the Ring finger domain is altered, strongly inhibited HShh-C degradation (Fig. 7 A). Although overexpression of the wild-type p97 ATPase did not delay degradation of HShh-C, the catalytically inactive p97-QQ mutant was strongly inhibitory (Fig. 7 B). ERAD was similarly inhibited by the overexpression of catalytically inactive Ubc6e (Ubc6e-C91S) or by the overexpression of a GFP fusion of the SEL1L-interacting protein UbxD8 (UbxD8-GFP; Fig. 7 C; Lilley and Ploegh, 2004; Mueller et al., 2008).
Figure 7.
Dominant-negative ERAD components inhibit HShh-C degradation. (A) Cells stably expressing the HShh-HA precursor were transfected with a catalytically inactive Myc-tagged Hrd1 (Hrd1-C291A) or with an empty vector. The fate of HShh-HA was followed after addition of cycloheximide (CHX), by SDS-PAGE and immunoblotting for HA; immunoblotting for p97 served as a loading control. All samples were also analyzed by SDS-PAGE followed by immunoblotting for Myc (Hrd1-C291A) and for glyceraldehyde 3-phosphate dehydrogenase (GAPDH; loading control). The right graph shows the quantification of the HShh-C in the experiment. n = 4 time points. (B) As in A, but with transfection of either wild-type p97 (p97-WT), a catalytically inactive p97 mutant (p97-QQ), or with an empty vector. The HA blot was stripped and reprobed with anti-GAPDH antibodies (loading control). Endogenous and overexpressed p97 were detected on the same gel by immunoblotting with anti-p97 antibodies. n = 3 time points. (C) HShh-HA was transiently expressed in 293T cells together with dominant-negative Ubc6e (Ubc6e-C91S), control vector, or UbxD8-GFP. Greater than 90% of the cells showed a strong GFP signal by live-cell fluorescence microscopy (not depicted). Protein synthesis was inhibited with cycloheximide, and the fate of HShh-HA was followed by SDS-PAGE and immunoblotting with HA antibodies. Ponceau S staining of the blot is shown to demonstrate the loading of equal amounts of protein. Molecular masses are given in kilodaltons.
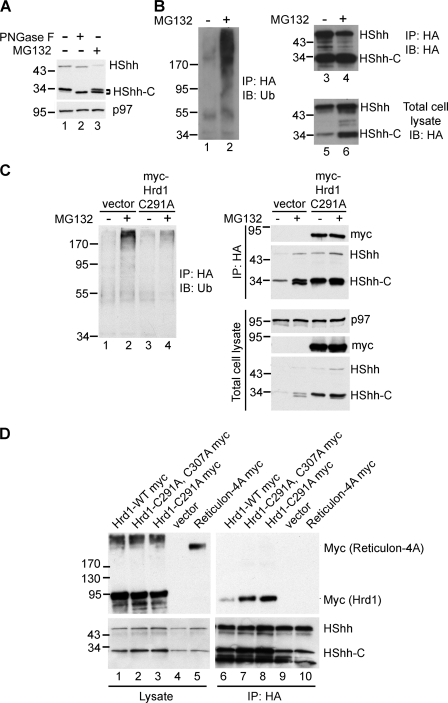
As with other ERAD substrates that are deglycosylated when arriving in the cytosol, we found that the major species of HShh-C accumulating in the presence of MG132 migrated slightly faster in SDS gels than the glycosylated fragment (Figs. 3, A and E; and 8 A). This band is indeed deglycosylated, as treatment of the glycosylated fragment with protein N-glycanase F generated a species of the same size (Fig. 8 A). As expected, depletion of ERAD components that block dislocation from the ER led to the accumulation of glycosylated HShh-C (Fig. 6).
Figure 8.
Cytoplasmic events preceding HShh-C proteolysis. (A) To test for deglycosylation of HShh-C, HShh-HA was stably expressed in 293T cells. A cycloheximide chase was performed for 2 h in the presence or absence of the proteasome inhibitor MG132. Cell lysates were incubated in the absence or presence of protein N-glycanase F (PNGase F) as indicated. Samples were analyzed by SDS-PAGE and immunoblotting with HA antibodies. Immunoblotting with p97 antibodies served as a loading control. (B) To test for polyubiquitination of HShh, cells stably expressing HShh-HA were incubated in the absence or presence of MG132 for 2 h. Extracts were subjected to immunoprecipitation (IP) with HA antibodies, and the proteins were analyzed by SDS-PAGE and immunoblotting (IB) with HA-antibodies (lanes 3 and 4) or ubiquitin (Ub) antibodies (lanes 1 and 2). Lanes 5 and 6 show blots of the extract before immunoprecipitation. (C) To test whether Hrd1 polyubiquitinates HShh, an experiment as in B was performed except that, where indicated, cells were transfected with a Myc-tagged dominant-negative Hrd1 mutant (Myc-Hrd1-291A). To test for the presence of the Hrd1 mutant, the samples were also analyzed by blotting with Myc antibodies. Immunoblotting for p97 served as a loading control. (D) Cells stably expressing HShh-HA were transfected with Myc-tagged wild-type Hrd1 (Hrd1-WT) or catalytically inactive Hrd1 mutants (Hrd1 C291A or Hrd1 C291A-C307A). As a control, a Myc-tagged version of reticulon 4A was used. Cell extracts were either analyzed directly (lanes 1–4) or subjected to immunoprecipitation with HA antibodies (lanes 5–8). All samples were analyzed by SDS-PAGE and immunoblotting with HA or Myc antibodies. Molecular masses are given in kilodaltons.
To test whether HShh-C was polyubiquitinated, we subjected cell extracts expressing HShh-HA to immunoprecipitation with HA antibodies followed by SDS-PAGE and immunoblotting with ubiquitin antibodies (Fig. 8 B). Polyubiquitinated HShh-HA was detected in cells treated with the proteasome inhibitor MG132 (Fig. 8 B, lane 2) but not in untreated cells even though equal amounts of HA-tagged protein were precipitated (Fig. 8 B, lanes 3 and 4). The specificity of the immunoprecipitation was demonstrated by a control IgG pull-down (Fig. S4). Polyubiquitination of HShh was dependent on Hrd1, as it was inhibited by the expression of a dominant-negative Hrd1 mutant (Fig. 8 C).
To further confirm the role of Hrd1, we tested whether it interacts with its HShh substrate. Myc-tagged wild-type or dominant-negative Hrd1 were introduced into cells stably expressing HShh-HA. Cell extracts were subjected to immunoprecipitation with HA antibodies, and precipitated proteins were analyzed by SDS-PAGE and immunoblotting with Myc antibodies. These experiments showed that HShh-HA precipitated all versions of Hrd1-Myc (Fig. 8 D, lanes 6–8); less precipitation was seen with wild-type Hrd1-Myc (Fig. 8 D, lane 6). An unrelated ER protein, Myc-tagged reticulon, did not interact with HShh-HA (Fig. 8 D, lane 5). Together, these experiments indicate that the ubiquitin ligase Hrd1 interacts with HShh substrates undergoing ERAD.
Finally, because the full-length Hh precursor is also degraded by ERAD (Figs. 3, B and C; and 6 E), we tested whether it requires the same components as HShh-C. Indeed, ERAD of the processing-defective HShh C198S mutant stably expressed in 293T cells was inhibited by depleting the same components, i.e., OS9, Hrd1, Sel1, and p97 (Fig. S5). These results show that the Hh precursor undergoes two competing reactions in the ER, cholesterol-dependent processing and degradation by an ERAD pathway identical to that used by HShh-C.
Discussion
The main result of the present study is that processing of the Hh precursor takes place in the ER. This conclusion is based on several observations: (a) the insensitivity of Hh processing to inhibition of vesicular transport out of the ER, (b) the requirement for disulfide bridge formation and reduction for Hh processing, (c) the involvement of the ER luminal PDI in the disulfide remodeling of Hh, (d) the degradation of the Hh precursor and its C-terminal fragment by ERAD, and (e) microscopic visualization of Hh degradation in the ER. Processing of the Hh precursor in the ER is somewhat unexpected given the relatively low concentration of cholesterol in the ER membrane. In fact, it was previously speculated that Hh processing might occur in another, cholesterol-rich compartment of the secretory pathway (Maity et al., 2005). How cholesterol modification can occur at the low cholesterol concentrations of the ER remains unclear, but one possibility is that an additional factor in the ER facilitates the reaction.
We show that a disulfide bond needs to form between the catalytic cysteine and the only other cysteine in the C-terminal domain, a disulfide bond that is conserved among all Hh proteins. Again, this is a surprising result because, in the next step of Hh processing, the catalytic cysteine needs to be reduced to initiate cleavage of the polypeptide backbone. Why then form a disulfide bridge in the first place? We speculate that the Hh precursor requires a disulfide bridge for proper folding, which in turn is required for the autocatalytic activity of the C-terminal domain. Both the formation and reduction of the essential disulfide bridge require the activity of a member of the PDI family. Although the oxidation of cysteines to form disulfide bridges is a common protein modification in the ER, net disulfide bond reduction is less common. One example is provided by the reduction of a disulfide bond that links the A1 and A2 chains of cholera toxin by PDI itself; the A1 chain thus freed from the rest of the toxin is retrotranslocated into the cytosol (Tsai et al., 2001).
Our results show that PDI and the closely related PDIp protein are involved in the remodeling of the conserved disulfide bridge in Hh. The specificity of these enzymes is indicated by the fact that seven other tested ER-localized family members do not interact and that only one of the two catalytic CXXC motifs of PDI and PDIp forms a mixed disulfide intermediate with Hh. Whether these enzymes are involved in the formation of the disulfide bridge, its reduction, or both is unclear. However, our observation that a mixed disulfide adduct between Hh and PDI forms rapidly and undergoes the intein reaction suggests that PDI reduces the disulfide bridge in Hh and remains bound to the noncatalytic cysteine while the Hh precursor is processed. Thus, disulfide bridge reduction by PDI might be mechanistically coupled to Hh processing and attachment of cholesterol.
Consistent with our conclusion that Hh processing takes place in the ER, we find that the resulting C-terminal fragment is degraded by ERAD. The degradation of the C-terminal fragment generated in the ER lumen requires several of the previously identified ERAD components. Our data suggest the following series of events in the degradation of the C-terminal fragment. The process probably begins with recognition of the single carbohydrate chain in the C-terminal fragment, as suggested by the requirement for either of the two lectins implicated in ERAD, OS9 and XTP-3B. The C-terminal fragment is also likely recognized by Sel1, similarly to substrate recognition by the yeast homologue Hrd3p (Denic et al., 2006; Gauss et al., 2006). The next step in the process involves the ubiquitin ligase Hrd1, which forms a complex with Sel1. Once the substrate is polyubiquitinated on the cytosolic side of the ER membrane, the p97 ATPase complex moves the C-terminal fragment from the membrane into the cytosol. The protein is also deglycosylated, likely by the cytoplasmic glycanase (Hirsch et al., 2003). Finally, the protein is degraded by the proteasome. As a result of ERAD, the C-terminal fragment never leaves the ER and is not secreted, in contrast to the N-terminal fragment, the Hh ligand. Given that the C-terminal fragment is not secreted even when stabilized by proteasome inhibitors, it appears that it is actively retained in the ER, possibly by its association with PDI.
ERAD is normally used to degrade misfolded ER proteins. In addition, there are several native proteins, such as HMG CoA reductase (Chin et al., 1985; Gil et al., 1985; Hampton and Rine, 1994), that are degraded in a regulated manner by ERAD. However, to our knowledge, there is only one native protein, Ubc6p, that is a constitutive ERAD substrate (Walter et al., 2001). This yeast membrane protein is exposed to the cytosol and is continuously ubiquitinated by the Doa10p ubiquitin ligase (Swanson et al., 2001). The C-terminal fragment of Hh is the first example of a native luminal ER protein that is constitutively degraded by ERAD. Whereas it is conceivable that HMG CoA reductase is induced to unfold by binding of cholesterol and that Ubc6p is not properly folded, the C-terminal fragment of Hh must be properly folded to allow the self-cleavage reaction to happen. This hypothesis is supported by the fact that noncleavable versions of the full-length Hh precursor are degraded by ERAD at least as fast as the C-terminal fragment and by the same ERAD components. Apparently, the rapid degradation of the full-length Hh precursor by ERAD is competing with its proper processing. This can explain why HShh mutants that are defective in precursor processing are causing holoprosencephaly, a frequent congenital brain malformation; these mutant proteins would be quickly degraded in the ER, and no active ligand would be secreted. Thus, our results suggest that ERAD plays a critical role in birth defects caused by Hh precursor mutations. Even for the wild-type Hh precursor, ERAD might play a role in determining how much active ligand is generated; perhaps, the concentration of cholesterol in the ER determines the balance between degradation and processing of the full-length precursor. Interestingly, the C-terminal fragment expressed by itself is also degraded but more slowly than the C-terminal fragment generated through processing. Perhaps, when the fragment is generated close to the ER membrane, there is a more efficient handover to the ERAD machinery. The signal that targets Hh to the ERAD pathway remains unclear, but it could be related to the hydrophobic properties of the C terminus.
Why is the posttranslational processing of Hh proteins so complicated? Much of the complexity of the processing mechanism seems to originate from the requirement for cholesterol modification of the Hh ligand. Cholesterol attachment necessitates the autocatalytic intein-like reaction, which in turn requires the C-terminal domain to be properly folded. The formation of a critical disulfide bond involving the catalytic cysteine ensures that proper folding precedes catalysis. Once the C terminus has done its job, however, it becomes dispensable, explaining why it is cleared by ERAD. Despite the progress, it remains to be clarified why the Hh ligand is modified by cholesterol, how cholesterol attachment changes its membrane association, and how the cholesterol-modified Hh ligand is ultimately released from cells.
Materials and methods
Materials
The following materials were used in this study: MG132 (Enzo Life Sciences, Inc.), bortezomib (gift from A. Goldberg, Harvard Medical School, Boston, MA), epoxomicin (Enzo Life Sciences, Inc.), diamide (TCI America), rat anti-HA antibody (3F10; Roche), mouse anti-Myc antibody (9E10; Roche), rabbit antiubiquitin antibody (Enzo Life Sciences, Inc.), rabbit anticalnexin antibody (Abcam), rabbit antigigantin antibody (Abcam), rabbit anti-Shh (Cell Signaling Technology), protein G–agarose (GE Healthcare), and mouse anti-FLAG M2 agarose (Sigma-Aldrich). Stealth siRNA duplexes were custom synthesized by Invitrogen.
Protein purification and in vitro DHh-processing assays
A fragment of DHh comprising amino acids 244–471 was expressed in bacteria as an MBP fusion (MBP-DHh). The soluble MBP-DHh was purified on amylose beads (New England Biolabs, Inc.) according to the manufacturer’s instructions. Point mutations in DHh were generated using a site-directed mutagenesis kit (QuikChange; Agilent Technologies), confirmed by DNA sequencing, and expressed and purified as MBP fusions as for the wild-type protein. The purified proteins were concentrated to 2–5 mg/ml and were stored at −80°C.
Processing reactions contained 0.2–0.5 mg/ml MBP-DHh in incubation buffer (20 mM Hepes, pH 7.5, 50 mM NaCl, and 0.1% Triton X-100) with or without a reducing agent (DTT or glutathione) and with or without cholesterol (250 µM final concentration added from a stock solution in DMSO). The reactions were incubated at room temperature and were stopped at the indicated times by the addition of SDS-PAGE sample buffer (with or without DTT). The samples were boiled and separated by SDS-PAGE, and the proteins were visualized by staining with GelCode blue reagent (Thermo Fisher Scientific).
To assay cysteine modification by Mal-PEG, 35S-labeled MBP-DHh proteins (wild type and mutants) were generated by in vitro translation in reticulocyte lysates and were dialyzed overnight against PBS to remove small molecule thiols. The proteins were incubated with or without 5 mM tris(2-carboxyethyl)phosphine for 10 min followed by incubation with 7 mM Mal-PEG at 5 kD (Laysan Bio, Inc.) for 30 min at room temperature. The reaction was terminated with 100 mM DTT, and the samples were separated by SDS-PAGE followed by autoradiography.
To determine whether MBP-DHh is modified with cholesterol, processing reactions were performed in the presence of 0.5 µCi/µl radioactive cholesterol (1,2,3,6,7-3H-cholesterol; 100 mCi/mmol; American Radiolabeled Chemicals, Inc.). The samples were separated by SDS-PAGE, and radioactive proteins were visualized by fluorography (Bonner and Laskey, 1974).
Hh processing in Xenopus egg extracts
XShh was cloned into the pCS2+ vector (Rupp et al., 1994), and radioactive XShh was generated by in vitro translation (TNT SP6 Coupled Reticulocyte Lysate System; Promega) in the presence of [35S]methionine (New England Nuclear). Xenopus egg extracts were prepared as previously described (Salic et al., 2000). A typical XShh-processing reaction contained 1 µl of in vitro translated protein in 14 µl Xenopus egg extract supplemented with 100 µg/ml cycloheximide. The processing reactions were incubated at room temperature, and aliquots were removed at the times indicated in the figures and mixed with SDS-PAGE sample buffer. The samples were boiled and separated by SDS-PAGE, and radioactive proteins were visualized by autoradiography. Triton X-114–partitioning experiments were performed as previously described (Bordier, 1981) using radioactive proteins incubated with or without Xenopus egg extract for 1 h at room temperature. Point mutants of XShh were generated using the QuikChange kit and were confirmed by DNA sequencing.
Cell culture and generation of stable cell lines
Human 293T cells were grown in DME supplemented with 10% fetal bovine serum, penicillin, and streptomycin. NIH-3T3 cells were grown in DME with 10% bovine calf serum, penicillin, and streptomycin. To generate stable cell lines, constructs encoding full-length HShh C-terminally tagged with an HA epitope (HShh-HA) or fused with mCherry (HShh-mCherry) were cloned into the retroviral vector pLHCX (Takara Bio Inc.), and retroviruses were produced in 293T cells. The retroviruses were used to infect NIH-3T3 or 293T cells, and cells stably expressing HShh-HA or HShh-Cherry were generated by hygromycin selection. Expression of HShh-HA or HShh-mCherry was confirmed by Western blotting and by immunofluorescence.
Immunofluorescence
Cultured cells were fixed in PBS with 4% formaldehyde and were permeabilized with TBST (10 mM Tris, pH 7.5, 150 mM NaCl, and 0.2% Triton X-100). Antibodies against HA (3F10, rat monoclonal; Roche), calnexin (rabbit polyclonal; Abcam), and gigantin (rabbit polyclonal; Abcam) were used at a final concentration of 0.25, 0.5, and 0.5 µg/ml, respectively. Alexa Fluor 594– and Alexa Fluor 488–conjugated secondary antibodies (Invitrogen) were used at a final concentration of 1 µg/ml. The immunostained cells were imaged by epifluorescence microscopy on an inverted microscope (TE2000U; Nikon) equipped with a digital camera (OrcaER; Hamamatsu) and a 100× Plan Apochromat 1.4 NA oil objective (Nikon). Images were collected using Metamorph image acquisition software (Applied Precision).
293T and NIH-3T3 cells stably expressing HShh-HA were incubated for 3 h in the presence of control media or the proteasome inhibitors MG132 (10 µM), bortezomib (1 µM), or epoxomicin (1 µM). The cells were fixed and processed for immunofluorescence to detect the HA epitope and calnexin.
DNA constructs for mammalian cell transfection
Full-length HShh, tagged at the C terminus with one copy of the HA epitope (HShh-HA), was cloned into pIRES2-EGFP (Takara Bio Inc.). The C-terminal fragment of HShh (HShh-C; amino acids 198–462 HA tagged at the C terminus) was cloned into pIRES2-EGFP behind a sequence encoding the signal sequence of CD5. Site-directed mutagenesis of HShh was performed using the QuikChange kit. Wild-type and catalytically inactive p97 (p97-WT and p97-QQ) were provided by Y. Ye (National Institutes of Health, Bethesda, MD); Myc-tagged wild type and the two dominant-negative mutants of Hrd1 (Hrd1-WT, Hrd1-C291A, and Hrd1-C291A-C307A) were provided by E. Wiertz (Leiden University, Leiden, Netherlands); Myc-tagged reticulon 4A was obtained from S.M. Strittmatter (Yale University, New Haven, CT); and FLAG-tagged wild type and the two inactive mutants of ERdj5 (ERdj5-WT, ERdj5-HQ, and ERdj5-SS) were provided by K. Nagata (Kyoto University, Kyoto, Japan). The Ubc6e-C91S and UbxD8-GFP expression constructs were described previously (Lilley and Ploegh, 2004; Mueller et al., 2008). The expression constructs for the mutant ER-localized Trx-like proteins were described previously (Schulman et al., 2010).
Transfection of plasmids and siRNAs into cultured cells
Plasmids were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. siRNA duplexes were transfected using a transfection reagent (TransIT-siQUEST; Mirus) at a final concentration of 50 nM siRNA according to the manufacturer’s instructions. siRNA transfection was performed twice, on days 1 and 3 of the experiment. On day 5, the cells were treated with 50 µM cycloheximide or 50 µM cycloheximide plus MG132 for the period of time indicated in the figures, after which cells were harvested, and proteins were analyzed by SDS-PAGE and immunoblotting. siRNA sequences are shown in Table S1.
Quantitative RT-PCR analysis
Total RNA was extracted with TRIZOL reagent, and cDNA was synthesized with reverse transcriptase (ImProm-IITM; Promega). Quantitative PCR was performed on an ABI Prism 7900 cycler using SYBR Green PCR Master Mix (Applied Biosystems). The degree of siRNA knockdown was calculated relative to HPRT1 mRNA levels. The primers used to quantify mRNA knockdown are shown in Table S2.
Immunoblotting
Cells were lysed on ice for 20 min in TBS (10 mM Tris, pH 7.5, and 150 mM NaCl) supplemented with protease inhibitors and 1% Triton X-100. The lysate was centrifuged for 30 min at 4°C and 20,000 g. The supernatant was collected, mixed with SDS-PAGE sample buffer with DTT (50 mM final concentration), and separated by SDS-PAGE followed by immunoblotting. Unless otherwise specified, p97 was used as a loading control by probing the p97 portion of the blot in parallel with the portion containing HShh and HShh-C.
Immunoprecipitation and ubiquitination of HShh-HA
293T cells expressing HShh-HA were treated with or without 50 µM MG132 for 2 h, and immunoprecipitation with anti-HA (3F10) antibody was performed as previously described (Mueller et al., 2008). The precipitated proteins were separated by SDS-PAGE, and ubiquitin conjugates were detected by immunoblotting with antiubiquitin antibodies.
Pulse-chase assays
Pulse-chase experiments were performed as previously described (Mueller et al., 2006). 293T cells were detached from plates and were incubated in suspension in methionine- and cysteine-free DME for 1 h at 37°C. The cells were then labeled for 3 min at 37°C with 300 µCi/ml [35S]methionine and [35S]cysteine (35S-Protein Express Labeling Mix; New England Nuclear). The cellular density during labeling was 107 cells/ml. The chase was started by adding cold methionine and cysteine at a final concentration of 5 mM and 1 mM, respectively. Aliquots of the cell suspension were removed at different time points, and cellular pellets were frozen. Cellular lysates were subjected to denaturing immunoprecipitation with HA antibodies. The precipitated proteins were separated on SDS-PAGE and were visualized by autoradiography.
Cycloheximide chase assays
293T cells stably expressing HShh-HA were incubated with 50 µg/ml cycloheximide in Opti-MEM (Invitrogen), in agitated suspension, at 37°C. At the times indicated in the figures, aliquots of the cell suspension were removed and HShh-HA was detected by immunoblotting.
To determine the effect of various dominant-negative constructs on HShh processing, 293T cells expressing HShh-HA were transfected with expression constructs for Derlin1-GFP, UbxD8-GFP, or dominant-negative Ubc6e as previously described (Mueller et al., 2006, 2008) followed by cycloheximide chase 24 h after transfection.
Screening for Trx-like enzymes involved in Hh processing
We used a collection of FLAG-tagged CXXA mutants representing nine different human ER-localized Trx-like proteins (Schulman et al., 2010). Each construct was coexpressed in 293T cells with either the wild-type HShh-HA or the processing-defective C198A mutant. 24 h later, the cells were harvested and lysed, and the lysate was subjected to denaturing immunoprecipitation with HA and FLAG antibodies as previously described (Schulman et al., 2010). The precipitated proteins were separated by SDS-PAGE under either reducing or nonreducing conditions followed by immunoblotting with FLAG or HA antibodies.
Analyzing Hh secretion
293T cells stably expressing HShh-HA were incubated for 12 h in DME containing 0.5% fetal bovine serum with or without epoxomicin added for the last 3 h of the incubation. The cells were harvested and lysed while the protein in the culture medium was precipitated with trichloracetic acid. HShh-N and HShh-C were analyzed by SDS-PAGE followed by immunoblotting with Shh antibodies (Cell Signaling Technology) and HA antibodies (Roche).
Online supplemental material
Fig. S1 shows a schematic of the Hh proteins used in this study and the ERAD of the HShh-C fragment expressed in isolation. Fig. S2 shows the dispersion of the Golgi by brefeldin A, the specificity of the HShh-PDI mixed disulfide detection, and the degradation of HShh-C by the proteasome. Fig. S3 shows knockdown of components that had no effect on HShh-C ERAD. Fig. S4 shows the specificity of the immunoprecipitation of polyubiquitinated HShh. Fig. S5 shows knockdown of ERAD components required for the degradation of the processing-defective precursor mutant HShh-C198S. Table S1 lists sequences of siRNA duplexes used in this study. Table S2 lists the primers used for quantitative RT-PCR. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201008090/DC1.
Acknowledgments
We thank Y. Ye, E. Wiertz, S. Strittmatter, and K. Nagata for providing reagents. We thank Pedro Carvalho for reading the manuscript.
H. Tukachinsky and C. Jao are supported by predoctoral fellowships from the American Heart Association and from the National Science Foundation, respectively. T.A. Rapoport is supported by the National Institutes of Health grant GM052586 and is a Howard Hughes Medical Institute Investigator. X. Chen is supported by funds from the National Research Program for Genomic Medicine, National Science Council, and National Health Research Institutes (Taiwan). A. Salic acknowledges the support from the Rita Allen Foundation and the Beckman Foundation.
Footnotes
Abbreviations used in this paper:
- DHh
- Drosophila Hh
- ERAD
- ER-associated degradation
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- Hh
- Hedgehog
- HShh
- human Shh
- Mal-PEG
- maleimide-polyethylene glycol
- MBP
- maltose-binding protein
- PDI
- protein disulfide isomerase
- Shh
- Sonic Hh
- Trx
- thioredoxin
- XShh
- Xenopus Shh
References
- Bays N.W., Gardner R.G., Seelig L.P., Joazeiro C.A., Hampton R.Y. 2001a. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat. Cell Biol. 3:24–29 10.1038/35050524 [DOI] [PubMed] [Google Scholar]
- Bays N.W., Wilhovsky S.K., Goradia A., Hodgkiss-Harlow K., Hampton R.Y. 2001b. HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol. Biol. Cell. 12:4114–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi R., Galli C., Calanca V., Nakajima T., Molinari M. 2010. Stringent requirement for HRD1, SEL1L, and OS-9/XTP3-B for disposal of ERAD-LS substrates. J. Cell Biol. 188:223–235 10.1083/jcb.200910042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner W.M., Laskey R.A. 1974. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur. J. Biochem. 46:83–88 10.1111/j.1432-1033.1974.tb03599.x [DOI] [PubMed] [Google Scholar]
- Bordallo J., Plemper R.K., Finger A., Wolf D.H. 1998. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol. Biol. Cell. 9:209–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordier C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256:1604–1607 [PubMed] [Google Scholar]
- Bumcrot D.A., Takada R., McMahon A.P. 1995. Proteolytic processing yields two secreted forms of sonic hedgehog. Mol. Cell. Biol. 15:2294–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P., Goder V., Rapoport T.A. 2006. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 126:361–373 10.1016/j.cell.2006.05.043 [DOI] [PubMed] [Google Scholar]
- Chamoun Z., Mann R.K., Nellen D., von Kessler D.P., Bellotto M., Beachy P.A., Basler K. 2001. Skinny hedgehog, an acyltransferase required for palmitoylation and activity of the hedgehog signal. Science. 293:2080–2084 10.1126/science.1064437 [DOI] [PubMed] [Google Scholar]
- Chin D.J., Gil G., Faust J.R., Goldstein J.L., Brown M.S., Luskey K.L. 1985. Sterols accelerate degradation of hamster 3-hydroxy-3-methylglutaryl coenzyme A reductase encoded by a constitutively expressed cDNA. Mol. Cell. Biol. 5:634–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson J.C., Shaler T.A., Tyler R.E., Kopito R.R. 2008. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol. 10:272–282 10.1038/ncb1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V., Quan E.M., Weissman J.S. 2006. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 126:349–359 10.1016/j.cell.2006.05.045 [DOI] [PubMed] [Google Scholar]
- Elbein A.D. 1991. Glycosidase inhibitors: inhibitors of N-linked oligosaccharide processing. FASEB J. 5:3055–3063 [DOI] [PubMed] [Google Scholar]
- Fang S., Ferrone M., Yang C., Jensen J.P., Tiwari S., Weissman A.M. 2001. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 98:14422–14427 10.1073/pnas.251401598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner R.G., Swarbrick G.M., Bays N.W., Cronin S.R., Wilhovsky S., Seelig L., Kim C., Hampton R.Y. 2000. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J. Cell Biol. 151:69–82 10.1083/jcb.151.1.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss R., Jarosch E., Sommer T., Hirsch C. 2006. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat. Cell Biol. 8:849–854 10.1038/ncb1445 [DOI] [PubMed] [Google Scholar]
- Gil G., Faust J.R., Chin D.J., Goldstein J.L., Brown M.S. 1985. Membrane-bound domain of HMG CoA reductase is required for sterol-enhanced degradation of the enzyme. Cell. 41:249–258 10.1016/0092-8674(85)90078-9 [DOI] [PubMed] [Google Scholar]
- Hall T.M., Porter J.A., Young K.E., Koonin E.V., Beachy P.A., Leahy D.J. 1997. Crystal structure of a Hedgehog autoprocessing domain: homology between Hedgehog and self-splicing proteins. Cell. 91:85–97 10.1016/S0092-8674(01)80011-8 [DOI] [PubMed] [Google Scholar]
- Hampton R.Y., Rine J. 1994. Regulated degradation of HMG-CoA reductase, an integral membrane protein of the endoplasmic reticulum, in yeast. J. Cell Biol. 125:299–312 10.1083/jcb.125.2.299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassink G., Kikkert M., van Voorden S., Lee S.J., Spaapen R., van Laar T., Coleman C.S., Bartee E., Früh K., Chau V., Wiertz E. 2005. TEB4 is a C4HC3 RING finger-containing ubiquitin ligase of the endoplasmic reticulum. Biochem. J. 388:647–655 10.1042/BJ20041241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch C., Blom D., Ploegh H.L. 2003. A role for N-glycanase in the cytosolic turnover of glycoproteins. EMBO J. 22:1036–1046 10.1093/emboj/cdg107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch C., Gauss R., Horn S.C., Neuber O., Sommer T. 2009. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 458:453–460 10.1038/nature07962 [DOI] [PubMed] [Google Scholar]
- Hosokawa N., Wada I., Nagasawa K., Moriyama T., Okawa K., Nagata K. 2008. Human XTP3-B forms an endoplasmic reticulum quality control scaffold with the HRD1-SEL1L ubiquitin ligase complex and BiP. J. Biol. Chem. 283:20914–20924 10.1074/jbc.M709336200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Kamiya Y., Kamiya D., Kato K., Nagata K. 2009. Human OS-9, a lectin required for glycoprotein endoplasmic reticulum-associated degradation, recognizes mannose-trimmed N-glycans. J. Biol. Chem. 284:17061–17068 10.1074/jbc.M809725200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosch E., Taxis C., Volkwein C., Bordallo J., Finley D., Wolf D.H., Sommer T. 2002. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat. Cell Biol. 4:134–139 10.1038/ncb746 [DOI] [PubMed] [Google Scholar]
- Kalderon D. 2005. The mechanism of hedgehog signal transduction. Biochem. Soc. Trans. 33:1509–1512 10.1042/BST20051509 [DOI] [PubMed] [Google Scholar]
- Kokame K., Agarwala K.L., Kato H., Miyata T. 2000. Herp, a new ubiquitin-like membrane protein induced by endoplasmic reticulum stress. J. Biol. Chem. 275:32846–32853 10.1074/jbc.M002063200 [DOI] [PubMed] [Google Scholar]
- Lai C.J., Ekker S.C., Beachy P.A., Moon R.T. 1995. Patterning of the neural ectoderm of Xenopus laevis by the amino-terminal product of hedgehog autoproteolytic cleavage. Development. 121:2349–2360 [DOI] [PubMed] [Google Scholar]
- Lee J.J., Ekker S.C., von Kessler D.P., Porter J.A., Sun B.I., Beachy P.A. 1994. Autoproteolysis in hedgehog protein biogenesis. Science. 266:1528–1537 10.1126/science.7985023 [DOI] [PubMed] [Google Scholar]
- Lilley B.N., Ploegh H.L. 2004. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 429:834–840 10.1038/nature02592 [DOI] [PubMed] [Google Scholar]
- Lum L., Beachy P.A. 2004. The Hedgehog response network: sensors, switches, and routers. Science. 304:1755–1759 10.1126/science.1098020 [DOI] [PubMed] [Google Scholar]
- Maity T., Fuse N., Beachy P.A. 2005. Molecular mechanisms of Sonic hedgehog mutant effects in holoprosencephaly. Proc. Natl. Acad. Sci. USA. 102:17026–17031 10.1073/pnas.0507848102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marigo V., Davey R.A., Zuo Y., Cunningham J.M., Tabin C.J. 1996. Biochemical evidence that patched is the Hedgehog receptor. Nature. 384:176–179 10.1038/384176a0 [DOI] [PubMed] [Google Scholar]
- Mueller B., Lilley B.N., Ploegh H.L. 2006. SEL1L, the homologue of yeast Hrd3p, is involved in protein dislocation from the mammalian ER. J. Cell Biol. 175:261–270 10.1083/jcb.200605196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller B., Klemm E.J., Spooner E., Claessen J.H., Ploegh H.L. 2008. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc. Natl. Acad. Sci. USA. 105:12325–12330 10.1073/pnas.0805371105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden S.K., Ascano M., Jr, Stegman M.A., Robbins D.J. 2004. Regulation of Hedgehog signaling: a complex story. Biochem. Pharmacol. 67:805–814 10.1016/j.bcp.2004.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter J.A., von Kessler D.P., Ekker S.C., Young K.E., Lee J.J., Moses K., Beachy P.A. 1995. The product of hedgehog autoproteolytic cleavage active in local and long-range signalling. Nature. 374:363–366 10.1038/374363a0 [DOI] [PubMed] [Google Scholar]
- Porter J.A., Ekker S.C., Park W.J., von Kessler D.P., Young K.E., Chen C.H., Ma Y., Woods A.S., Cotter R.J., Koonin E.V., Beachy P.A. 1996a. Hedgehog patterning activity: role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell. 86:21–34 10.1016/S0092-8674(00)80074-4 [DOI] [PubMed] [Google Scholar]
- Porter J.A., Young K.E., Beachy P.A. 1996b. Cholesterol modification of hedgehog signaling proteins in animal development. Science. 274:255–259 10.1126/science.274.5285.255 [DOI] [PubMed] [Google Scholar]
- Rabinovich E., Kerem A., Fröhlich K.U., Diamant N., Bar-Nun S. 2002. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol. Cell. Biol. 22:626–634 10.1128/MCB.22.2.626-634.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E., El-Jaick K.B., Dubourg C., Vélez J.I., Solomon B.D., Pineda-Alvarez D.E., Lacbawan F., Zhou N., Ouspenskaia M., Paulussen A., et al. 2009. The mutational spectrum of holoprosencephaly-associated changes within the SHH gene in humans predicts loss-of-function through either key structural alterations of the ligand or its altered synthesis. Hum. Mutat. 30:E921–E935 10.1002/humu.21090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp R.A., Snider L., Weintraub H. 1994. Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev. 8:1311–1323 10.1101/gad.8.11.1311 [DOI] [PubMed] [Google Scholar]
- Salic A., Lee E., Mayer L., Kirschner M.W. 2000. Control of beta-catenin stability: reconstitution of the cytoplasmic steps of the wnt pathway in Xenopus egg extracts. Mol. Cell. 5:523–532 10.1016/S1097-2765(00)80446-3 [DOI] [PubMed] [Google Scholar]
- Schulman S., Wang B., Li W., Rapoport T.A. 2010. Vitamin K epoxide reductase prefers ER membrane-anchored thioredoxin-like redox partners. Proc. Natl. Acad. Sci. USA. 107:15027–15032 10.1073/pnas.1009972107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze A., Standera S., Buerger E., Kikkert M., van Voorden S., Wiertz E., Koning F., Kloetzel P.M., Seeger M. 2005. The ubiquitin-domain protein HERP forms a complex with components of the endoplasmic reticulum associated degradation pathway. J. Mol. Biol. 354:1021–1027 10.1016/j.jmb.2005.10.020 [DOI] [PubMed] [Google Scholar]
- Stagg H.R., Thomas M., van den Boomen D., Wiertz E.J., Drabkin H.A., Gemmill R.M., Lehner P.J. 2009. The TRC8 E3 ligase ubiquitinates MHC class I molecules before dislocation from the ER. J. Cell Biol. 186:685–692 10.1083/jcb.200906110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone D.M., Hynes M., Armanini M., Swanson T.A., Gu Q., Johnson R.L., Scott M.P., Pennica D., Goddard A., Phillips H., et al. 1996. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. 384:129–134 10.1038/384129a0 [DOI] [PubMed] [Google Scholar]
- Swanson R., Locher M., Hochstrasser M. 2001. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 15:2660–2674 10.1101/gad.933301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traiffort E., Dubourg C., Faure H., Rognan D., Odent S., Durou M.R., David V., Ruat M. 2004. Functional characterization of sonic hedgehog mutations associated with holoprosencephaly. J. Biol. Chem. 279:42889–42897 10.1074/jbc.M405161200 [DOI] [PubMed] [Google Scholar]
- Tsai B., Rapoport T.A. 2002. Unfolded cholera toxin is transferred to the ER membrane and released from protein disulfide isomerase upon oxidation by Ero1. J. Cell Biol. 159:207–216 10.1083/jcb.200207120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai B., Rodighiero C., Lencer W.I., Rapoport T.A. 2001. Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell. 104:937–948 10.1016/S0092-8674(01)00289-6 [DOI] [PubMed] [Google Scholar]
- Ushioda R., Hoseki J., Araki K., Jansen G., Thomas D.Y., Nagata K. 2008. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 321:569–572 10.1126/science.1159293 [DOI] [PubMed] [Google Scholar]
- Walter J., Urban J., Volkwein C., Sommer T. 2001. Sec61p-independent degradation of the tail-anchored ER membrane protein Ubc6p. EMBO J. 20:3124–3131 10.1093/emboj/20.12.3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiertz E.J.H.J., Jones T.R., Sun L., Bogyo M., Geuze H.J., Ploegh H.L. 1996. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell. 84:769–779 10.1016/S0092-8674(00)81054-5 [DOI] [PubMed] [Google Scholar]
- Xie W., Ng D.T. 2010. ERAD substrate recognition in budding yeast. Semin. Cell Dev. Biol. 21:533–539 10.1016/j.semcdb.2010.02.007 [DOI] [PubMed] [Google Scholar]
- Ye Y., Meyer H.H., Rapoport T.A. 2001. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 414:652–656 10.1038/414652a [DOI] [PubMed] [Google Scholar]
- Ye Y., Shibata Y., Yun C., Ron D., Rapoport T.A. 2004. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 429:841–847 10.1038/nature02656 [DOI] [PubMed] [Google Scholar]