Rad54’s ATPase activity does not affect accumulation of homologous recombination proteins in repair foci, but influences its dissociation and that of Rad51.
Abstract
Rad54, a member of the SWI/SNF protein family of DNA-dependent ATPases, repairs DNA double-strand breaks (DSBs) through homologous recombination. Here we demonstrate that Rad54 is required for the timely accumulation of the homologous recombination proteins Rad51 and Brca2 at DSBs. Because replication protein A and Nbs1 accumulation is not affected by Rad54 depletion, Rad54 is downstream of DSB resection. Rad54-mediated Rad51 accumulation does not require Rad54’s ATPase activity. Thus, our experiments demonstrate that SWI/SNF proteins may have functions independent of their ATPase activity. However, quantitative real-time analysis of Rad54 focus formation indicates that Rad54’s ATPase activity is required for the disassociation of Rad54 from DNA and Rad54 turnover at DSBs. Although the non–DNA-bound fraction of Rad54 reversibly interacts with a focus, independent of its ATPase status, the DNA-bound fraction is immobilized in the absence of ATP hydrolysis by Rad54. Finally, we show that ATP hydrolysis by Rad54 is required for the redistribution of DSB repair sites within the nucleus.
Introduction
To preserve the integrity of their genome, cells have evolved several pathways to deal with DNA damage that is created by both endogenous sources, such as some byproducts of cellular metabolism like oxygen radicals, and exogenous sources, including ultraviolet and ionizing radiation (Friedberg et al., 2004). Among different kinds of lesions, DNA double-strand breaks (DSBs) present a special challenge to the cells because both strands of the double helix are affected. If misrepaired, DSBs can cause genome rearrangements such as translocations and deletions that can result in development of cancer (Hoeijmakers, 2001; Bassing and Alt, 2004; Agarwal et al., 2006). Thus, it is paramount that DSBs are repaired precisely and in a timely fashion.
Homologous recombination is an error free, high-fidelity pathway that repairs DSBs by using an undamaged homologous DNA molecule, usually the sister chromatid, as a template to repair the broken molecule (Wyman and Kanaar, 2006). The process is performed by the Rad52 epistasis group proteins, identified by the genetic analyses of ionizing radiation–sensitive Saccharomyces cerevisiae mutants (Game and Mortimer, 1974; Symington, 2002). Several Rad52 group proteins, including Rad51 and Rad54, are conserved in mammals, as is the core mechanism of homologous recombination (Wyman and Kanaar, 2004). The central protein of homologous recombination is Rad51, which mediates the critical step of homologous pairing and DNA strand exchange between the broken DNA molecule and the homologous intact repair template. Once a DSB occurs, it is processed to single-stranded DNA tails with a 3′ polarity, onto which Rad51 promoters assemble into a nucleoprotein filament. This nucleoprotein filament is the active molecular entity in recognition of homologous DNA and the subsequent exchange of DNA strands. An extensive number of mediator and/or accessory proteins are implicated in assisting Rad51 at various stages of recombination (Sung et al., 2003), one of which is Rad54.
RAD54, first identified in S. cerevisiae, is conserved in vertebrates (Kanaar et al., 1996; Bezzubova et al., 1997; Essers et al., 1997). Rad54 is a member of the SWI2/SNF2 family of double-strand DNA-stimulated ATPases that modulate protein–DNA interactions (Pollard and Peterson, 1998). Rad54−/− mouse embryonic stem (ES) cells are ionizing radiation–sensitive, display reduced level of homologous recombination, and exhibit defects in repair of DSBs (Essers et al., 1997; Dronkert et al., 2000). A plethora of biochemical activities of Rad54 have been uncovered that have the potential to augment the central function of Rad51 in homologous recombination (Tan et al., 2003; Heyer et al., 2006). First, Rad54 physically interacts with Rad51, both in vitro and in vivo (Jiang et al., 1996; Clever et al., 1997; Golub et al., 1997; Tan et al., 1999). Interestingly, in mammalian cells, the interaction between the proteins can only be detected in cells that have been challenged with DNA-damaging agents, which suggests that Rad54 interacts with the Rad51 nucleoprotein filament rather than Rad51 protomers that are not engaged in recombination (Tan et al., 1999; Essers et al., 2002a). Second, the interaction is not only physical but also functional, as Rad54 stimulates Rad51 mediated D-loop formation, i.e., the generation of a joint between homologous DNA molecules (Petukhova et al., 1998). Third, Rad54 has a potent ATPase activity that is triggered specifically by double-stranded DNA (Petukhova et al., 1998; Swagemakers et al., 1998). Fourth, the protein uses energy gained from ATP hydrolysis to translocate along the DNA double helix (Van Komen et al., 2000; Ristic et al., 2001; Amitani et al., 2006). Fifth, presumably through its DNA translocase activity, Rad54 can affect the interaction of proteins with DNA. Specifically, it can influence the position of histones on DNA and remove Rad51 nucleoprotein filaments from double-stranded DNA (Alexiadis and Kadonaga, 2002; Solinger et al., 2002; Alexeev et al., 2003; Jaskelioff et al., 2003; Li and Heyer, 2009; Li et al., 2009). Sixth, its translocase activity also allows the protein to perturb DNA structures. Rad54 can promote branch migration, thereby affecting the processing of the Holliday junction, which is a four-way DNA junction that can arise as intermediates at sites where the recombination partners are physically joined (Bugreev et al., 2006). Many of the biochemical activities of Rad54 are affected by abrogating its ATPase activity. Hence, the proper functioning of Rad54 depends on its ability to harness the energy from ATP hydrolysis, and this in turn is responsible for augmenting the role of Rad51. However, Rad51 nucleoprotein filament stabilization by Rad54, which is probably required before joint molecule formation occurs, turned out to be independent of its ATPase activity (Mazin et al., 2003).
A striking characteristic of several proteins involved in homologous recombination, including Rad51 and Rad54, is their ability to accumulate at a high local concentration into nuclear foci (Haaf et al., 1995; Tan et al., 1999; Essers et al., 2006). This occurs spontaneously, that is in the absence of exogenously induced DNA damage, in a low percentage of cells in S phase (Tashiro et al., 2000; Tarsounas et al., 2003). Upon the induction of DNA damage to cells, the majority of cells display colocalizing Rad51 and Rad54 foci at sites of DNA damage (Tan et al., 1999; Tashiro et al., 2000; Aten et al., 2004). Although the nature, composition, and requirement for foci formation is not apparent from a biochemical point, it is clear that the foci, particularly of Rad51, are biologically relevant because mutant cells that cannot form them are DNA damage–sensitive and display spontaneous chromosomal aberrations (Thacker and Zdzienicka, 2004; van Veelen et al., 2005). The nature of these foci with respect to protein composition is highly dynamic. Photobleaching experiments have shown that Rad51 and Rad54 dissociate and associate with foci, with each protein having a characteristic dwell time (Essers et al., 2002b).
Here, we studied the ATPase function of Rad54 in homologous recombination in vivo. To this end we compared knockin mouse ES cell lines that express GFP-tagged wild-type Rad54 protein with cell lines expressing ATP hydrolysis–defective Rad54 proteins using advanced live-cell confocal microscopy. Time-lapse imaging and fluorescence correlation spectroscopy (FCS) of individual cells and foci allowed quantitative analysis of the Rad54 in live cells. We measured protein concentrations and discovered that DSB repair foci relocalize to the nuclear periphery upon DNA damage induction. In addition, we used fluorescence photobleaching techniques to show that defective ATPase activity renders a small fraction of the Rad54 molecules in a focus immobile. This suggests that only a minority of molecules present in foci are functional in DNA DSB repair and that the ATPase activity of Rad54 is required for the release of protein from DNA damage–induced structures on chromatin.
Results
The ATPase activity of Rad54 contributes to DNA damage resistance and homologous recombination
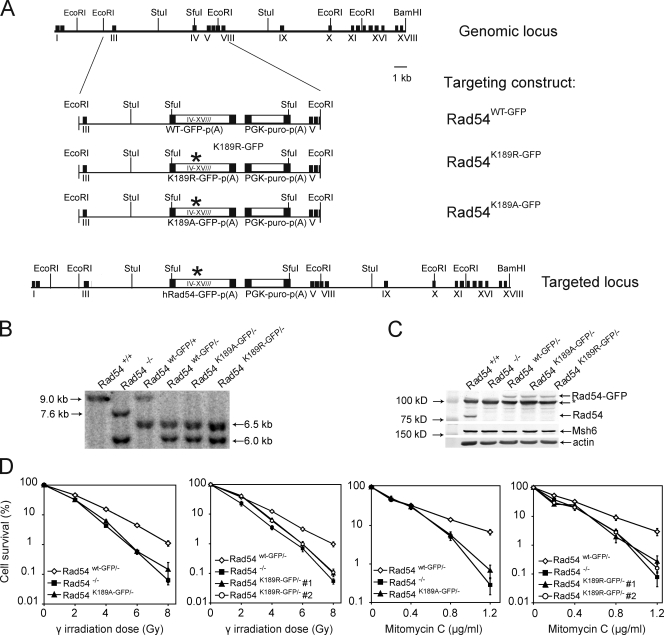
To study the effect of the ATPase activity of Rad54 at the cellular level, mouse ES cells were generated that express wild-type and ATPase-defective versions of GFP-fused Rad54 from the endogenous Rad54 locus. A targeting construct, consisting of the human RAD54 cDNA exons IV–XVIII fused to a GFP coding sequence or containing a point mutation in the Walker A ATPase domain (Fig. 1 A), was electroporated into ES cells of the genotype Rad54wt-HA/−, in which one Rad54 allele is inactivated (Tan et al., 1999). Two different mutant constructs were used, one in which the lysine at position 189 was replaced by arginine, which is indicated by K189R and one in which the lysine is replaced by alanine, the K189A mutation. The ATPase activity of the purified Rad54K189R and Rad54K189A proteins was reduced >100-fold in comparison to the wild-type protein (Swagemakers et al., 1998 and unpublished data). Clones carrying a homologously integrated knockin construct were identified by DNA blot analysis. A probe that detects exons VII and VIII was used in combination with genomic DNA digested with StuI, which yielded the expected doublet of bands ∼6.5 kb for the Rad54 knockin allele, whereas a 6.0-kb band was observed that is diagnostic for the Rad54 knockout allele (Fig. 1 B). Proper expression of the full-length wild-type or mutant Rad54–GFP fusion proteins was confirmed by immunoblot analysis (Fig. 1 C). In the subsequent studies, two independent clones for Rad54K189R-GFP/− and one for Rad54K189A-GFP/− were used. As a positive control for all experiments, knockin Rad54wt-GFP/− ES cells were used; these cells express wild-type Rad54 fused to GFP from the endogenous Rad54 locus. The function of Rad54 is not affected by its fusion to GFP because Rad54wt-GFP/− cells are not DNA damage sensitive (unpublished data).
Figure 1.
Characterization of mouse ES cells carrying ATPase-defective Rad54–GFP alleles. (A) Schematic representation of the mouse Rad54 locus and the gene-targeting constructs. The top line represents a 30-kb portion of endogenous Rad54 locus, where black boxes indicate exons I–XVIII. The middle line shows the linearized targeting construct, containing the human RAD54 cDNA sequence spanning exons IV–XVIII fused to the GFP coding sequence. The K189R and K189A mutations in the Walker A ATPase domain are indicated by the asterisks. The construct contains a gene encoding for puromycin resistance as a selectable marker. The targeting construct will replace the regions between exons III and VIII when correctly integrated to generate the targeted allele, as shown in the targeted locus. Homologous integration results in the expression of full-length, GFP-tagged Rad54 from its endogenous promoter. (B) DNA blot analysis of ES cells carrying the knockin contructs. DNA blot analysis was performed using genomic DNA purified from puromycin-resistant clones and digested with StuI. Detection of bands was performed using a probe that recognized exons VII/VIII. Restriction of the wild-type allele by StuI, (indicated by “+”), yields a 9.0-kb band after hybridization with an exon VII/VIII probe. Diagnostic bands for the neomycin-resistant knockout alleles, indicated by “−“, are 7.6 kb for a hygromycin-resistant allele and 6.0 kb for a neomycin-resistant allele. Knockin alleles are characterized by a doublet of bands ∼6.5 kb. (C) Immunoblot analysis of proteins produced by the Rad54–GFP knockin and -out alleles. Whole cell extracts of ES cells with the indicated genotypes were probed with affinity purified anti–human Rad54 antibodies. The position of Rad54 and Rad54–GFP are indicated. The arrowhead indicates a nonspecific signal. Probing against Msh6 and actin was used to confirm equal protein loading. (D) Ionizing radiation and mitomycin C survivals. ES cells of the indicated genotypes were tested for their ability to survive treatments with increasing doses of ionizing radiation (γ irradiation) or mitomycin C using clonogenic survival assays. The assays were performed in triplicate and the error bars indicated the standard error of the mean.
Mouse Rad54−/− ES cells are hypersensitive to ionizing radiation and the interstrand DNA cross-linker mitomycin C (Essers et al., 1997). Therefore, we investigated the effect of these DNA-damaging agents on Rad54K189R-GFP/− and Rad54K189A-GFP/− ES cells. Cells expressing ATPase-defective versions of Rad54 were hypersensitive to ionizing radiation and mitomycin C compared with isogenic control cells; this hypersensitivity was similar to that demonstrated by cells lacking Rad54 protein altogether (Fig. 1 D). Next we tested the effect of Rad54 ATPase activity on homologous recombination. As a measure of homologous recombination efficiency, we determined the efficiency of homologous gene targeting (Niedernhofer et al., 2001; Essers et al., 2002a). ES cells of the genotypes indicated in Table I were electroporated with a linearized targeting construct for the Rb locus that carried a hygromycin-selectable marker gene. Genomic DNA was isolated from individual clones and analyzed by DNA blotting to discriminate between homologous and random integration events. Homologous recombination efficiency was measured as the percentage of clones containing the homologously integrated targeting construct relative to the total number of drug-resistant clones analyzed (Table I). The homologous targeting efficiency of ∼32% in Rad54wt-GFP/− ES cells was reduced to ∼1% in Rad54K189R-GFP/− and Rad54K189A-GFP/− ES cells. A similar reduction in homologous recombination efficiency was observed in the absence of Rad54. We conclude that the ATPase activity of Rad54 is essential for its DNA repair and recombination functions in vivo. In these assays, the physical presence of ATPase-defective Rad54 or the complete absence of the protein results in indistinguishable phenotypes.
Table I.
Efficiency of homologous recombination in Rad54 ATPase-defective cells
| Genotype | Targeting efficiency at Rb locus |
| Rad54WT-GFP/- | 31.9% (16 out of 46) |
| Rad54K189R-GFP/- | 0.95% (1 out of 105) |
| Rad54K189A-GFP/- | 1.2% (1 out of 81) |
| Rad54−/− | <1% (0 out of 82) |
ES cells with the indicated genotypes were electroporated with a linearized Rb-Hyg construct (Niedernhofer et al., 2001). Hygromycin-resistant clones were expanded, genomic DNA was purified, and samples were subjected to DNA blot analysis to distinguish between randomly and homologously integrated events. Values indicate the percentage of clones that contain the homologously integrated targeting construct relative to the total number of clones analyzed. Absolute numbers are indicated in parentheses. The differences in recombination efficiency between Rad54WT-GFP/− cells and cells of all other genotypes listed are significant (P > 0.001), whereas the difference between the mutant genotypes is not.
ATP hydrolysis by Rad54 affects foci behavior in unchallenged cells
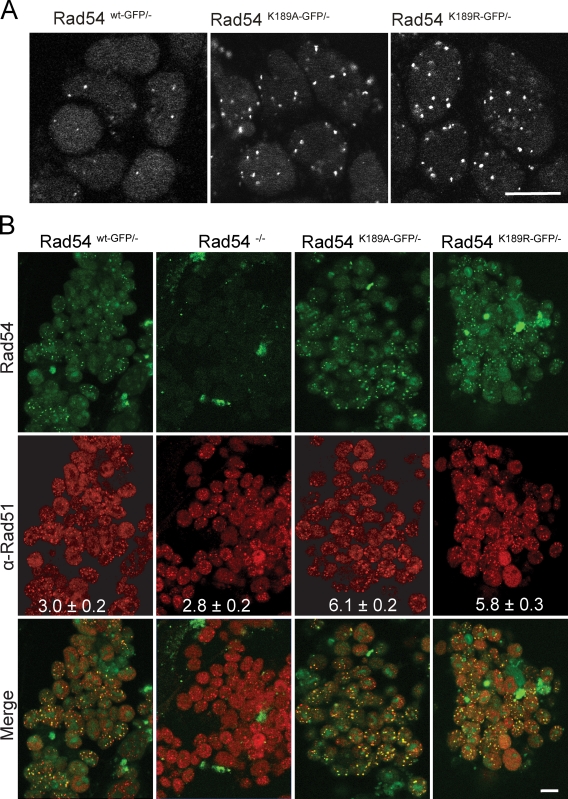
Several proteins involved in the cellular response to DNA damage and repair are known to accumulate in nuclear foci at sites of DNA damage (Wyman and Kanaar, 2006). We analyzed the ATPase-defective Rad54 mutant cells for accumulation of Rad54 foci in the absence of exogenously induced DNA damage. Observation of living cells using a confocal microscope revealed an increase in the amount of foci present in cells containing ATPase-deficient Rad54K189R and Rad54K189A protein compared with wild-type Rad54 protein (Fig. 2 A). It should be noted that the increase in spontaneous foci is only observed when all Rad54 molecules in the cell are ATPase defective, as such an increase is absent in Rad54K189R-GFP/+ and Rad54K189A-GFP/+ cells (unpublished data).
Figure 2.
Effect of Rad54 ATPase activity on focus formation. (A) Shown are confocal images of untreated living ES cells expressing wild-type or ATPase-defective Rad54–GFP. The mean number of spontaneous nuclear foci in cells expressing either version of ATPase-defective Rad54 is considerably greater compared with cells expressing wild-type Rad54. (B) Rad51 immunostaining in untreated ES cells of the indicated genotypes. The top shows confocal images of Rad54 as detected by GFP fluorescence. The middle shows the Rad51 staining pattern as detected by anti-Rad51 antibody staining. The merged images are shown on the bottom. The number of Rad51 foci per cell is indicated (mean ± SD). The difference in number of Rad51 foci per cell between Rad54wt-GFP/− and Rad54−/− ES cells and the difference between Rad54K189R-GFP/− and Rad54K189A-GFP/− ES cells is not significant, whereas the difference between these two groups is (P < 0.0001), as determined by the one-way analysis of variance (ANOVA) and a Student’s t test. Bars, 10 µm.
Rad54 is an accessory protein for Rad51 that performs the core reaction of homologous recombination, homologous DNA pairing, and DNA strand exchange (Wyman and Kanaar, 2006). The proteins physically interact and work closely together in several biochemical assays (Sung et al., 2003). At the cellular level, both proteins colocalize in DNA damage–induced foci (Tan et al., 1999). We analyzed Rad51 foci in the mutant cells to determine whether the ATPase activity of Rad54 impacted the behavior of Rad51 in vivo. Unchallenged ES cells were fixed and stained with an antibody against Rad51, and both Rad51 and Rad54–GFP were detected by confocal microscopy (Fig. 2 B). Compared with cells expressing wild-type Rad54–GFP or lacking Rad54, cells expressing the ATPase-defective variants of Rad54–GFP displayed a statistically significant twofold increase in the number of “spontaneous” Rad51 foci. Furthermore, almost all Rad54 foci detected (>90%), including those of Rad54K189R-GFP and Rad54K189A-GFP, colocalized with Rad51 (Fig. 2 B). Interestingly, although the number of Rad51 foci per confocal slice was elevated in cells expressing the ATPase-defective Rad54 mutants, it was not increased in cells completely lacking Rad54 (Fig. 2 B).
The increase in spontaneous Rad54 foci does not represent increased endogenous DNA damage
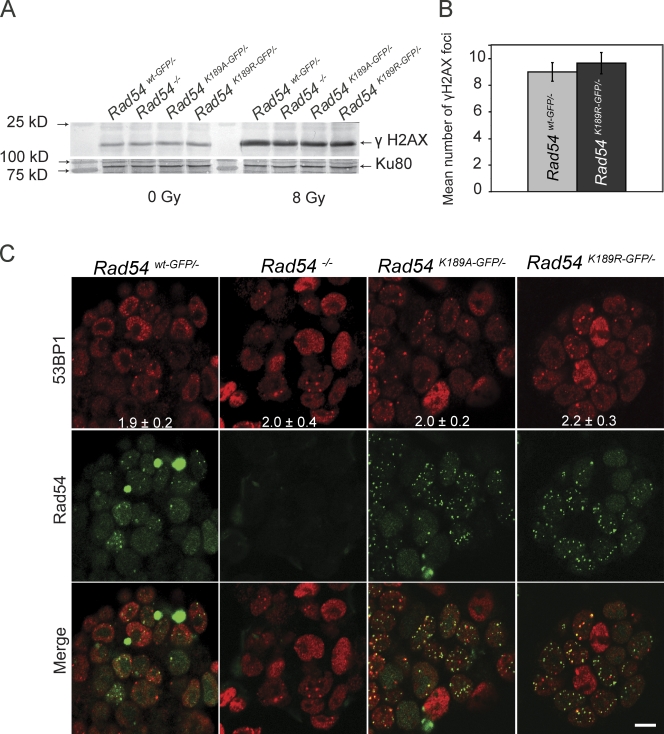
Spontaneous foci, including Rad51 and Rad54, that are observed in unchallenged cells are thought to be present at sites of spontaneous DSBs such as those that might occur at impaired DNA replication forks (Cox et al., 2000; Tarsounas et al., 2003; Budzowska and Kanaar, 2009). Therefore, we asked whether the increased number of spontaneous foci detected in cells was caused by accumulated unrepaired DNA damage in these cells by determining whether the foci colocalized with DNA damage. First, we investigated the levels of γH2AX, an early marker for DSBs, in whole cell extracts from wild-type and mutant cell lines. Anti-γH2AX antibodies recognize a specific phosphorylation on the histone variant H2AX that is triggered by certain types of DNA damage, including DSBs (Rogakou et al., 1998). However, no increase in the level of H2AX phosphorylation was detected by immunoblotting in populations of unchallenged Rad54K189R-GFP/− and Rad54K189A-GFP/− compared with Rad54wt-GFP/− ES cells (Fig. 3 A, left). The cells expressing ATPase-defective Rad54 were able to increase H2AX phosphorylation upon treatment with ionizing radiation (Fig. 3 A, right). In addition, we analyzed H2AX phosphorylation by immunofluorescence experiments (Fig. 3 B). Untreated Rad54wt-GFP/− and Rad54K189R-GFP/− ES cells displayed similar levels of γH2AX foci, which is consistent with the γH2AX immunoblotting results. In addition to γH2AX, we also did not observe an increase in the number of foci for the DNA damage marker 53BP1 (Fig. 3 C). We conclude that the increase in spontaneous Rad54 foci in unchallenged Rad54K189R-GFP/− and Rad54K189A-GFP/− ES cells is unlikely to be caused by a dramatically increased level of unrepaired DNA damage.
Figure 3.
Analysis of γH2AX and 53BP1 in Rad54 ATPase-defective ES cells. (A) Whole cell extracts of ES cells with the indicated genotype were either not treated (left) or harvested 1 h after irradiation with 8 Gy (right) and analyzed by immunoblotting using an anti-γH2AX antibody. Antibodies against Ku80 were used to confirm equal loading (bottom). (B) Quantification of the mean number of γH2AX foci in untreated Rad54wt-GFP/− and Rad54K189R-GFP/− ES cells. Error bars indicates standard error of the mean. (C) Immunofluorescence detection of 53BP1 in untreated ES cells. The top shows 53BP1 staining in Rad54wt-GFP/−, Rad54K189R-GFP/−, and Rad54K189A-GFP/− ES cells, the middle shows the GFP staining, and the panel shows the merged images. Bar, 10 µm.
Lack of ATPase activity of Rad54 affects dynamic interaction with nuclear foci
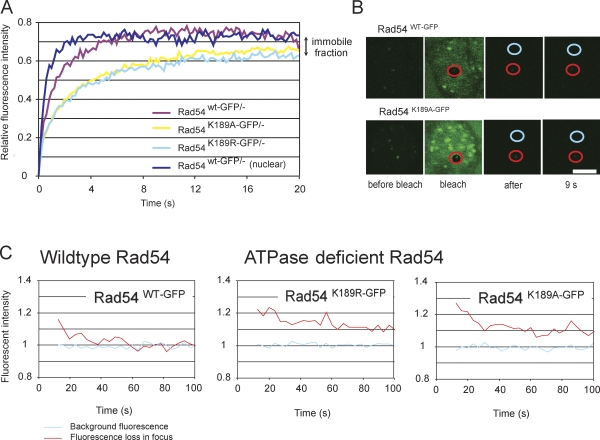
A remarkable feature of DNA damage–induced foci is their highly dynamic nature. We previously showed that these accumulations of proteins are not static but that their components dynamically interact with the structures, with residence times ranging from a couple of seconds for Rad54 to several minutes for Rad51 (Essers et al., 2002b). To determine whether the ATPase activity of Rad54 influences this mobility, we analyzed Rad54 interaction with foci using photobleaching techniques. For this purpose, we used a spot-FRAP protocol in which a small square encompassing a single Rad54-containing focus was bleached and subsequently monitored. We quantified and compared the fluorescence recovery of ATPase-proficient and defective Rad54 foci (Fig. 4 A). The bleaching protocol led to the irreversible bleaching of ∼20% of the total pool of fluorescent RAD54 in the nucleus. In the experiment shown in Fig. 4 A, the final measured fluorescence intensity was normalized to the prebleach pulse fluorescence intensity. The fluorescence in the bleached area recovered to ∼80% for the wild-type Rad54–GFP protein in the bleached focus. In contrast, ATPase-defective Rad54K189R-GFP was present in foci in two distinct kinetic pools; a transiently immobile fraction (∼90%) similar to wild-type Rad54–GFP (Misteli, 2001), and a permanently immobilized fraction (∼10%), not observed in wild type. The t1/2 of ATPase proficient foci was 0.9 ± 0.06 s, which represents a faster recovery as compared with Rad54K189A-GFP/− and Rad54K189R-GFP/− (1.3 ± 0.10 s).
Figure 4.
Photobleaching analysis of Rad54 in foci. (A) Spot-FRAP analysis of Rad54 in foci. A small square containing an individual Rad54 focus was bleached and monitored for fluorescence recovery for each indicated genotype (n = 35). As a control, the fluorescence recovery of non–foci-associated nuclear Rad54 was quantitated (dark blue line). (B) Visualization of iFRAP analysis of Rad54 in foci. The whole cell was bleached excluding a small circular area containing an individual Rad54 focus (red circle). Fluorescence depletion of the nonbleached focus was monitored and compared with the fluorescence level of an unbleached region without a focus (blue circle) for Rad54wt-GFP and Rad54K189A-GFP. Shown here are four frames: before, during, directly after, and 9 s after bleaching. Bar, 5 µm. (C) Quantification of the iFRAP experiment described in B. Graphs represent the fluorescent depletion over time and are based on five individual cells.
To obtain independent confirmation of the dynamic behavior of the wild-type Rad54 and Rad54K189R-GFP proteins in the DNA damage–induced structures, we examined them using inversed FRAP (iFRAP; Houtsmuller and Vermeulen, 2001). In these experiments, the laser pulse used for bleaching fluorescence was aimed to photobleach the fluorescence of a complete nucleus with the exception of one individual focus and a region of similar size in which no focus was present. The fluorescence intensity in the two regions (indicated in Fig. 4 B, red circle and blue circle, respectively) in the nucleus was measured after the bleach pulse. The observed loss of fluorescence of the unbleached focus (Fig. 4 B, red circle) was then monitored and compared with the fluorescence level of the unbleached region without a focus (blue circle). Measurement of the residence time of Rad54 in the nuclear focus revealed a complete exchange of wild-type Rad54 molecules in a focus but a stably associated fraction for the Rad54K189R-GFP mutant, confirming the spot-FRAP measurements (Fig. 4 C). Thus, although the mobility of the vast majority is hardly affected, a fraction of RAD54 proteins is permanently immobilized if it cannot hydrolyze ATP.
The Rad54 protein, but not its ATPase activity, affects Rad51 recruitment to DSBs
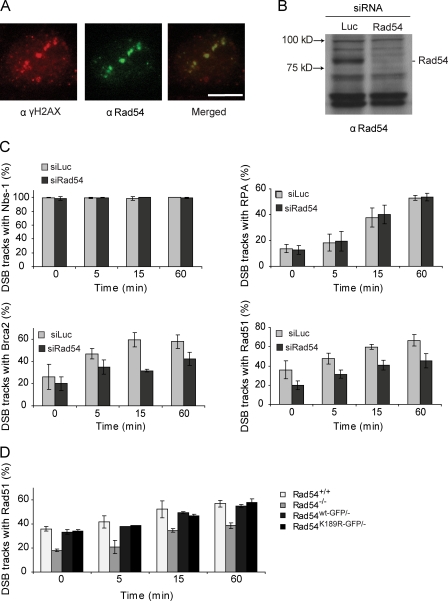
Biochemical experiments have demonstrated that Rad54 stimulates D-loop formation by Rad51 (Petukhova et al., 1998). These and other results have led to the suggestion that Rad54 is important to target the Rad51 nucleoprotein filament to homologous duplex DNA, where it will then engage its ATPase activity to promote repair of the DSB (Heyer et al., 2006). To test whether Rad54 is involved in targeting of Rad51 to the site of DSBs in vivo, we subjected a human osteosarcoma cell line (U2Os) to local α particle irradiation using an 241Am source (Aten et al., 2004; Stap et al., 2008). α particle irradiation can be especially useful to analyze the initial response to DNA damage because it can be used to induce DNA damage locally and in a defined pattern, allowing a clear distinction between already existing foci and local protein accumulations caused by the induced DNA damage. Straight tracks of DSBs due to the interaction of the α particle with chromatin can be visualized using antibodies against γH2AX (Rogakou et al., 1998) or 53BP1 (Bekker-Jensen et al., 2006). Using this method, we showed that Rad54 localized to the DSB sites (Fig. 5 A). Next, we wanted to analyze the effect of the accumulation of other repair proteins to the α tracks in the presence and absence of Rad54. Rad54 was depleted from U2Os cells (Fig. 5 B), and cells were subsequently irradiated with α particles. Over time, there was an increased localization of repair proteins acting early in homologous recombination, including Nbs1 and replication protein A (RPA), as well as Rad51 and BRCA2, to the sites of damage (Fig. 5 C). Upon Rad54 depletion, we observed a transient but considerable delay or partial impairment in the recruitment of Rad51 and BRCA2, but not of RPA and Nbs1, indicating at the cellular level that Rad54 affects a specific step in the progression of homologous recombination. This delayed accumulation of Rad51 at DSBs was also observed in ES cells deficient in Rad54 (Rad54−/−) but, remarkably, was independent of Rad54 ATPase activity, as both Rad54wt-GFP/− and mutant Rad54K189A-GFP/− knockin ES cells showed similar kinetics for Rad51 accumulation at sites of DNA damage as wild-type ES cells (Fig. 5 D). These results show that timely accumulation of Rad51 in foci at sites of DNA damage is dependent on the Rad54 protein but not on its ATPase activity.
Figure 5.
The Rad54 protein, but not its ATPase activity, affects Rad51 recruitment to sites of DSBs. Accumulation of DSB repair proteins at α particle–induced DSB tracks. (A) Localization of Rad54 to the α particle–induced double-stranded break colocalizing with DSB marker γH2Ax. Bar, 5 µm. (B) RAD54 protein levels in U2Os cells transfected with indicated siRNAs. Cell lysates were analyzed by immunoblotting with antibodies against RAD54. Equal sample loading was verified by the equal presence of nonspecific bands. (C) Quantification of accumulation of Nbs1, RPA, Rad51, and BRCA2 at α particle–induced tracks of DSBs 0, 5, 15, and 60 min after irradiation in the presence or absence Rad54. U2Os cells were stained for either γH2Ax (Nbs1 and Rad51) or 53BP1 (RPA and Brca2) as a DSB marker and for one of the indicated repair proteins at t = 0, 5, 15, and 60 min after irradiation. t = 0 indicates the first time point after α particle irradiation. Graphs represent mean percentage of positive DSB tracks with a repair protein. 100 cells containing α particle–induced tracks were scored per experiment. Error bars represent the range of percentages obtained from three independent experiments. (D) Quantification of Rad51 accumulation at DSB sites 0, 5, 15, and 60 min after α particle irradiation in Rad54+/+, Rad54−/−, Rad54wt-GFP/−, and Rad54K189R-GFP/− ES cells. Graphs represent mean percentage of Rad51-positive tracks per γH2Ax track. 100 cells containing damage induced by α particles were scored per experiment. Error bars represent the range of percentages obtained from two independent experiments.
Real-time analysis of Rad54–GFP foci
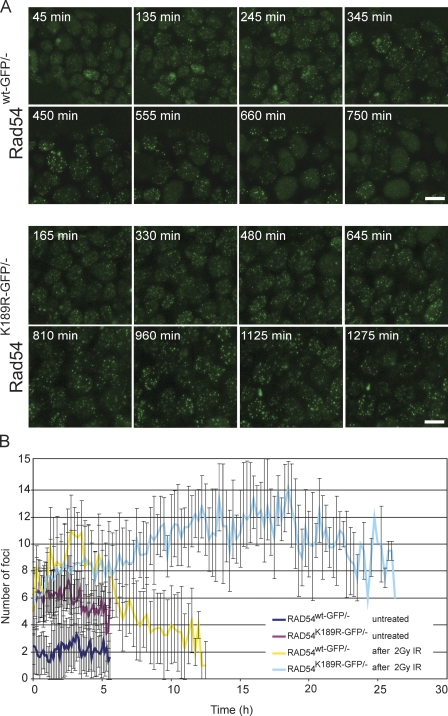
To investigate whether the ATPase activity of Rad54 influenced the kinetics of Rad54 foci, we used a global irradiation protocol using a 137CS source followed by prolonged time-lapse analyses of irradiated cells. To this end, time-lapse movies were made of Rad54wt-GFP/− and Rad54K189R-GFP/− ES cells starting 45 min after irradiation with 2 Gy (Fig. 6 A and Material and methods). In wild-type Rad54wt-GFP/− cells, the number of Rad54 foci per cell increased fivefold upon induction of DNA damage in the first 2 h and then declined. 12 h after irradiation, the number of Rad54 foci was back at the baseline level (Fig. 6 B). Interestingly, the kinetics of ATPase-defective Rad54 foci disappearance was reduced compared with wild-type Rad54 foci, as indicated by a comparison of the time required for a twofold reduction in the number of Rad54 foci from their peak values. This twofold reduction occurred ∼6 h after the irradiation in Rad54wt-GFP/− cells, whereas it took at least 25 h in Rad54K189R-GFP/− ES cells. The number of foci in Rad54 ATPase-defective cells also increased upon irradiation, but over time stayed at much higher levels. This indicates that Rad54’s ATPase affects the chromatin association of the protein and shows that Rad54 ATPase activity specifically influenced its dissociation from foci.
Figure 6.
Quantification of Rad54 foci over time in response to irradiation. (A) Time-lapse imaging of irradiated Rad54wt-GFP/− and Rad54K189R-GFP/− ES cells. Cells were treated with 2 Gy and imaged every 15 min starting 45 min after irradiation. Each picture represents a frame in the resulting movie at the indicated time point. (B) Quantification of the number of foci per cell over time based on the movies represented in A. Quantification was performed using ImageJ as described in the Materials and methods. Error bars indicate SD. Bars, 10 µm.
Quantification of the number of Rad54–GFP molecules
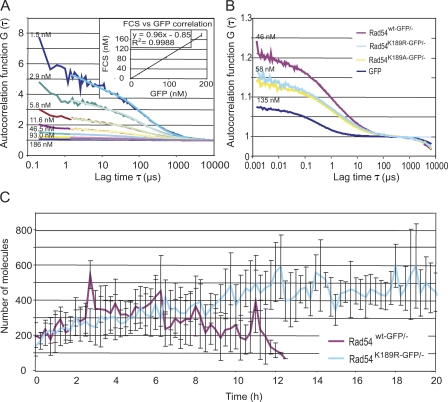
The knockin ES cells expressing the Rad54–GFP fusion protein from the endogenous Rad54 locus allow accurate quantification of the endogenous concentration of Rad54 molecules in an individual cell by FCS and an estimation of the number of molecules per focus. First, we calibrated the instrument using a dilution series of purified GFP protein in solution (Fig. 7 A). We then measured autocorrelation function in cells expressing GFP itself, Rad54wt-GFP, and Rad54K189A-GFP, and determined actual concentration by curve fitting (Fig. 7 B). Subsequently, we determined a mean ES cell nuclear volume and determined the total number of molecules per cell. Next, we used the time-lapse movies of Rad54wt-GFP/− and Rad54K189R-GFP/− ES cells, in which the cells were treated with a dose of 2 Gy and subsequently followed up to 25 h to analyze the mean number of molecules in DNA damage–induced foci (Fig. 6 A). The ratio between free molecules and molecules in foci was determined using a calculated thresholding as described in the Materials and methods to establish the number of molecules in foci and nucleoplasm. The mean number of molecules per focus over time was then plotted against the time period for which the cells could be followed (Fig. 7 C). This analysis revealed that the mean number of molecules in an individual focus varies between 100 and 600 molecules. In Rad54wt-GFP/− cells, the number of molecules increased after damage induction and declined over time. In contrast, the number of molecules also increased in the Rad54K189R-GFP/− mutant cells but then failed to decrease over time.
Figure 7.
FCS concentration measurement. (A) Autocorrelation function G (τ) measured by FCS of increasing concentrations of GFP. Inset indicates GFP concentration measurement by FCS of purified GFP in solution plotted as FCS concentration (nM) versus GFP concentration (nM). (B) Autocorrelation function G (τ) measured by FCS in Rad54wt-GFP/−, Rad54K189R-GFP/−, and Rad54K189A-GFP/− ES cells. As a control, cells expressing free untagged GFP (GFP) were used. (C) Quantification of the number of Rad54–GFP molecules after treatment of cells with 2 Gy, either wild type or K189R mutant, in a single focus over time. Error bars indicate SD.
Rad54 influences relocalization of DSBs to the nuclear periphery
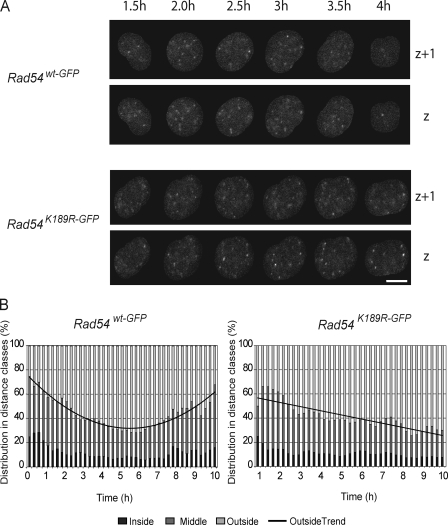
In yeast, persistent DNA breaks are fixed over time at the nuclear periphery (Kalocsay et al., 2009; Oza et al., 2009; Oza and Peterson, 2010). Our targeted knockin approach allowed tracking of the nuclear localization of Rad54 foci over time to determine their cellular localization in mammalian stem cells. To this end, we used the time-lapse movies recorded after DNA damage induction by ionizing radiation described and analyzed in Fig. 6 (A and B). In cells followed for at least 15 consecutive time points, the relocalization of the DNA damage–induced foci was analyzed (Fig. 8 A). For each focus, the center of mass was determined in 2D. Next, the distance of the focus location to the center of mass of the cell was calculated. The foci were then classified in three distance classes: “inside,” distance <2 µm from the center of cell; “middle,” distance between 2 and 4 µm from the center of the cell; and “outside,” the distance >4 µm from the center of the cell (Fig. 8 B). Dividing the cell into circles with those radii results in mean volumes of 34 µm3 (inside), 235 µm3 (middle), and 181 µm3 (outside). In wild-type cells, the number of foci in the outside distance class increases temporarily during the repair process, as indicated by the observation that Rad54 foci relocalized to the nuclear periphery and back to their starting positions over time (Fig. 8 B). In contrast, ATPase-defective Rad54 foci localized to the nuclear periphery but persisted at this position, which indicates impaired relocalization of DSB repair. However, this could also indicate the persistence of foci in less accessible peripherally localized heterochromatin.
Figure 8.
Nuclear relocalization of Rad54–GFP foci in response to DNA damage. (A) Grayscale representation of the nuclear redistribution of Rad54 foci in a wild-type or ATPase-defective Rad54–GFP cell after treatment with ionizing radiation using a 137Cs source. Shown are a subset of time points in 2 (out of 10) z axes of a single cell. Bar, 5 µm. (B) Quantification of A. The relative distribution of foci in distance classes; inside (0–2 µm), middle (2–4 µm), and outside (> 4 µm) followed over time. Graph indicates the best fitting distribution curve of outside foci in Rad54wt-GFP/− and Rad54K189R-GFP/− cells.
Discussion
Rad54 is a multifunctional protein that possesses several different activities that promote the progression of homologous recombination, an accurate pathway of repairing DSBs (Tan et al., 2003). In addition to a close functional interaction with Rad51, the central protein of homologous recombination, Rad54 also displays potent ATPase activity, which allows it to translocate along DNA. This allows the protein to change the conformation of the template DNA, thereby perturbing DNA structures and influencing protein–DNA interactions (Heyer et al., 2006). Here, we have investigated the effect of a mutation in the ATPase domain of Rad54 on its cellular behavior.
The primary results of this study are as follows. First, the ATPase activity of Rad54 influences the number of spontaneous Rad54 foci in unchallenged cells, as ATPase-defective Rad54 cells contain more of them compared with cells expressing wild-type Rad54. Second, the Rad54 ATPase-defective cells also display an increase in spontaneous Rad51 foci. However, the increase of foci containing homologous recombination proteins does not correspond to an increase in DNA damage. Third, the ATPase activity of Rad54 is not required for the formation of foci induced by DNA damaging agents. Fourth, Rad54 but not its ATPase activity is required to accumulate Rad51 at sites of DSBs in a timely fashion. Fifth, an immobile fraction of ATPase-defective Rad54 molecules occurs in foci. Sixth, time-lapse studies revealed that the disappearance of DNA damage–induced foci is delayed when Rad54’s ATPase activity is attenuated, and these ATPase-defective Rad54 DNA repair foci are stuck at the nuclear periphery.
The ATPase activity of Rad54 is essential for its DNA repair function in vivo
To address the importance of the ATPase activity of Rad54, we generated mouse ES cells that express ATPase-defective mutants from the endogenous Rad54 locus fused to a carboxy-terminal GFP tag (Fig. 1), ensuring physiological levels of mutant protein (Fig. 1 C). The DNA damage sensitivity profiles of the ATPase-defective Rad54 mutants are similar to cells lacking Rad54 altogether in terms of hypersensitivity to mitomycin C and ionizing irradiation (Fig. 1 D). The damage hypersensitivities of the Rad54 ATPase-defective cells probably result from defective homologous recombination because Rad54 ATPase-defective and Rad54−/− ES cells are equally impaired in homologous recombination (Table I). This indicates that the Rad54 ATPase activity is required for the type of recombination that is used to repair breaks induced by irradiation and mitomycin C, as well to effectively homologously integrate a linear piece of DNA into the genome. Furthermore, this indicates that both binding and hydrolysis of ATP is essential because ATPase mutants (K189A and K189R) display similar phenotypes. These experiments reveal the essential role of the Rad54 ATPase function in mammalian cells and its in vivo importance for DNA repair.
Differential cellular behavior of ATPase-proficient and -defective Rad54
The remarkable feature of Rad54K189R-GFP/− and Rad54K189A-GFP/− cells is the presence of an elevated number of spontaneous, “uninduced” Rad54 foci in their nuclei compared with Rad54wt-GFP/− cells (Fig. 2 A). A clear phenotypic difference in the cell biology of cells lacking Rad54 altogether and cells that express ATPase mutants is the corresponding elevated number of Rad51 foci in ATPase mutant cells (Fig. 2 B). Thus, in addition to causing an increase in its own foci in unchallenged cells, the inability of Rad54 to hydrolyze ATP effectively also causes an increase in the foci of its partner protein, Rad51. Thus it is possible that the ATPase activity of Rad54 is involved in turning over Rad51 in the foci, which is consistent with biochemical experiments demonstrating that Rad54 can displace Rad51 from double-stranded DNA (Solinger et al., 2002). Yet, in the absence of Rad54, no increase in Rad51 is detected, although its stability is affected (Tan et al., 1999; van Veelen et al., 2005). It is possible that when Rad54 is completely absent, a redundant protein can act at the site of the DSB, which might take over Rad54 function with respect to removal of Rad51 but not with respect to DSB repair, as this is still impaired in knockout cells. A candidate protein for this function is the Rad54 paralogue Rad54B (Wesoly et al., 2006). The presence of the ATPase-defective Rad54 protein would then dominantly affect this aspect of Rad54B activity.
The increase in the number of Rad54–GFP foci does not correlate with an increase in spontaneous DNA damage in Rad54 ATPase-defective cells, as no increase in the DNA damage marker γH2AX can be detected in Rad54 ATPase-defective cells versus Rad54 ATPase-proficient cells (Fig. 3 A). Consistently, when analyzed by immunofluorescence, no increase in the number of 53BP1 and γH2AX foci can be detected (Fig. 3, B and C). Thus, within the limitations of these techniques, the level of DNA damage is not significantly different in mutant, wild-type, and knockout cells, showing that the elevated number of spontaneous foci is not caused by an increased number of unrepaired breaks. We cannot exclude the possibility that replication-associated DSBs generated because of endogenous damage are efficiently repaired by Rad51, but the complex of Rad54 remains attached after the repair has been completed and therefore Rad54 and Rad51 foci persist. Our data are consistent with the absence of an overt proliferation defect as well as unaffected genomic stability of the cells expressing ATPase-defective Rad54 and cells lacking Rad54 (unpublished data). In addition, the immunofluorescence experiments show that the ATPase activity of Rad54 is not important for localization of Rad54 and Rad51 to sites of DNA damage. Untreated ES cells contain many more γH2AX foci than Rad54 foci and therefore not all γH2AX colocalize with Rad54. However, most if not all Rad54 foci, including those containing ATPase-defective Rad54, are at sites marked by γH2AX (unpublished data).
ATPase-deficient Rad54 is partially immobilized in nuclear DNA repair foci
The nature and function of foci is still ambiguous. Many models of the composition of a focus have been postulated. It is possible that, as has been put forth for yeast, the mammalian foci are also a reflection of the so-called “repair centers” (Lisby et al., 2004), but this premise lacks clear-cut evidence in mammalian cells. Foci have also been assumed to represent the one or all of the various stages of recombination, and are therefore not a clear method of distinguishing between the different stages of recombination. It is still not clear why such a high local concentration of protein is required at the site of damage, especially because in biochemical studies, the optimal ratios of Rad51 and Rad54 are not necessarily 1:1 (Jiang et al., 1996; Clever et al., 1997; Golub et al., 1997). However, the presence and quantification of foci have been used as an indication of repair activity because foci form within a short time in response to DNA-damaging agents, and decrease in number over time. A focus is a highly dynamic structure, with active and rapid association and disassociation of proteins, particularly Rad54 (Essers et al., 2002b). Our FCS analysis revealed that the mean number of molecules in an individual focus varies between 100 and 600 molecules (Fig. 7). This number, however, is higher than expected based on the few Rad54 molecules predicted to be necessary to repair a single DSB. These results implicate that not all molecules present in a foci are necessary to repair the break. Attracting and maintaining Rad54 molecules at the site of the break apparently continues until the break is repaired, as seen by the damage-induced increase in the number of molecules over time (Fig. 7). These foci might simply continue growing because the signal that locates Rad54 to the site of damage continues to signal. Furthermore, our FRAP data show that a small fraction of the molecules in the mutant are stably bound to the chromatin (the immobile fraction) (Fig. 4), which suggests that only a minority of molecules present in foci are functional in DNA DSB repair. Several interesting questions remain. Why are there 10-fold more molecules present in foci than necessary for repair? What role do the excess molecules have? Is it simply a mechanism of trial and error, the higher the local concentration the higher the efficiency of repair, or is there an underlying reason that remains to be discovered in the future?
The molecular processes required for the accumulation and disassembly of homologous recombination proteins in foci at sites of DNA damage are not well understood. Cytological studies in yeast, chicken DT-40 cells, and mouse ES cells suggest that Rad54 is not necessary for the formation of Rad51 foci (Shinohara et al., 2000; Takata et al., 2000; Lisby et al., 2004; Miyazaki et al., 2004; van Veelen et al., 2005). We show that the Rad54 but not its ATPase activity is required to accumulate Rad51 and BRCA2 into foci in a timely fashion (Fig. 5). However, Rad54 does not influence the accumulation of Nbs1 and RPA. These results establish the role of Rad54 downstream of break resection and indicate that Rad54 is mainly involved in stages of recombination performed by Rad51 and BRCA2. These results resemble the biochemical properties of Rad54. We know from biochemical experiments that Rad54 can load Rad51 on the filaments. This activity is independent of Rad54 ATPase activity, as shown in yeast experiments (Wolner and Peterson, 2005). Genetic studies have demonstrated that the ATPase activity is crucial for in vivo Rad54 function, and we have shown that Rad54 K189R mutants display DNA damage sensitivities equivalent to the deletion mutant (Fig. 1). In contrast, the disassembly process is affected by Rad54-mediated hydrolysis of ATP (Fig. 7). Clearance of foci induced by DNA damage takes about twice as long in Rad54K189R-GFP/− cells compared with Rad54wt-GFP/−. These foci especially persist close to the nuclear periphery (Fig. 8). The temporal resolution we used in these time-lapse experiments was too low to actually track individual foci. The temporal resolution could not be increased because of the sensitivity of ES cells to laser irradiation and bleaching of the fluorescent signal. We can therefore not distinguish whether DNA damaged–induced relocalization of foci over time is impaired or whether DNA damage–induced foci are immobile and persist longer closer to the nuclear periphery in ATPase-deficient cells (Soutoglou et al., 2007). ATPase-dependent Rad54 translocation might be required for local chromatin decondensation observed around DSBs (Kruhlak et al., 2006; Kim et al., 2007) because compact heterochromatin structure at the nuclear periphery could inhibit access of repair proteins to DSBs or DNA strand exchange. However, because the persisting foci are found in the vicinity of the nuclear border, it is also possible that the transient increase in Rad54 foci in this area reflects migration of damaged DNA toward low-density chromatin, resulting in concentration at the border of condensed DNA, as is also the case for γH2AX foci in ataxia telangiectasia mutated (ATM)-deficient cells (Goodarzi et al., 2008). This regulation of DSB relocalization to the nuclear periphery has also been observed for yeast Rad51 (Kalocsay et al., 2009; Oza et al., 2009; Oza and Peterson, 2010) and indicates that the prolonged presence of mutant Rad54 at the periphery reflects the presence of delayed repair of DSBs. It is not unexpected for a protein in the SWI2/SNF2 family to affect this feature of foci. Just as genuine chromatin remodeling motor proteins affect histone DNA interactions, Rad54’s motor activity, in conjunction with its direct interaction with Rad51, might be well suited to deal with accumulations of homologous recombination proteins on chromatin after the repair process has been completed.
Mutations that attenuate the ATPase activity of Rad54 are separation-of-functions alleles
The ATPase activity of Rad54 is essential for many of its biochemical activities (Tan et al., 2003), but its effect on the cellular behavior of the protein is unknown. Using photobleaching experiments in living cells, we determined previously that all of the homogenously distributed Rad54–GFP in the nucleoplasm is highly mobile (Essers et al., 2002b). The combination of the time-lapse and photobleaching experiments provides additional insight into foci biology. The experiments presented in Fig. 4 reveal that although the ATPase activity of Rad54 slightly affects its effective diffusion rate, it renders a fraction of ∼10% of molecules immobile in Rad54K189A-GFP/− and Rad54K189R-GFP/− cells compared with Rad54wt-GFP/− cells. The result of this is that it takes about twice as long to repopulate the entire focus. A DNA damaged–induced Rad54 focus disappears half as fast, and the time it takes to repopulate a focus takes twice as long when Rad54 cannot hydrolyze ATP. The net effect is that on average, the same number of Rad54 molecules associated with a single focus over time in wild-type and mutant cells, even though DNA repair is inoperative in the mutant cells.
Our study shows that Rad54’s ATPase activity is important for DNA repair and recombination, but, interestingly, that it also affects its cellular behavior. The ATPase activity is required for release of the protein from DNA damaged–induced structures on chromatin, not only of itself but also of Rad51. The fact that the ATPase activity of Rad54 affects its cellular behavior is interesting because this activity is only triggered when it is bound to DNA. Mutations that attenuate the ATPase activity of Rad54 are likely to be separation-of-function alleles that differentially affect the behavior of the pool of Rad54 in a focus that is bound to DNA versus the pool that is not bound to DNA. Rad54 molecules that are not bound to DNA are not hydrolyzing ATP, are therefore not actively engaged in repair, and can still reversibly interact with the focus. However, the Rad54 molecules bound to DNA but no longer capable of hydrolyzing ATP appear to lose the ability to quickly turn over in the focus. Our observations are of interest in the context of the number of each homologous recombination protein required for DNA repair, which is much less than are present in a focus based on biochemical experiments. Thus, our experiments reveal the need to develop a cellular system that allows for identification and tracking of individual molecules in a crowd, and the ability to separate those that do the work from those that do not.
Materials and methods
Cell culture
The mouse ES cells used to generate cells expressing ATPase-defective Rad54 had the genotype Rad54wt-HA/−, where one allele is disrupted and the other expresses HA-tagged Rad54 from the endogenous Rad54 locus (Tan et al., 1999). ES cells were cultured on gelatin-coated dishes in a 1:1 mixture of DME and buffalo rat liver (BRL)-conditioned medium, supplemented with 10% (vol/vol) FBS (Thermo Fisher Scientific), 0.1 mM nonessential amino acids (Biowhittaker; Lonza), 50 mM β-mercaptoethanol (Sigma-Aldrich), and 500 U ml−1 leukemia inhibitory factor. U2Os cells were cultured in a 1:1 mixture of DME and Ham’s F10, supplemented with 10% (vol/vol) fetal calf serum (Thermo Fisher Scientific) and streptomycin/penicillin at 37°C in an atmosphere containing 5% CO2.
Antibodies
The primary antibodies used in this study were: anti-Rad51 (rabbit polyclonal; van Veelen et al., 2005), anti-Rad54 (rabbit polyclonal; Essers et al., 1997), anti-γH2AX (Millipore), anti-53BP1 (rabbit polyclonal; Novus Biologicals), anti-Rad54 (goat polyclonal, D-18; Santa Cruz Biotechnology, Inc.), anti-NBS1 (goat polyclonal, C-19; Santa Cruz Biotechnology, Inc.), anti-RPA34 (mouse monoclonal, Ab-2; Oncogene), and anti-BRCA2 (mouse monoclonal, Ab-1; EMD). The secondary antibodies conjugated with alkaline phosphatase were obtained from Roche. The horseradish peroxidase–conjugated antibodies were obtained from Jackson ImmunoResearch Laboratories, and relevant Alexa Fluor secondary antibodies were obtained from Invitrogen.
Generation of ES cells carrying knockin alleles expressing ATPase-defective Rad54 protein
The wild-type and ATPase-deficient Rad54–GFP knockin constructs were designed to obtain expression of tagged Rad54 from the endogenous promoter upon homologous integration. The wild-type construct was generated by fusing exons IV–XVII of the hRAD54 cDNA, but with the omission of the STOP codon, to a DNA encoding a 3′-terminal GFP tag, followed by a poly(A) signal (p(A)) and a phosphoglycerate kinase (PGK) promoter–driven puromycin selectable marker gene. This fragment was subcloned into exon IV of a 9-kb EcoRI genomic Rad54 fragment. Using linkers, a downstream SfuI site was introduced. Digestion of this construct with SfuI yielded a fragment containing the 3′-terminal part of the Rad54–GFP cDNA spanning exons IV–XVII and the puromycin gene. This fragment was subcloned into the unique SfuI site in exon IV of a 9-kb EcoRI fragment of the mouse Rad54 genomic sequence containing exons IV–VII in pBluescript II KS. The Rad54 mutant constructs expressing the K189R and K189A mutation were generated in a similar way after introduction of these mutations in the wild-type hRAD54 cDNA using the following primers: 5′-GGGCCTAGGAAGGACGCT-3′ (K189R) and 5′-GGGCCTAGGAGCAACGCT-3′ (underlines indicate the mutant codon in Rad54). The constructs are schematically depicted in Fig. 1 A. Targeting constructs bearing either the K189A or K189R mutation (Fig. 1) were purified as plasmids and linearized with PvuI, then purified using electro-elution, phenol extraction, and ethanol precipitation. These linearized constructs were then electroporated into Rad54wt-HA/− cells, using a 2-mm cuvette, at 117 V, 1200 μF, and left for 10 ms in an ECM 830 electroporator (BTX). Replacement of the Rad54HA locus would generate ES cells with genotypes Rad54K189A-GFP/− or Rad54K189R-GFP/−. 24 h after electroporation, cells were subjected to puromycin selection (1 µg/ml). 100 puromycin resistant colonies were isolated for each construct and their DNA was analyzed for homologous integration of the knockin constructs in the Rad54HA locus by DNA blotting using a probe recognizing exons VII and VIII. Genomic sequence analysis was performed to confirm the correct integration and presence of mutations in the Rad54 locus, and protein expression was subsequently analyzed by immunoblotting. DNA damage sensitivities of the cells were assessed by performing clonogenic survival assays as described previously (Essers et al., 1997). For ionizing radiation, dishes were treated right away with the indicated dosage. For the rest, cells were allowed to attach for 12–16 h before treatment. Mitomycin C was added for 1 h before washing. Finally, as a measure of homologous recombination efficiency, the frequency of homologous versus random integration of gene targeting constructs in the Rb locus was determined as described previously (Niedernhofer et al., 2001).
Ionizing radiation and immunofluorescence
ES cells were seeded on a feeder layer of lethally irradiated (70–80%) confluent mouse embryonic fibroblasts and left to attach overnight. Cells were irradiated with the indicated doses of ionizing radiation using a 137Cs source and left to recover for the indicated amount of time. α particle irradiation was performed using a 241Am source as described previously (Stap et al., 2008). In brief, U2Os cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions with siRNA against luciferase (5′-CGUACGCGGAAUACUUCGAdTdT-3′) or Rad54 (5′-GAACUCCCAUCCAGAAUGAUU-3′) for 48 h. After 24 h, cells were plated on a 1.8 µm-thick polyester membrane (Mylar), and transfection was repeated. In the case of ES cells, Mylar dishes were coated with carbon atoms (SC500 sputter coater; EMscope) and gelatin, and cells were subsequently plated 24 h before irradiation. Cells were irradiated with α particles and subsequently fixed and stained for immunofluorescence at indicated time points as described previously (Aten et al., 2004). In short, cell cultures were fixed by incubation in a 2% paraformaldehyde (Electron Microscopy Sciences) solution in PBS for 15 min at 21°C and washed three times with PBS. Before immunochemical staining, the cells were incubated for 30 min in TNBS (PBS supplemented with 0.1% Triton X-100 and 1% fetal calf serum) to improve their permeability. Cells were then incubated for 90 min in PBS+ (PBS supplemented with 1% fetal calf serum) containing the primary antibodies. After washing twice with TNBS for 5 min, cells were incubated in PBS+ containing the secondary antibodies, after which they were stained with Hoechst 33342 in a final concentration of 5 µg/ml for 10 min. After washing again with TNBS, a droplet of Vectashield (Invitrogen) was placed on top of the stained cells, and the cells were covered with a coverslip. The piece of Mylar containing the stained cells was then cut out and was placed, together with the coverslip, on a slide. The piece of Mylar with the coverslip on top of it was glued to the slide using rubber cement. In case of Rad51 staining, preextraction for 1 min with Triton X-100 buffer (0.5% Triton X-100, 20 mM Hepes-KOH, pH 7.9, 50 mM NaCl, 3 mM MgCl2, and 300 mM sucrose) was performed (Petrini, 2000). α particle tracks were visualized with a confocal laser scanning microscope (LSM 510 META; Carl Zeiss, Inc.) consisting of an inverted microscope (Axiovert 100) equipped with an Argon gas laser (visualizing Alexa Fluor 488, green) and a helium neon laser (visualizing Alexa Fluor 543, red). Images were taken with 63× Plan-Apochromat 1.4 NA oil immersion lens (Carl Zeiss, Inc), on a single plane of ∼1 µm thickness, through the middle of the cell.
Immunoblotting
Whole cell extracts were prepared by lysing cells with SDS sample buffer (2% SDS, 10% glycerol, and 60 mM Tris-HCl, pH 6.8) 48 h after transfection. After the protein concentration was determined by a Lowry protein assay, extracts were supplemented with 0.5% β-mercaptoethanol and 0.02% bromophenol blue. After fractionation by SDS-PAGE, proteins were transferred to polyvinylidene fluoride membrane. The blots were blocked with PBS/3% skimmed milk/0.1% Tween 20 and probed with primary antibodies. After washing with PBS/0.1% Tween 20, the membranes were probed with relevant secondary antibodies and developed with ECL Western blotting detection reagents (GE Healthcare).
Live cell imaging, semi-automated foci counting, and foci tracking
ES cells were grown overnight on lethally irradiated MEF feeder layers on 24-mm round coverslips. Cells that grew to an ∼70% confluent monolayer were irradiated with 2 Gy and transferred to a specially adapted chamber fitted to the confocal microscope 45 min after irradiation, where they could be maintained at 37°C with 5% CO2. Using a macro for automated time-lapse imaging, the cells were imaged taking 10 z slices (covering 9 µm in total) every 15 min. Movies were analyzed in the AIM image browser (Carl Zeiss, Inc.) and exported for detailed analysis using ImageJ software (http://rsb.info.nih.gov/ij/).
Time-lapse imaging of individual cells
The low-contrast in the images of the Rad54 knockin cells between green nuclei and background prohibited automated segmentation of the ES cells. A custom made ContourJ selection tool was used to select all individual cells by hand (available at http://www.imagescience.org/meijering/software/contourj/). Then, the center of mass (x,y location) for each cell was determined. When the distance of the center of mass in two consecutive time points was <3 µm, the cell was regarded as being the same cell. In this way cells could be followed in time. Those cells that could be tracked for at least 15 time points were selected for detailed foci analyses. Each selected cell was then processed to determine the number of foci in the 3D stack. This number per cell was determined as described previously (van Royen et al., 2007). In brief, for each cell, the mean intensity and SD was determined. Foci were selected based on fluorescence intensity values above mean + 1.5× the SD. Visual inspection of the images showed that this method detected the majority of the foci. From the resulting image, the number of foci was counted using the particles analysis function of ImageJ. Duplicate foci in different z planes were automatically removed from the counting. The data thus obtained was plotted as number of foci per cell against the time during which the cells could be followed.
FCS experiments
A microscope system (LSM510 confocor 2; Carl Zeiss, Inc.) was used for the FCS experiments. Data were analyzed with the SSTC data processor (Scientific Software Technologies Center). Every cell was measured five times for 20 s. The raw data were autocorrelated, and the autocorrelation curves were analyzed as either a one-component free diffusion triplet state model (GFP in solution) or a two-component free diffusion triplet state model (Rad54–GFP) in cells. The models were used to determine the diffusion time, i.e., the time it takes a molecule to move through the confocal laser spot and the total number of molecules in the diffraction limited spot. To be able to estimate the concentration of RAD54 molecules in a cell nucleus, the volume of the diffraction limited spot was determined using standard Rhodamine 6G (Invitrogen) and GFP solutions with known concentrations (Weisshart et al., 2004). A mean ES cell nuclear volume was determined by measuring two perpendicular diameters in the confocal plain with the largest size of the nucleus (n = 37; 9.5 ± 1.2 [mean ± SD] µm/cell). The volume of individual cells was estimated by regarding the nucleus as an ellipsoid object of which the volume was determined using the following formula: volume = (π × d1 × d2 × (d1 + d2)/2)/6, in which d1 and d2 are the two perpendicular diameters. The mean volume of the nuclei was 453 ± 190 µm3 (mean ± SD, n = 39). Using the FCS determined concentration and the estimated volume, the total number of molecules per nucleus was calculated. From the confocal planes, we determined the ratio between the fluorescent signal coming from the foci and the nucleus in total. With the total number of molecules per cell known and the percentage of the signal coming from the foci, the number of molecules per focus was calculated (plotted in Fig. 7 C).
Photobleaching experiments
To determine the mobility of free Rad54 protein, FRAP experiments were performed where fluorescence of individual nuclear foci was bleached as described previously (Essers et al., 2002b). To determine the dynamic interaction of the Rad54 protein with nuclear foci in the ES cells, an area of 25 × 25 pixels containing a focus was bleached at 80% laser intensity, and fluorescence recovery was measured in the area at 0.2-s intervals for 20 s. 60 cells were monitored for each genotype in three independent experiments. Wild-type ES cells stably transfected with pGK-GFP-p(A) were used as a positive control for fluorescence recovery, where cells with comparable fluorescence level as the knockin cells were chosen and treated as detailed for the Rad54 knockin cells.
Complementary iFRAP experiments were performed on a confocal microscope (TCS SP5; Leica) with a 63× oil Plan-Apochromat 1.4 NA oil immersion lens (Dundr et al., 2002). All fluorescence in an individual nucleus, with exception of a small region containing a single focus, was bleached, and the decrease in fluorescence in the nonbleached focus was recorded. For each time point, the relative intensity was calculated as follows: Irel;t = (It − BG)/(I0 − BG), where I0 is the mean intensity of the region of interest before bleaching and BG is the background signal.
Acknowledgments
We thank Nicole van Vliet for expert technical assistance.
This work was supported by a grant from the Dutch Cancer Society and a TOP grant from the Netherlands Organization for Scientific Research (NWO) and the Netherlands Genomics Initiative/NWO.
Footnotes
Abbreviations used in this paper:
- DSB
- double-strand break
- ES
- embryonic stem
- FCS
- fluorescence correlation spectroscopy
- iFRAP
- inversed FRAP
- RPA
- replication protein A
References
- Agarwal S., Tafel A.A., Kanaar R. 2006. DNA double-strand break repair and chromosome translocations. DNA Repair (Amst.). 5:1075–1081 10.1016/j.dnarep.2006.05.029 [DOI] [PubMed] [Google Scholar]
- Alexeev A., Mazin A., Kowalczykowski S.C. 2003. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat. Struct. Biol. 10:182–186 10.1038/nsb901 [DOI] [PubMed] [Google Scholar]
- Alexiadis V., Kadonaga J.T. 2002. Strand pairing by Rad54 and Rad51 is enhanced by chromatin. Genes Dev. 16:2767–2771 10.1101/gad.1032102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amitani I., Baskin R.J., Kowalczykowski S.C. 2006. Visualization of Rad54, a chromatin remodeling protein, translocating on single DNA molecules. Mol. Cell. 23:143–148 10.1016/j.molcel.2006.05.009 [DOI] [PubMed] [Google Scholar]
- Aten J.A., Stap J., Krawczyk P.M., van Oven C.H., Hoebe R.A., Essers J., Kanaar R. 2004. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 303:92–95 10.1126/science.1088845 [DOI] [PubMed] [Google Scholar]
- Bassing C.H., Alt F.W. 2004. The cellular response to general and programmed DNA double strand breaks. DNA Repair (Amst.). 3:781–796 10.1016/j.dnarep.2004.06.001 [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S., Lukas C., Kitagawa R., Melander F., Kastan M.B., Bartek J., Lukas J. 2006. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 173:195–206 10.1083/jcb.200510130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezzubova O., Silbergleit A., Yamaguchi-Iwai Y., Takeda S., Buerstedde J.M. 1997. Reduced X-ray resistance and homologous recombination frequencies in a RAD54-/- mutant of the chicken DT40 cell line. Cell. 89:185–193 10.1016/S0092-8674(00)80198-1 [DOI] [PubMed] [Google Scholar]
- Budzowska M., Kanaar R. 2009. Mechanisms of dealing with DNA damage-induced replication problems. Cell Biochem. Biophys. 53:17–31 10.1007/s12013-008-9039-y [DOI] [PubMed] [Google Scholar]
- Bugreev D.V., Mazina O.M., Mazin A.V. 2006. Rad54 protein promotes branch migration of Holliday junctions. Nature. 442:590–593 10.1038/nature04889 [DOI] [PubMed] [Google Scholar]
- Clever B., Interthal H., Schmuckli-Maurer J., King J., Sigrist M., Heyer W.D. 1997. Recombinational repair in yeast: functional interactions between Rad51 and Rad54 proteins. EMBO J. 16:2535–2544 10.1093/emboj/16.9.2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox M.M., Goodman M.F., Kreuzer K.N., Sherratt D.J., Sandler S.J., Marians K.J. 2000. The importance of repairing stalled replication forks. Nature. 404:37–41 10.1038/35003501 [DOI] [PubMed] [Google Scholar]
- Dronkert M.L., Beverloo H.B., Johnson R.D., Hoeijmakers J.H., Jasin M., Kanaar R. 2000. Mouse RAD54 affects DNA double-strand break repair and sister chromatid exchange. Mol. Cell. Biol. 20:3147–3156 10.1128/MCB.20.9.3147-3156.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M., Hoffmann-Rohrer U., Hu Q., Grummt I., Rothblum L.I., Phair R.D., Misteli T. 2002. A kinetic framework for a mammalian RNA polymerase in vivo. Science. 298:1623–1626 10.1126/science.1076164 [DOI] [PubMed] [Google Scholar]
- Essers J., Hendriks R.W., Swagemakers S.M., Troelstra C., de Wit J., Bootsma D., Hoeijmakers J.H., Kanaar R. 1997. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell. 89:195–204 10.1016/S0092-8674(00)80199-3 [DOI] [PubMed] [Google Scholar]
- Essers J., Hendriks R.W., Wesoly J., Beerens C.E., Smit B., Hoeijmakers J.H., Wyman C., Dronkert M.L., Kanaar R. 2002a. Analysis of mouse Rad54 expression and its implications for homologous recombination. DNA Repair (Amst.). 1:779–793 10.1016/S1568-7864(02)00110-6 [DOI] [PubMed] [Google Scholar]
- Essers J., Houtsmuller A.B., van Veelen L., Paulusma C., Nigg A.L., Pastink A., Vermeulen W., Hoeijmakers J.H., Kanaar R. 2002b. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J. 21:2030–2037 10.1093/emboj/21.8.2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers J., Houtsmuller A.B., Kanaar R. 2006. Analysis of DNA recombination and repair proteins in living cells by photobleaching microscopy. Methods Enzymol. 408:463–485 10.1016/S0076-6879(06)08029-3 [DOI] [PubMed] [Google Scholar]
- Friedberg E.C., McDaniel L.D., Schultz R.A. 2004. The role of endogenous and exogenous DNA damage and mutagenesis. Curr. Opin. Genet. Dev. 14:5–10 10.1016/j.gde.2003.11.001 [DOI] [PubMed] [Google Scholar]
- Game J.C., Mortimer R.K. 1974. A genetic study of x-ray sensitive mutants in yeast. Mutat. Res. 24:281–292 10.1016/0027-5107(74)90176-6 [DOI] [PubMed] [Google Scholar]
- Golub E.I., Kovalenko O.V., Gupta R.C., Ward D.C., Radding C.M. 1997. Interaction of human recombination proteins Rad51 and Rad54. Nucleic Acids Res. 25:4106–4110 10.1093/nar/25.20.4106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi A.A., Noon A.T., Deckbar D., Ziv Y., Shiloh Y., Löbrich M., Jeggo P.A. 2008. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell. 31:167–177 10.1016/j.molcel.2008.05.017 [DOI] [PubMed] [Google Scholar]
- Haaf T., Golub E.I., Reddy G., Radding C.M., Ward D.C. 1995. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl. Acad. Sci. USA. 92:2298–2302 10.1073/pnas.92.6.2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer W.D., Li X., Rolfsmeier M., Zhang X.P. 2006. Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res. 34:4115–4125 10.1093/nar/gkl481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers J.H. 2001. Genome maintenance mechanisms for preventing cancer. Nature. 411:366–374 10.1038/35077232 [DOI] [PubMed] [Google Scholar]
- Houtsmuller A.B., Vermeulen W. 2001. Macromolecular dynamics in living cell nuclei revealed by fluorescence redistribution after photobleaching. Histochem. Cell Biol. 115:13–21 [DOI] [PubMed] [Google Scholar]
- Jaskelioff M., Van Komen S., Krebs J.E., Sung P., Peterson C.L. 2003. Rad54p is a chromatin remodeling enzyme required for heteroduplex DNA joint formation with chromatin. J. Biol. Chem. 278:9212–9218 10.1074/jbc.M211545200 [DOI] [PubMed] [Google Scholar]
- Jiang H., Xie Y., Houston P., Stemke-Hale K., Mortensen U.H., Rothstein R., Kodadek T. 1996. Direct association between the yeast Rad51 and Rad54 recombination proteins. J. Biol. Chem. 271:33181–33186 10.1074/jbc.271.52.33181 [DOI] [PubMed] [Google Scholar]
- Kalocsay M., Hiller N.J., Jentsch S. 2009. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol. Cell. 33:335–343 10.1016/j.molcel.2009.01.016 [DOI] [PubMed] [Google Scholar]
- Kanaar R., Troelstra C., Swagemakers S.M., Essers J., Smit B., Franssen J.H., Pastink A., Bezzubova O.Y., Buerstedde J.M., Clever B., et al. 1996. Human and mouse homologs of the Saccharomyces cerevisiae RAD54 DNA repair gene: evidence for functional conservation. Curr. Biol. 6:828–838 10.1016/S0960-9822(02)00606-1 [DOI] [PubMed] [Google Scholar]
- Kim J.A., Kruhlak M., Dotiwala F., Nussenzweig A., Haber J.E. 2007. Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. J. Cell Biol. 178:209–218 10.1083/jcb.200612031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak M.J., Celeste A., Dellaire G., Fernandez-Capetillo O., Müller W.G., McNally J.G., Bazett-Jones D.P., Nussenzweig A. 2006. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J. Cell Biol. 172:823–834 10.1083/jcb.200510015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Heyer W.D. 2009. RAD54 controls access to the invading 3′-OH end after RAD51-mediated DNA strand invasion in homologous recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 37:638–646 10.1093/nar/gkn980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Stith C.M., Burgers P.M., Heyer W.D. 2009. PCNA is required for initiation of recombination-associated DNA synthesis by DNA polymerase delta. Mol. Cell. 36:704–713 10.1016/j.molcel.2009.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M., Barlow J.H., Burgess R.C., Rothstein R. 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 118:699–713 10.1016/j.cell.2004.08.015 [DOI] [PubMed] [Google Scholar]
- Mazin A.V., Alexeev A.A., Kowalczykowski S.C. 2003. A novel function of Rad54 protein. Stabilization of the Rad51 nucleoprotein filament. J. Biol. Chem. 278:14029–14036 10.1074/jbc.M212779200 [DOI] [PubMed] [Google Scholar]
- Misteli T. 2001. Protein dynamics: implications for nuclear architecture and gene expression. Science. 291:843–847 10.1126/science.291.5505.843 [DOI] [PubMed] [Google Scholar]
- Miyazaki T., Bressan D.A., Shinohara M., Haber J.E., Shinohara A. 2004. In vivo assembly and disassembly of Rad51 and Rad52 complexes during double-strand break repair. EMBO J. 23:939–949 10.1038/sj.emboj.7600091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer L.J., Essers J., Weeda G., Beverloo B., de Wit J., Muijtjens M., Odijk H., Hoeijmakers J.H., Kanaar R. 2001. The structure-specific endonuclease Ercc1-Xpf is required for targeted gene replacement in embryonic stem cells. EMBO J. 20:6540–6549 10.1093/emboj/20.22.6540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oza P., Peterson C.L. 2010. Opening the DNA repair toolbox: localization of DNA double strand breaks to the nuclear periphery. Cell Cycle. 9:43–49 10.4161/cc.9.1.10317 [DOI] [PubMed] [Google Scholar]
- Oza P., Jaspersen S.L., Miele A., Dekker J., Peterson C.L. 2009. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 23:912–927 10.1101/gad.1782209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrini J.H. 2000. The Mre11 complex and ATM: collaborating to navigate S phase. Curr. Opin. Cell Biol. 12:293–296 10.1016/S0955-0674(00)00091-0 [DOI] [PubMed] [Google Scholar]
- Petukhova G., Stratton S., Sung P. 1998. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 393:91–94 10.1038/30037 [DOI] [PubMed] [Google Scholar]
- Pollard K.J., Peterson C.L. 1998. Chromatin remodeling: a marriage between two families? Bioessays. 20:771–780 [DOI] [PubMed] [Google Scholar]
- Ristic D., Wyman C., Paulusma C., Kanaar R. 2001. The architecture of the human Rad54-DNA complex provides evidence for protein translocation along DNA. Proc. Natl. Acad. Sci. USA. 98:8454–8460 10.1073/pnas.151056798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858–5868 10.1074/jbc.273.10.5858 [DOI] [PubMed] [Google Scholar]
- Shinohara M., Gasior S.L., Bishop D.K., Shinohara A. 2000. Tid1/Rdh54 promotes colocalization of rad51 and dmc1 during meiotic recombination. Proc. Natl. Acad. Sci. USA. 97:10814–10819 10.1073/pnas.97.20.10814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinger J.A., Kiianitsa K., Heyer W.D. 2002. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol. Cell. 10:1175–1188 10.1016/S1097-2765(02)00743-8 [DOI] [PubMed] [Google Scholar]
- Soutoglou E., Dorn J.F., Sengupta K., Jasin M., Nussenzweig A., Ried T., Danuser G., Misteli T. 2007. Positional stability of single double-strand breaks in mammalian cells. Nat. Cell Biol. 9:675–682 10.1038/ncb1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stap J., Krawczyk P.M., Van Oven C.H., Barendsen G.W., Essers J., Kanaar R., Aten J.A. 2008. Induction of linear tracks of DNA double-strand breaks by alpha-particle irradiation of cells. Nat. Methods. 5:261–266 10.1038/nmeth.f.206 [DOI] [PubMed] [Google Scholar]
- Sung P., Krejci L., Van Komen S., Sehorn M.G. 2003. Rad51 recombinase and recombination mediators. J. Biol. Chem. 278:42729–42732 10.1074/jbc.R300027200 [DOI] [PubMed] [Google Scholar]
- Swagemakers S.M., Essers J., de Wit J., Hoeijmakers J.H., Kanaar R. 1998. The human RAD54 recombinational DNA repair protein is a double-stranded DNA-dependent ATPase. J. Biol. Chem. 273:28292–28297 10.1074/jbc.273.43.28292 [DOI] [PubMed] [Google Scholar]
- Symington L.S. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66:630–670 10.1128/MMBR.66.4.630-670.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M., Sasaki M.S., Sonoda E., Fukushima T., Morrison C., Albala J.S., Swagemakers S.M., Kanaar R., Thompson L.H., Takeda S. 2000. The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol. Cell. Biol. 20:6476–6482 10.1128/MCB.20.17.6476-6482.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan T.L., Essers J., Citterio E., Swagemakers S.M., de Wit J., Benson F.E., Hoeijmakers J.H., Kanaar R. 1999. Mouse Rad54 affects DNA conformation and DNA-damage-induced Rad51 foci formation. Curr. Biol. 9:325–328 10.1016/S0960-9822(99)80142-0 [DOI] [PubMed] [Google Scholar]
- Tan T.L., Kanaar R., Wyman C. 2003. Rad54, a Jack of all trades in homologous recombination. DNA Repair (Amst.). 2:787–794 10.1016/j.dnarep.2003.04.001 [DOI] [PubMed] [Google Scholar]
- Tarsounas M., Davies D., West S.C. 2003. BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene. 22:1115–1123 10.1038/sj.onc.1206263 [DOI] [PubMed] [Google Scholar]
- Tashiro S., Walter J., Shinohara A., Kamada N., Cremer T. 2000. Rad51 accumulation at sites of DNA damage and in postreplicative chromatin. J. Cell Biol. 150:283–291 10.1083/jcb.150.2.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker J., Zdzienicka M.Z. 2004. The XRCC genes: expanding roles in DNA double-strand break repair. DNA Repair (Amst.). 3:1081–1090 10.1016/j.dnarep.2004.04.012 [DOI] [PubMed] [Google Scholar]
- Van Komen S., Petukhova G., Sigurdsson S., Stratton S., Sung P. 2000. Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol. Cell. 6:563–572 10.1016/S1097-2765(00)00055-1 [DOI] [PubMed] [Google Scholar]
- van Royen M.E., Cunha S.M., Brink M.C., Mattern K.A., Nigg A.L., Dubbink H.J., Verschure P.J., Trapman J., Houtsmuller A.B. 2007. Compartmentalization of androgen receptor protein-protein interactions in living cells. J. Cell Biol. 177:63–72 10.1083/jcb.200609178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veelen L.R., Essers J., van de Rakt M.W., Odijk H., Pastink A., Zdzienicka M.Z., Paulusma C.C., Kanaar R. 2005. Ionizing radiation-induced foci formation of mammalian Rad51 and Rad54 depends on the Rad51 paralogs, but not on Rad52. Mutat. Res. 574:34–49 [DOI] [PubMed] [Google Scholar]
- Weisshart K., Jüngel V., Briddon S.J. 2004. The LSM 510 META - ConfoCor 2 system: an integrated imaging and spectroscopic platform for single-molecule detection. Curr. Pharm. Biotechnol. 5:135–154 10.2174/1389201043376913 [DOI] [PubMed] [Google Scholar]
- Wesoly J., Agarwal S., Sigurdsson S., Bussen W., Van Komen S., Qin J., van Steeg H., van Benthem J., Wassenaar E., Baarends W.M., et al. 2006. Differential contributions of mammalian Rad54 paralogs to recombination, DNA damage repair, and meiosis. Mol. Cell. Biol. 26:976–989 10.1128/MCB.26.3.976-989.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolner B., Peterson C.L. 2005. ATP-dependent and ATP-independent roles for the Rad54 chromatin remodeling enzyme during recombinational repair of a DNA double strand break. J. Biol. Chem. 280:10855–10860 10.1074/jbc.M414388200 [DOI] [PubMed] [Google Scholar]
- Wyman C., Kanaar R. 2004. Homologous recombination: down to the wire. Curr. Biol. 14:R629–R631 10.1016/j.cub.2004.07.049 [DOI] [PubMed] [Google Scholar]
- Wyman C., Kanaar R. 2006. DNA double-strand break repair: all’s well that ends well. Annu. Rev. Genet. 40:363–383 10.1146/annurev.genet.40.110405.090451 [DOI] [PubMed] [Google Scholar]