Abstract
Trim5α is a host antiviral protein that recognizes incoming retroviral capsids in the cytoplasm and prevents productive infections. Although present in most mammals, the state of the Trim5 gene is dynamic in that primates have one copy with several splice variants, while rodents and cows have multiple copies. Mouse Trim30 (one of the mouse Trim5α homologs) has been shown to negatively regulate NF-kappaB activation by targeting upstream signaling intermediates TAB2 and TAB3 for degradation. We show that human Trim5α also affects levels of TAB2, resulting in abrogation of TAB2-dependent NF-kappaB activation. Surprisingly, unlike mouse Trim30, human and rhesus Trim5α are able to activate NF-kappaB-driven reporter gene expression in a dose-dependent manner. We show that Trim5α uses distinct domains for the distinct abilities of affecting TAB2 levels, regulating NF-kappaB, and recognizing retroviral capsids. Our results demonstrate functions of Trim5α that are not dependent on recognizing the retroviral capsid.
Keywords: Trim5, Trim5alpha, Trim30, TAB2, NF-kappaB, tripartite motif, retrovirus, restriction, innate immunity
INTRODUCTION
Trim5α is a host antiviral protein initially identified as a factor in rhesus macaque monkeys that blocks human immunodeficiency virus-1 (HIV-1) infection during an early post-entry stage (Stremlau et al., 2004), (reviewed in Towers, 2005). Proteins such as Trim5α that are part of the interface between host and pathogen undergo adaptive evolution which manifests itself in the form of a higher rate of non-synonymous changes compared to synonymous changes (Holmes, 2004). Analysis of rapidly evolving codons among primate Trim5α orthologs has identified amino acids that determine the specificity of Trim5α towards retroviral capsids (Sawyer et al., 2005). These amino acids are mostly located in the PRY-SPRY domain due to interactions between this domain and retroviral capsids (Sawyer et al., 2005), (Liu et al., 2005), (Stremlau et al., 2004), (Sebastian and Luban, 2005).
While the PRY-SPRY domain has been rapidly evolving, the other domains of the protein including the RING, Bbox, and coiled-coil (RBCC) domains (Nisole et al., 2005), (Reymond et al., 2001) show more sequence conservation (Sawyer et al., 2005), (Tareen et al., 2009), (Sawyer et al., 2007), (Johnson and Sawyer, 2009). The reasons for the evolutionary constraint in these more conserved domains could be due to maintaining structural integrity, catalytic functions, and/or other unknown functions. For example, the RING and Bbox domains of Trim5α perform effector functions and contain characteristic zinc-finger motifs (Stremlau et al., 2004; Perez-Caballero et al., 2005), while the coiled-coil domain allows multimerization (Mische et al., 2005). Although some of the constraint in the RBCC domains of Trim5α could be explained by the need to maintain functional zinc-finger motifs in the RING and Bbox, and by the need to maintain the ability to multimerize through the coiled coil domain, the question still remains whether Trim5α harbors other unknown functions in addition to its role as an antiviral restriction factor.
The Trim5 locus has undergone expansions on more than one occasion, such that in cows and rodents there have been two independent paralogous expansions of Trim5 genes (reviewed in Johnson and Sawyer, 2009). Cows have up to five Trim5 genes (Sawyer et al., 2007; Si et al., 2006), while rats have three and mice have up to eight (Tareen et al., 2009). Two of the mouse Trim5 genes were previously known as Trim12 and Trim30. However, phylogenetic analysis of these two genes and their paralogs (named Trim12-1, 12-2, and Trim30-1, 30-2, 30-3, 30-4) revealed that they all are, in fact, homologs of Trim5 (Tareen et al., 2009). One of these mouse Trim5 genes, known as Trim30, has been shown to negatively regulate Toll-like receptor (TLR)-mediated nuclear factor-kappaB (NF-kappaB) activation by targeting TAB2 and TAB3 for degradation (Shi et al., 2008). Although at the time, it was not appreciated that mouse Trim30 was a homolog of primate Trim5α (Bowie, 2008; Shi et al., 2008), this result suggested that Trim5α might have additional roles in innate immunity besides direct recognition of retroviral capsids.
TAB2 and TAB3 are two adaptor proteins that play a key role in activation of a kinase called TAK1 (TGF-beta activated kinase 1) (Cheung et al., 2004; Takaesu et al., 2000; Yamaguchi et al., 1995), resulting in downstream activation of members of the mitogen activated protein kinase (MAPK) family that is indispensable for several signaling pathways such as NF-kappaB, IL-1beta, TNF, and TLR signaling (reviewed in Chen et al., 2006; Delaney and Mlodzik, 2006). Upstream ligand-receptor interactions are transmitted via TAK1 to downstream MAPKs as well as to NF-kappaB through phosphorylation of its inhibitor IkB kinase (IKK) (Chen, 2005; Shim et al., 2005). Expression of mouse Trim30 targets both TAB2 and TAB3, thus abrogates the activation of TAK1 and the subsequent downstream activation of NF-kappaB (Shi et al., 2008). These observations raise the question whether this function is conserved across Trim5α homologs from all mammals.
Here, we find that the ability to affect levels of TAB2 is a conserved function of Trim5α in mice and humans. Use of proteasomal inhibitors as well as RING domain zinc finger mutants of Trim5α demonstrate that Trim5α affects TAB2 levels through a mechanism that is independent of E3 ubiquitin ligase activity. The ability to affect TAB2 levels is present in human and mouse Trim5α, but not in rhesus Trim5α. However, unlike mouse Trim5α that abrogates TAB2-dependent NF-kappaB activation, both human and rhesus Trim5α by themselves are able to activate NF-kappaB-driven transcription. By defining the domains of human Trim5α necessary for these functions, we were able to genetically separate the ability of Trim5α to affect TAB2 levels from its ability to activate NF-kappaB. These independent abilities of Trim5α did not require the adaptively evolving retroviral capsid recognizing PRY-SPRY domain. Our results demonstrate functions of Trim5α that are not dependent on recognizing the retroviral capsid and may point to an independent role of Trim5α in the innate immune response to pathogens.
RESULTS
A conserved function for murine Trim5α paralogs and Human Trim5α: Affecting levels of TAB2
The mouse Trim5 locus consists of eight Trim5 genes, five of which are predicted to encode the full-length splice variant homologous to Trim5α in humans. These full-length genes are known as Trim12-1, Trim12-2, Trim30, Trim30-2 and Trim30-3 (Tareen et al., 2009). Of these we have been able to clone and express three from mouse RNA, namely Trim12-2, Trim30, and Trim30-3. Since Trim30 has been shown to target TAB2 and TAB3 for degradation (Shi et al., 2008), we asked if this function is conserved across other mouse Trim5α paralogs.
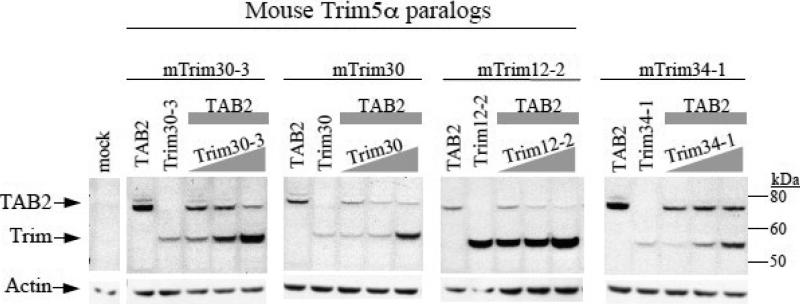
We co-transfected 293T cells with constant amounts of TAB2 alone or in the presence of increasing amounts of the TRIM gene being tested (Figure 1). Consistent with what has previously been shown (Shi et al., 2008), we found that the levels of murine TAB2 decrease with increasing amounts of Trim30. Trim12-2 and Trim30-3 behaved the same way such that increasing amounts of Trim12-2 or Trim30-3 resulted in a decrease in the levels of TAB2 (Fig. 1). This concentration-dependent manner in which TAB2 levels are affected is specific to mouse Trim5α paralogs since Trim34-1, the next closest related gene that sits near the Trim5 locus but is not one of the Trim5α genes (Tareen et al., 2009), was not able to affect the levels of TAB2 (Fig. 1, right side). These results show that affecting the levels of TAB2 is a conserved function of mouse Trim5α paralogs.
Figure 1. Targeting TAB2 for degradation is a conserved among mouse Trim5α paralogs.
293T cells were co-transfected with constant amounts of mouse TAB2 (indicated by gray rectangle) and with increasing amounts of either of the mouse Trim5α paralogs (Trim12-2, Trim30, or Trim30-3) or of mouse Trim34-1 (indicated by gray triangle). Total amount of DNA transfected was normalized using empty vector DNA. All constructs contain HA tags. Lysates were probed with anti-HA antibodies, then stripped and re-probed with anti-actin. Mouse Trim is abbreviated as mTrim.
Since mouse Trim5α paralogs are able to affect protein levels of TAB2, we asked if this activity is also present in human and rhesus Trim5α proteins. Co-transfecting 293T cells with human TAB2 in the presence of increasing amounts of human Trim5α or empty vector (LPCX) showed that increasing amounts of human Trim5α results in lower levels of human TAB2 (Fig. 2A). However, a closely related TRIM protein in humans, Trim22, was not able to affect TAB2 levels, even in exceedingly large amounts of Trim22 being expressed (Fig. 2A). Human Trim5α was also able to affect the levels of mouse TAB2 (Fig. 2A, right side) and rhesus TAB2 (data not shown), even though rhesus Trim5α was unable to affect the levels of either rhesus or human TAB2 (Fig. 2A). These results show that targeting TAB2 is present among human and murine Trim5α homologs, but may not be a universal property of Trim5 since we did not observe it for the rhesus homolog..
Figure 2. Human Trim5α specifically affects levels of TAB2.
(A) 293T cells were co-transfected with constant amounts of either mouse TAB2 (0.2ug DNA transfected per lane), human or rhesus TAB2 (2ug DNA per lane). Amount of TAB2 transfected varied between species to match protein levels. Increasing amounts of either human Trim5α or Trim22 or rhesus Trim5α or with empty vector LPCX were transfected (indicated by gray triangle; increasing gradient of 0.06ug, 0.2ug, 0.6ug DNA transfected per lane). (B) 293T cells were co-transfected with constant amounts of human TAB2 and with increasing amounts empty vector LPCX or with increasing amounts of the C35A mutant of Trim5α which has a cysteine to alanine substitution in its zinc finger of the RING domain. DNA transfected per lane is same as in Fig2A. (C) 293T cells were co-transfected with constant levels of human TAB2 alone or with increasing amounts of human Trim5α in the presence or absence of either proteasomal inhibitors (10uM MG132) or lysosomal inhibitors (20mM NH4CL and 0.1mM Leupeptin). Drugs were added 24 hours after transfection and cells were treated for 2 hours before they were lysed. All constructs contain HA tags. DNA transfected per lane is same as in Fig2A. Lysates were probed with anti-HA antibodies, then stripped and reprobed with anti-actin. Human, rhesus and mouse are abbreviated as Hu, Rh, and Mus, respectively.
To understand the mechanism of how human Trim5α affects TAB2 levels, we asked if a proteasomal pathway requiring E3 ubiquitin ligase activity of Trim5α was involved. We generated a Trim5α protein that contains a mutated zinc finger in its RING domain by substituting cysteine at position 35 to alanine (Trim5α-C35A). Trim5α-C35A was still able to affect TAB2 levels (Fig. 2B) indicating that an intact zinc finger within the RING domain of Trim5α is not required for affecting TAB2, thus making it unlikely that the effect is through an E3 ubiquitin ligase activity of Trim5. We further assessed this by . treating cells with either proteasomal inhibitors or lysosomal inhibitors. We found that the ability for Trim5α to affect TAB2 levels was not affected by proteasomal inhibitors even though levels of Trim5 were increased (Fig. 2C). We observed a very slight effect of lysosomal inhibitors compared to no drug control, is consistent with mouse Trim30 which also targets mouse TAB2 through the lysosome (Shi et al., 2008), but this effect did not seem to be consistent over a range of Trim5 levels. (Fig. 2C). Taken together, these findings suggest that the ability of Trim5α to target TAB2 for degradation is conserved across mice and humans (although not present in the rhesus homolog of Trim5) , and that the ability to affect TAB2 levels is not linked to proteasomal degradation.
Human and rhesus Trim5α induces NF-kappaB reporter activity
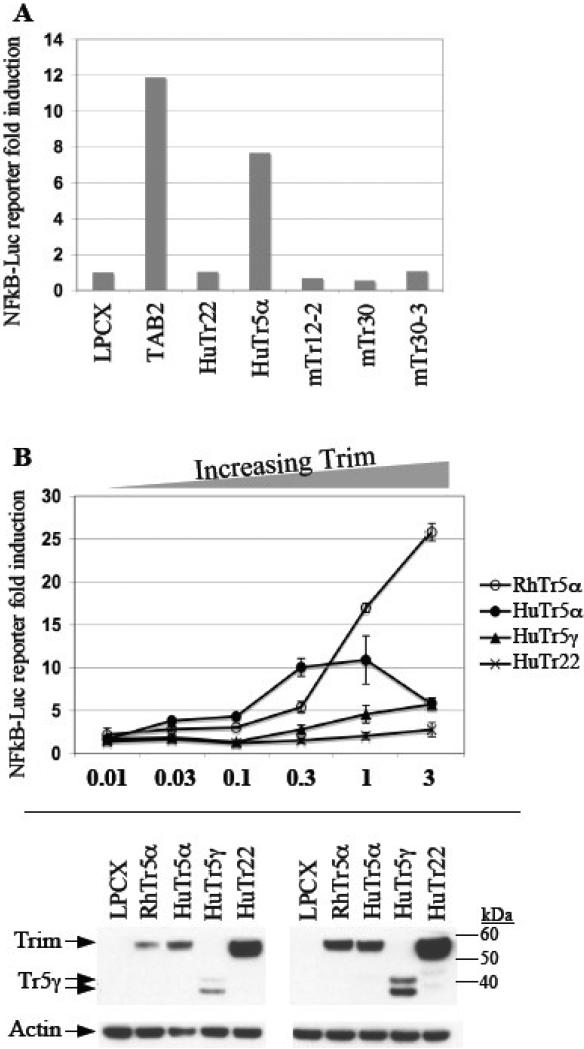
A downstream result of TAB2 signaling is activation of NF-kappaB. Mouse Trim30, one of the mouse Trim5α paralogs, was previously shown to negatively regulate TAB2-dependent NF-kappaB activation by targeting TAB2 for degradation (Shi et al., 2008). We asked whether the conserved ability of human and mouse Trim5α to target TAB2 also resulted in similar outcomes when it came to regulation of NF-kappaB. Using an NF-kappaB inducible luciferase reporter gene we assessed the effects of overexpressing human or mouse Trim5α homologs in 293T cells. As expected, expressing TAB2 by itself was sufficient to activate NF-kappaB-driven transcription (Fig. 3A). Surprisingly however, expression of human Trim5α by itself resulted in activation of NF-kappaB-driven transcription, which was not the case with the mouse Trim5α paralogs we tested (mouse Trim12-2, Trim30, Trim30-3) (Fig. 3A). Furthermore, the closely related human Trim22 failed to activate the NF-kappaB inducible reporter gene (Fig. 3A).
Figure 3. Primate but not mouse Trim5α activates NF-kappaB-driven reporter expression.
(A) 293T cells were co-transfected with an NF-kappaB-driven Luciferase reporter construct (NF-kappaB-LUC, 0.5ug DNA transfected per lane) together with 0.5 ug DNA of either empty vector LPCX, human TAB2, human Trim5α, human Trim22, or one of the mouse Trim5α paralogs (Trim12-2, Trim30, Trim30-3). (B) 293T cells were co-transfected with an NF-kappaB-driven Luciferase reporter construct (0.5ug DNA per lane) together with increasing amounts of human Trim5α, or human Trim5γ, or human Trim22, or rhesus Trim5α, or empty vector LPCX. Luciferase expression is plotted on the y-axis as fold induction relative to reporter gene plus empty vector LPCX. The DNA transfection ratio of LPCX or TRIM relative to NF-kappaB-LUC is plotted on the x-axis (3-fold means 1.5ug DNA of TRIM vs. 0.5ug of TAB2, and so on.) Protein expression was compared using anti-HA antibody. The immunoblots on the left and the right are to compare protein expression for the x-axis transfection ratios of 1 and 3, respectively. The doublet band of Trim5γ has been reported before (Stremlau et al., 2004).
Since human, but not mouse Trim5α, is able to activate the NF-kappaB inducible reporter gene expression, we asked if this ability is conserved for other primate Trim5α orthologs. We found that both human and rhesus Trim5α are able to activate NF-kappaB in a dose dependent manner (Fig. 3B) suggesting that this ability may be more important for primate Trim5α than is the ability to affect TAB2 levels. Interestingly, at higher concentrations human and rhesus Trim5α responded differently such that human, but not rhesus, Trim5α reached saturation and resulted in a drop in NF-kappaB activation (Fig. 3B). A naturally occurring splice variant of human Trim5 that lacks the retroviral capsid recognizing PRY-SPRY domain, Trim5γ (Trim5gamma), was also able to activate NF-kappaB, albeit not as strongly as human Trim5α. (Fig. 3B). As before, Trim22 expression had little to no effect on NF-kappaB regulation even though it was expressed at higher levels than Trim5α (Fig. 3B, bottom). Taken together, these findings indicate that activation of NF-kappaB-driven transcription is conserved among human and rhesus Trim5α orthologs, and most likely across primate Trim5α orthologs, and that the adaptively evolving PRY-SPRY domain of Trim5α is not required for this activity.
The ability of human Trim5α to regulate TAB2 levels, to activate NF-kappaB, and to recognize retroviral capsids are genetically separable
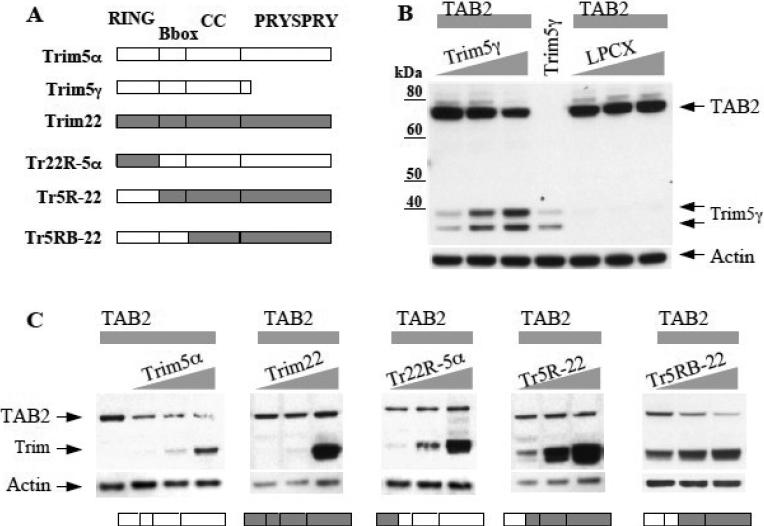
Trim5α as a restriction factor recognizes the viral capsid through its C-terminal PRY-SPRY domain. The genetic conflict arising from this interaction has caused adaptive evolution of Trim5α to take place mostly in its PRY-SPRY domain. We hypothesized that Trim5α must use distinct domains for its conserved functions of targeting TAB2 and for activating NF-kappaB, than for recognizing the retroviral capsid. First, we tested this hypothesis using Trim5γ, which encodes all but the PRY-SPRY domain of Trim5α, and is not able to restrict retroviral capsids (Stremlau et al., 2004). Increasing amounts of Trim5γ was able to affect TAB2 levels, compared to increasing amounts of empty vector LPCX (Fig. 4B). Thus, the domain necessary for recognition of retroviral capsids is not required for the activity of Trim5α in affecting TAB2 levels.
Figure 4. Human Trim5α affecting TAB2 levels is dependent on its RING and B-box domains, but not on its PRY-SPRY domain.
(A) Diagram showing domains of Trim5α, Trim5γ, Trim22, and the chimeras constructed between Trim5 and Trim22. Trim22R-5α indicates Trim22 RING in a Trim5α background. Trim5R-22 indicates Trim5 RING in a Trim22 background. Trim5RB-22 indicates a Trim5 RING and B-box in a Trim22 background. (B) 293T cells were co-transfected with constant amounts of human TAB2 (2ug DNA per lane) and with increasing amounts of either empty vector LPCX or with human Trim5γ (gradient of 0.06ug, 0.2ug, 0.6ug DNA per lane). The doublet band of Trim5γ has been reported before (Stremlau et al., 2004). (C) 293T cells were co-transfected with constant amounts of human TAB2 and with increasing amounts of either human Trim5α, or human Trim22, or the chimeras Trim22R-5α, Trim5R-22, or Trim5RB-22. DNA transfected same as Fig4B. Diagrams of each Trim construct is shown below their respective immunoblot for conveniance. All constructs contain HA tags. Lysates were probed with anti-HA antibodies, then stripped and re-probed with anti-actin.
To narrow in on the domains of Trim5α necessary for affecting TAB2 levels, we designed several chimeras between Trim5α and its close relative Trim22, which does not affect TAB2 levels (Fig. 4A). When the RING domain of Trim5α was substituted with the Trim22 RING (Trim22R-5α) the ability of Trim5α to affect TAB2 levels was lost (Fig. 4C), indicating that the RING domain of Trim5α is necessary. When the Trim5 RING was swapped into Trim22 in place of the Trim22 RING (Trim5R-22), this chimera did not gain the ability to affect TAB2 levels (Fig. 4C), indicating that the RING of Trim5α is not sufficient. Only when both the Trim5 RING and Bbox were swapped into Trim22 (Tr5RB-22) was the ability to affect TAB2 gained (Fig. 4C). These findings indicate that both the RING and Bbox of Trim5 are sufficient in a Trim22 backbone to gain the ability to affect TAB2 levels. Moreover, although the RING domain of Trim5 is necessary, E3 ubiquitin ligase activity and an intact zinc finger in the RING domain is not required as shown by the proteasomal inhibitors and the C35A mutant (Figures 2B and 2C). Therefore, despite the rapid evolution of PRY-SPRY, Trim5α appears to use its more conserved domains to maintain the ability to target TAB2 and affect its levels.
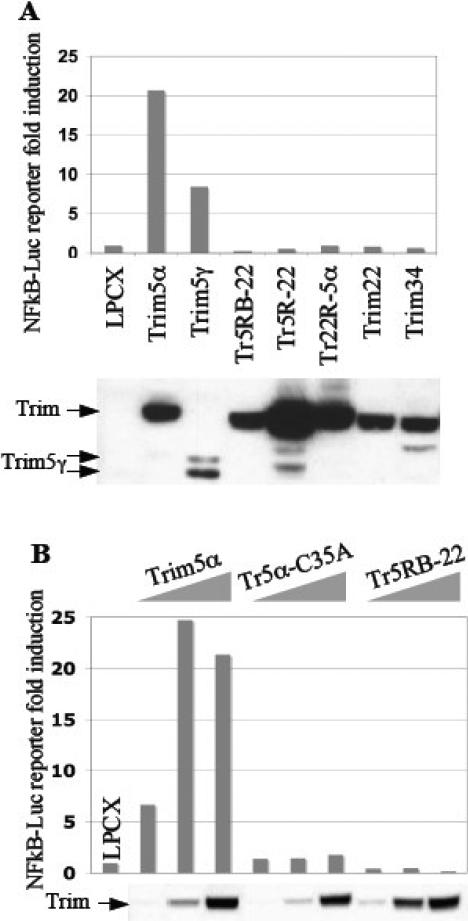
We then asked if the ability of Trim5α to affect TAB2 levels is independent of its ability to activate NF-kappaB by testing these same domain swaps for their ability to activate an NF-kappaB reporter. As described earlier (Fig. 3B), Trim5γ is able to activate NF-kappaB, albeit not as strongly as Trim5α (probably due to expression differences—see Westen blot below Fig. 5A). This indicates that the PRY-SPRY domain of Trim5α is not required to activate NF-kappaB-driven reporter gene expression (Fig. 5A). The Tr22R-5α chimera constructed between human Trim5α and Trim22 (Fig. 4A) showed that the RING domain of Trim5α was necessary to activate NF-kappaB (Fig. 5A). Interestingly however, while the RING and Bbox of Trim5α were sufficient to regulate TAB2, they were not sufficient to activate NF-kappaB (Tr5RB-22 chimera in Figures 5A and 5B). Furthermore, while the zinc finger of the Trim5α RING domain was dispensable for regulating TAB2, it was required for inducing NF-kappaB because the C35A mutant of Trim5α lost this ability (compare Fig. 5B with Fig. 2B). These results genetically separate the ability of Trim5α to affect TAB2 levels and to activate NF-kappaB-driven transcription.
Figure 5. The ability of human Trim5α to affect TAB2 levels and to activate NF-kappaB are genetically separable.
(A) 293T cells were co-transfected with an NF-kappaB-driven Luciferase reporter construct (0.5ug DNA per lane) together with either empty vector LPCX, human Trim5α, human Trim5γ, human Trim22, or either of the chimeras Trim22R-5α, Trim5R-22, or Trim5RB-22 (0.5ug DNA per lane). (B) 293T cells were co-transfected with an NF-kappaB-driven Luciferase reporter construct (0.5ug DNA per lane) together with increasing amounts of either human Trim5α, C35A mutant of human Trim5α, or the chimera Trim5RB-22 (gradient of 0.06ug, 0.2ug, 0.6ug DNA per lane). Luciferase expression was plotted on y-axis as fold induction relative to empty vector LPCX. Protein expression was compared using anti-HA antibody.
The interplay of the independent abilities of Trim5α to affect TAB2 levels and to activate NF-kappaB
We next investigated how the ability of Trim5α to reduce levels of TAB2 affects NF-kappaB activity. To do so we first asked if Trim5α, by affecting TAB2 levels, would abrogate TAB2-dependent NF-kappaB activation. Over-expression of either Trim5α or TAB2 by itself activates NF-kappaB-driven reporter gene expression, with higher TAB2-dependent activation (Fig. 6A). However, adding Trim5α together with TAB2 brings NF-kappaB activation back down to the levels of Trim5α only, consistent with the decrease in the protein levels of TAB2 (Fig. 6A). Therefore, human Trim5α, similar to mouse Trim30, abrogates TAB2-dependent NF-kappaB activation.
Figure 6. Interplay of the independent abilities of Trim5α to affect TAB2 levels and to activate NF-kappaB.
(A) 293T cells were co-transfected with an NF-kappaB-driven Luciferase reporter construct (0.5ug DNA per lane) together with either empty vector LPCX, or human Trim5α alone, or human TAB2 alone, or human TAB2 together with human Trim5α (0.5ug DNA per lane). Total amount of DNA transfected was normalized using empty vector LPCX. Luciferase expression is plotted on the y-axis as fold induction relative to reporter gene plus empty vector LPCX. Protein expression was compared using anti-HA antibody.
(B) 293T cells were co-transfected with an NF-kappaB-driven Luciferase reporter construct (0.5ug DNA per lane) together with constant amounts of human TAB2 (2ug DNA per lane) in the presence of increasing amounts of either empty vector LPCX (green line), or human Trim5α (red line), or rhesus Trim5α (blue line), or the human chimera Trim5RB-22 (orange line). The DNA transfection ratio of LPCX or TRIM relative to NF-kappaB-LUC is plotted on the x-axis (3-fold means 1.5ug DNA of TRIM vs. 0.5ug of TAB2, and so on). Protein expression was detected using anti-HA antibody, and is shown below the graph to represent the full transfection gradient (0.1 to 3-fold) for either [TAB2 plus LPCX] or [TAB2 plus Trim].
Dose-response curves showed that with increasing amounts of human Trim5α, the amount of NF-kappaB activation eventually began to decrease, although this did not occur with rhesus Trim5α (Fig. 3B). We hypothesized that this drop in NF-kappaB was due to human Trim5α being able to decrease endogenous levels of TAB2, thus affecting TAB2-dependent NF-kappaB activation. Accordingly, we would expect rhesus Trim5α, which has no effect on human TAB2 levels, to have no effect on TAB2-dependent NF-kappaB activity. We tested this hypothesis by looking at the dose-responsive effects of Trim5α on TAB2-dependent activation of NF-kappaB. We transfected a constant amount of a TAB2-expressing plasmid along with increasing amounts of Trim5α or empty vector and then measured the activity of an NF-kappaB reporter. We observed an additive effect of TAB2 and Trim5α on NF-kappaB activation at low Trim5α levels, followed by a drop in response to increasing amounts of Trim5α, resulting from more degradation of TAB2 and thus stronger abrogation of TAB2-dependent NF-kappaB activation (follow the red line in Fig. 6B). When we used increasing amounts of rhesus Trim5α that only activates NF-kappaB but does not specifically target TAB2 (performs like empty vector LPCX), we only observed an increase in NF-kappaB activation but no abrogation of TAB2-dependent NF-kappaB activation (follow the blue line in Fig. 6B). These results are consistent with what we observed earlier in the absence of exogenously expressed TAB2 (Fig. 3B). In contrast, with increasing amounts of the Tr5RB-22 chimera that affects TAB2 but does not activate NF-kappaB, we observed a dramatic drop in TAB2-dependent NF-kappaB activation (follow the orange line in Fig. 6B). These findings show that Trim5α regulates NF-kappaB activation by at least two independent mechanisms: activation of NF-kappaB by Trim5α itself, and abrogation of TAB2-dependent NF-kappaB activation due to Trim5α targeting TAB2 for lysosomal degradation.
DISCUSSION
Here we describe functions of human Trim5α that do not depend on recognition of retroviral capsids. We show that human Trim5α, similar to its mouse homolog Trim30, targets TAB2 for degradation, resulting in abrogation of TAB2-dependent NF-kappaB activation. However, unlike mouse Trim30, human and rhesus Trim5α are able to activate NF-kappaB-driven reporter gene expression in a dose-dependent manner. Using splice variants, chimeras and mutants we show that human Trim5α uses distinct domains for the distinct abilities of affecting TAB2 levels, regulating NF-kappaB, and recognizing retroviral capsids. We speculate that the induction of NF-kappaB is the more critical of these activities since it is common between the two primate Trim5 genes tested.
Viruses either escape from or harness innate immunity pathways in order to continue their life cycle. For example, NS1 protein of influenza A virus prevents NF-kappaB-dependent interferon signaling by binding viral dsRNA and preventing PKR activation (Wang et al., 2000). Based on work reported here, Trim5α, by regulating TAB2 levels or activating NF-kappaB, could be involved in controlling a response to viral infection at various stages. Implications of a TRIM protein in signaling events that affect viruses are not without precedent. For example Trim25, which also has RBCC and PRY-SPRY domains, is able to induce IFN signaling against RNA viruses by activation of RIG-I through K63 polyubiquitination (Gack et al., 2007). In this example, the antiviral properties of Trim25 appear to be directly linked to its role in RIG-I signaling. Interestingly, human T-cell lymphotropic virus type 1 (HTLV-1) appears to hijack TAB2-dependent signaling through induction of TAB2 levels using its Tax oncoprotein, resulting in transcriptional activation and transformation (Boxus et al., 2008; Suzuki et al., 2007; Yu et al., 2008).
It is unclear why a rapidly evolving antiviral factor such as Trim5α would be co-opted to play a key role in regulating signaling, or vice versa. One explanation may be that the ability of Trim5α to function as an antiviral factor is linked to its role in signaling, and that these two roles are inseparable even though we show here that genetically they are separable. The recent finding that restriction by Trim5α in the cytoplasmic bodies relies on the presence of p62/sequestosome-1, an interferon-inducible gene involved in signaling pathways such as TRAF6 and NF-kappaB (O'Connor et al., 2010), supports a role of cell signaling in the Trim5α restriction pathway. Furthermore, it is possible that Trim5α signaling and restriction functions are actually linked in some way since infection of cells with a retrovirus that is recognized by Trim5α leads to the degradation of Trim5α (Rold and Aiken, 2008). Thus, the signaling mechanisms induced by Trim5α could be a cellular response to a viral infection where the “recognition” event is the loss of Trim5α in the cell, which then leads to the absence of those constitutive Trim5α signals.
The ability of Trim5 to target TAB2 and to upregulate NF-kappaB appear to be independent of the PRY-SPRY domain (Figures 3 and 4), however we cannot rule out that the possibility that the PRY-SPRY domain might be involved in moderating these activities. The fact that the PRY-SPRY domain is used by Trim5α for recognizing and binding to the viral capsid, and that this domain contains the region under most positive selection, indicates that Trim5α is able to maintain its other functions while still being able to adaptively evolve in its PRY-SPRY domain. The RBCC domains of Trim5α appear to be involved in the TAB2 and NF-kappaB functions we describe here. Since these functions most likely involve the formation of complexes with other cellular proteins through protein-protein interactions, we would expect evolution to select against amino acid changes that could impede with these functions. Therefore, the functions of Trim5α in TAB2 and NF-kappaB signaling could maintain the evolutionary constraint seen in the RBCC domains of Trim5α. Interestingly, certain amino acids in parts of the RBCC domains are also under positive selection, although not as strongly and as dense as in the PRY-SPRY (reviewed in Johnson and Sawyer, 2009). Whether these amino acids specifically contribute to the abilities of Trim5α described here remain to be investigated. The inability of rhesus Trim5α to affect TAB2 levels, even though this ability is conserved in humans and mice, could indicate a loss in the ability of rhesus Trim5α to target TAB2, possibly due to amino acid differences in the RBCC domains between human and rhesus Trim5α. It is possible that polymorphisms in rhesus Trim5α will affect this ability, but we have not yet tested other alleles. Likewise, it is possible that human polymorphisms in Trim5α will also affect these activities.
Using chimeras and mutants we were able to separate the ability of human Trim5α to affect TAB2 levels, from its ability to activate NF-kappaB. The interplay of these two abilities (Figure 6) suggests that human Trim5α either upregulates NF-kappaB downstream of where it targets TAB2 in the signaling cascade, that Trim5α is able to upregulate NF-kappaB through a pathway that is independent of TAK1 activation. NF-kappaB is regulated through two pathways known as the canonical and non-canonical pathways (reviewed in Skaug et al., 2009). TAK1 is involved in the canonical pathway where, upon TNF or IL-1beta stimulation, TAK1 activates IKK, which then targets IkB proteins for degradation, thus allowing the p50/65 NF-kappaB dimmer to enter the nucleus. In the non-canonical pathway, CD40L stimulation results in the activation of the NF-kappaB inducing kinase (NIK) and the IKKalpha subunit. The latter, in turn, targets p100 for degradation, thus allowing the p52/Rel-B dimmer to enter the nucleus. The reporter construct we used in our experiments for NF-kappaB activation is sensitive to both of these pathways. It is also possible that the seeming degradation of TAB2 by human Trim5α is an epi-phenomenon that is simply a readout of a binding activity to TAB2 that is shared with rhesus Trim5α and which results in induction of NF-kappaB in both cases.
Primates, rabbits, cats and dogs have one copy of Trim5 whereas rodents and cows have multiple copies of Trim5 in their locus (Schaller et al., 2007; McEwan et al., 2009; Tareen et al., 2009; Sawyer et al., 2007; Si et al., 2006). This diverse nature of the Trim5 locus among mammals should make for interesting strategies in balancing the abilities of viral restriction with regulating TAB2 and NF-kappaB pathways. In the case of rodents and cows, paralogous Trim5 gene expansions may be selected for temporal and spatial sharing of duties; for example, certain Trim5 paralogs may be expressed during certain stages of development and take on functional responsibility. Alternatively, different Trim5 paralogs may complement one another by splitting different functions. Interestingly, two of the mouse Trim5 paralogs, namely Trim12 and Trim30-1, do not encode the PRY-SPRY domain (Tareen et al., 2009). However, our findings that the PRY-SPRY is not required suggests that mouse Trim12 and Trim30-1 may also be capable of negatively regulating NF-kappaB activation by targeting TAB2 and TAB3 for degradation, similar to Trim30. The fact that the functions for Trim5α we describe here are present in mice and humans suggests that either evolution has maintained this function since the divergence of these two species, or that, although less likely, convergent evolution explains these functions in mouse and human Trim5α.
One problem that rapidly evolving antiviral factors may face is how they are able to adaptively evolve while maintaining sequence conservation for conserved functions. Trim5α appears to have solved this problem by using distinct domains to recognize retroviral capsids versus regulating TAB2 and NF-kappaB signaling: the PRY-SPRY recognizes the capsid and is free to rapidly evolve, while the remaining domains are under evolutionary constraint in order to maintain the functions described here. These functions of Trim5α may have bestowed upon it multiple roles in innate immunity, thus possibly explaining its maintenance over millions of years throughout mammalian evolution even in the absence of retroviral recognition.
MATERIALS AND METHODS
Constructs
All constructs used for over-expression studies are N-terminally tagged with an HA epitope, and are cloned into retroviral mammalian expression vectors pLNCX or pLPCX. Mouse and human TAB2 and TAB3 cDNAs were obtained from Open Biosystems. Mouse Trim5 paralogs (namely Trim12-2, Trim30, Trim30-3) as well as mouse Trim34-1 have been cloned from RNA from NIH3T3 cells as described (Tareen et al., 2009). Rhesus Trim5α was obtained from Joseph Sodroski. Human Trim5α was obtained from the NIAID AIDS repository. Human Trim22 was previously described (Sawyer et al., 2007). Human Trim5γ and Trim34 have been cloned from RNA from HeLa cells. Chimeras of human Trim5 and Trim22 have been cloned by designing overlapping PCR primers annealing to either the end of the RING domain (for RING only chimeras as in Trim5RING22 or Trim22RING5) or the end of the Bbox domain (for RING and Bbox chimeras as in Trim5RBbox22). C35A mutant of Trim5α was constructed using Quickchange XL2 kit (Stratagene). Human Trim5α SNPs were previously described (Sawyer et al., 2006).
Transient transfections
293T cells were plated at 106 cells/ml/well in a 12 well plate. Next day, transfections were performed using 3ul of TransIT (MirusBio) per 1ug of DNA according to protocol. DNA in each well was kept constant by adding empty vector (LPCX). Due to the differences in turnover rates between the proteins being over-expressed, whenever possible the DNA amounts transfected were adjusted to match similar protein levels. Cells were lysed 24 hours after transfection and cytoplasmic extracts were made and prepared for Western blotting.
Proteasomal and lysosomal inhibitors
293T cells were transfected as described above. Proteasomal inhibitors (10uM MG132) or lysosomal inhibitors (20mM NH4CL and 0.1mM Leupeptin) were added 24 hours after transfection and cells were treated for 2 hours before they were lysed.
NF-kappaB-Luciferase reporter assays
293T cells were transfected (as described above) with appropriate constructs as well as with pCEP4-NF-kappaB-Luc reporter construct (Promega). 24 or 48 hours after transfection cells were trypsinized and then lysed with bright-glow luciferase reagent. Luciferase units were read on plate reader.
Antibodies and cell lysis
Anti-HA was obtained from Covance. Anti-Actin was obtained from Sigma. Cells were lysed using 1% NP40-DOC lysis buffer in the presence of 1x protease inhibitors EDTA free and 1x phosphatase inhibitors (Roche). Nuclei were spun out and lysates were quantified using Bradford reagent. NuPage Bis-Tris gels and NuPage transfers were used for Western transfers (Invitrogen).
ACKNOWLEDGEMENTS
We thank Jeremy Luban for discussions and sharing unpublished results, Alex Compton, Efrem Lim, Melody Li, and Nisha Duggal for comments on the manuscript, and Harshana De Silva for technical assistance. This work was supported by NIH grant R37 AI30937 (M.E.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of competing interests: The authors declare that they have no competing interests.
REFERENCES
- Anderson JL, Campbell EM, Wu X, Vandegraaff N, Engelman A, Hope TJ. Proteasome inhibition reveals that a functional preintegration complex intermediate can be generated during restriction by diverse TRIM5 proteins. J.virol. 2006;80:9754–9760. doi: 10.1128/JVI.01052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie AG. TRIM-ing down tolls. Nat.immunol. 2008;9:348–350. doi: 10.1038/ni0408-348. [DOI] [PubMed] [Google Scholar]
- Boxus M, Twizere JC, Legros S, Dewulf JF, Kettmann R, Willems L. The HTLV-1 tax interactome. Retrovirology. 2008;5:76. doi: 10.1186/1742-4690-5-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ. Ubiquitin signalling in the NF-kappaB pathway. Nat.cell biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Bhoj V, Seth RB. Ubiquitin, TAK1 and IKK: Is there a connection?. Cell death differ. 2006;13:687–692. doi: 10.1038/sj.cdd.4401869. [DOI] [PubMed] [Google Scholar]
- Cheung PC, Nebreda AR, Cohen P. TAB3, a new binding partner of the protein kinase TAK1. Biochem.J. 2004;378:27–34. doi: 10.1042/BJ20031794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney JR, Mlodzik M. TGF-beta activated kinase-1: New insights into the diverse roles of TAK1 in development and immunity. Cell.cycle. 2006;5:2852–2855. doi: 10.4161/cc.5.24.3558. [DOI] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- Holmes EC. Adaptation and immunity. PLoS biol. 2004;2:E307. doi: 10.1371/journal.pbio.0020307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Sawyer SL. Molecular evolution of the antiretroviral TRIM5 gene. Immunogenetics. 2009;61:163–176. doi: 10.1007/s00251-009-0358-y. [DOI] [PubMed] [Google Scholar]
- Liu HL, Wang YQ, Liao CH, Kuang YQ, Zheng YT, Su B. Adaptive evolution of primate TRIM5alpha, a gene restricting HIV-1 infection. Gene. 2005;362:109–116. doi: 10.1016/j.gene.2005.06.045. [DOI] [PubMed] [Google Scholar]
- McEwan WA, Schaller T, Ylinen LM, Hosie MJ, Towers GJ, Willett BJ. Truncation of TRIM5 in the feliformia explains the absence of retroviral restriction in cells of the domestic cat. J.virol. 2009;83:8270–8275. doi: 10.1128/JVI.00670-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mische CC, Javanbakht H, Song B, Diaz-Griffero F, Stremlau M, Strack B, et al. Retroviral restriction factor TRIM5alpha is a trimer. J.virol. 2005;79:14446–14450. doi: 10.1128/JVI.79.22.14446-14450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisole S, Stoye JP, Saib A. TRIM family proteins: Retroviral restriction and antiviral defence. Nat.rev.microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- O'Connor C, Pertel T, Gray S, Robia SL, Bakowska JC, Luban J, Campbell EM. p62/sequestosome-1 associates with and sustains the expression of retroviral restriction factor TRIM5alpha. J.virol. 84:5997–6006. doi: 10.1128/JVI.02412-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Caballero D, Hatziioannou T, Yang A, Cowan S, Bieniasz PD. Human tripartite motif 5alpha domains responsible for retrovirus restriction activity and specificity. J.virol. 2005;79:8969–8978. doi: 10.1128/JVI.79.14.8969-8978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole E, Groves I, MacDonald A, Pang Y, Alcami A, Sinclair J. Identification of TRIM23 as a cofactor involved in the regulation of NF-kappaB by human cytomegalovirus. J.virol. 2009;83:3581–3590. doi: 10.1128/JVI.02072-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rold CJ, Aiken C. Proteasomal degradation of TRIM5alpha during retrovirus restriction. PLoS pathog. 2008;4:e1000074. doi: 10.1371/journal.ppat.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Emerman M, Malik HS. Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals. PLoS pathog. 2007;3:e197. doi: 10.1371/journal.ppat.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Wu LI, Akey JM, Emerman M, Malik HS. High-frequency persistence of an impaired allele of the retroviral defense gene TRIM5alpha in humans. Curr.biol. 2006;16:95–100. doi: 10.1016/j.cub.2005.11.045. [DOI] [PubMed] [Google Scholar]
- Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc.natl.acad.sci.U.S.A. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller T, Hue S, Towers GJ. An active TRIM5 protein in rabbits indicates a common antiviral ancestor for mammalian TRIM5 proteins. J.virol. 2007;81:11713–11721. doi: 10.1128/JVI.01468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian S, Luban J. TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Deng W, Bi E, Mao K, Ji Y, Lin G, et al. TRIM30 alpha negatively regulates TLR-mediated NF-kappa B activation by targeting TAB2 and TAB3 for degradation. Nat.immunol. 2008;9:369–377. doi: 10.1038/ni1577. [DOI] [PubMed] [Google Scholar]
- Shim JH, Xiao C, Paschal AE, Bailey ST, Rao P, Hayden MS, et al. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes dev. 2005;19:2668–2681. doi: 10.1101/gad.1360605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si Z, Vandegraaff N, O'huigin C, Song B, Yuan W, Xu C, et al. Evolution of a cytoplasmic tripartite motif (TRIM) protein in cows that restricts retroviral infection. Proc.natl.acad.sci.U.S.A. 2006;103:7454–7459. doi: 10.1073/pnas.0600771103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-kappaB regulatory pathways. Annu.rev.biochem. 2009;78:769–796. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in old world monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Singhirunnusorn P, Mori A, Yamaoka S, Kitajima I, Saiki I, et al. Constitutive activation of TAK1 by HTLV-1 tax-dependent overexpression of TAB2 induces activation of JNK-ATF2 but not IKK-NF-kappaB. J.biol.chem. 2007;282:25177–25181. doi: 10.1074/jbc.C700065200. [DOI] [PubMed] [Google Scholar]
- Takaesu G, Kishida S, Hiyama A, Yamaguchi K, Shibuya H, Irie K, et al. TAB2, a novel adaptor protein, mediates activation of TAK1 MAPKKK by linking TAK1 to TRAF6 in the IL-1 signal transduction pathway. Mol.cell. 2000;5:649–658. doi: 10.1016/s1097-2765(00)80244-0. [DOI] [PubMed] [Google Scholar]
- Tareen SU, Sawyer SL, Malik HS, Emerman M. An expanded clade of rodent Trim5 genes. Virology. 2009;385:473–483. doi: 10.1016/j.virol.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers GJ. Control of viral infectivity by tripartite motif proteins. Hum.gene ther. 2005;16:1125–1132. doi: 10.1089/hum.2005.16.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li M, Zheng H, Muster T, Palese P, Beg AA, et al. Influenza A virus NS1 protein prevents activation of NF-kappaB and induction of alpha/beta interferon. J.virol. 2000;74:11566–11573. doi: 10.1128/jvi.74.24.11566-11573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Anderson JL, Campbell EM, Joseph AM, Hope TJ. Proteasome inhibitors uncouple rhesus TRIM5alpha restriction of HIV-1 reverse transcription and infection. Proc.natl.acad.sci.U.S.A. 2006;103:7465–7470. doi: 10.1073/pnas.0510483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Shirakabe K, Shibuya H, Irie K, Oishi I, Ueno N, et al. Identification of a member of the MAPKKK family as a potential mediator of TGF-beta signal transduction. Science. 1995;270:2008–2011. doi: 10.1126/science.270.5244.2008. [DOI] [PubMed] [Google Scholar]
- Yu Q, Minoda Y, Yoshida R, Yoshida H, Iha H, Kobayashi T, et al. HTLV-1 tax-mediated TAK1 activation involves TAB2 adapter protein. Biochem.biophys.res.commun. 2008;365:189–194. doi: 10.1016/j.bbrc.2007.10.172. [DOI] [PubMed] [Google Scholar]