Abstract
For pre-clinical bioluminescence or fluorescence optical tomography, the animal's surface topography and internal anatomy need to be estimated for improving the quantitative accuracy of reconstructed images. The animal's surface profile can be measured by all-optical systems, but estimation of the internal anatomy using optical techniques is non-trivial. A 3D anatomical mouse atlas may be warped to the estimated surface. However, fitting an atlas to surface topography data is challenging because of variations in the posture and morphology of imaged mice. In addition, acquisition of partial data (for example, from limited views or with limited sampling) can make the warping problem ill-conditioned. Here, we present a method for fitting a deformable mouse atlas to surface topographic range data acquired by an optical system. As an initialization procedure, we match the posture of the atlas to the posture of the mouse being imaged using landmark constraints. The asymmetric L2 pseudo-distance between the atlas surface and the mouse surface is then minimized in order to register two data sets. A Laplacian prior is used to ensure smoothness of the surface warping field. Once the atlas surface is normalized to match the range data, the internal anatomy is transformed using elastic energy minimization. We present results from performance evaluation studies of our method where we have measured the volumetric overlap between the internal organs delineated directly from MRI or CT and those estimated by our proposed warping scheme. Computed Dice coefficients indicate excellent overlap in the brain and the heart, with fair agreement in the kidneys and the bladder.
1. Introduction
Anatomical mouse atlases are detailed representations of normal morphology and physiology (Jacobs et al 1999, Segars et al 2004) and with appropriate co-registration schemes they can be useful tools for small animal studies involving modalities that are sub-optimal for imaging anatomy (MacKenzie-Graham et al 2004, Thompson and Toga 1997). Specifically, in fluorescence optical tomography (FOT) and bioluminescence tomography (BLT) studies of small animals, where estimation of the internal organ optical properties via all-optical techniques is difficult (Gibson et al 2005), deformable anatomical atlases may be used with published optical properties (Cherry 2004, Alexandrakis et al 2005, Chaudhari et al 2005, Wang et al 2006, Srinivasan et al 2007, Song et al 2007). The atlas must first be aligned with the optical images of the individual mouse being studied. Sufficient accuracy is necessary in this process because the optical properties of organs—the reduced scattering coefficient and the absorption coefficient μa—vary in different tissue types, and misalignment of internal organs can lead to source localization errors (Alexandrakis et al 2006, Han et al 2007). Differences in posture between the mouse being imaged and the atlas make registration particularly challenging. This problem can be ameliorated using a positioning device to ensure consistent posture (Kovacevic et al 2003, Chow et al 2006). Unfortunately, these positioning systems are often not suitable for optical imaging since they attenuate visible light.
The surface geometry and internal anatomy of the animal can be directly measured from computed tomography (CT) or magnetic resonance imaging (MRI) scans acquired before or after the optical scan without moving the animal (Ntziachristos et al 2002, Cherry 2004, Chaudhari et al 2005, Lv et al 2006, Joshi et al 2008, Li et al 2008). However, this requires that the CT or MRI scanner be located within the same facility as the optical instrument. Additionally, there are concerns about radiation dose from CT (especially for longitudinal scans), of viability and of associated cost (Ntziachristos et al 2004). This has motivated the development of all-optical techniques for estimating animal surface geometry. Approaches based on photogrammetric systems (Ripoll et al 2003), systems that project structured light on the animal surface (Rice et al 2006), shadowgrammetry (Meyer et al 2007) or 3D volume carving techniques (Lasser et al 2008) have been published. These systems typically produce a height map of the animal consisting of either discrete points (range data), contours or silhouettes which can then be used to generate a 3D representation of the animal surface. For a finite element method (FEM) solution to the diffusion equation for light propagation, a volumetric tessellation of the animal needs to be generated (Schweiger et al 1993, Arridge et al 2000, Joshi et al 2004). This process may be challenging if these range data are incomplete or under-sampled (Hoppe et al 1992). The animal volume can be assumed to be homogeneous (Rice et al 2001, Ntziachristos and Weissleder 2002). Using this approach, the optical forward propagation modeling of photons in tissue, subject to boundary conditions, can be solved either analytically using a simplified geometry (Rice et al 2001, Schulz et al 2004) or via FEM. However, the homogeneity assumption may lead to both localization and quantification errors (Roy et al 2003, Wang et al 2004, Chaudhari et al 2005). Robustness against optical property variability can be achieved for FOT by data normalization techniques (Soubret et al 2005, Swartling et al 2005). However, this alternative does not exist for BLT (Cong et al 2006).
Instead of simply assuming after surface extraction that the animal volume has homogeneous optical properties, the internal organs may be estimated by using a deformable mouse atlas without (Wang et al 2006) or with surface alignment constraints (Chaudhari et al 2007). Previously these methods had limited success because the deformation was either rigid or did not enforce any constraints on the movement of internal organs. In the past, we have used exact surface matching for registering the atlas to the animal (Chaudhari et al 2007). However, the intermediate coordinate system chosen in this case was initially developed for brain imaging (Joshi et al 2007) and was found to be less suitable for mouse imaging. Thus, inaccuracies in organ alignment prevailed. Here we describe an alternative formulation for atlas deformation with surface-matching constraints.
To register the atlas to the mouse using surface data, we must first define a distance measure between the two surfaces. Commonly used symmetric distance metrics such as L2 or Hausdorff distance are not suitable when one measured data set is incomplete. The data may be incomplete if, for example, the field-of-view of the imaging system is limited, or if data quality is poor (for example, in the head region where occlusions due to the ears or a nose cone that delivers gas anesthesia may occur). In this case, the absence of full correspondence between the two surfaces can cause minima in the distance metric to produce large distortions of the complete surface. On the other hand, when the asymmetric L2 pseudo-distance metric is used, local minima can occur when incomplete data match only the part of the complete data to which they correspond (Pelizzari et al 1989, Zhang 1994). A good initialization is important to obtain a reasonable result, which we provide by first performing posture correction.
In this paper, we describe a volumetric registration scheme (DigiWarp) where the Digimouse atlas (Dogdas et al 2007) is warped to the mouse being optically imaged using only the measured surface of the animal. We achieve this registration in two stages.
The Digimouse is repositioned and its posture is corrected to match the position and posture of the mouse in the acquired data set. This is done using landmark constraints. The warped surface is then used to elastically deform the internal anatomy of the atlas.
The posture-matched atlas is then warped to the available surface topographic data using asymmetric L2 pseudo-distance. The internal anatomy of the posture-corrected atlas is transformed elastically to match its deformed surface.
2. Materials and methods
2.1. The Digimouse atlas
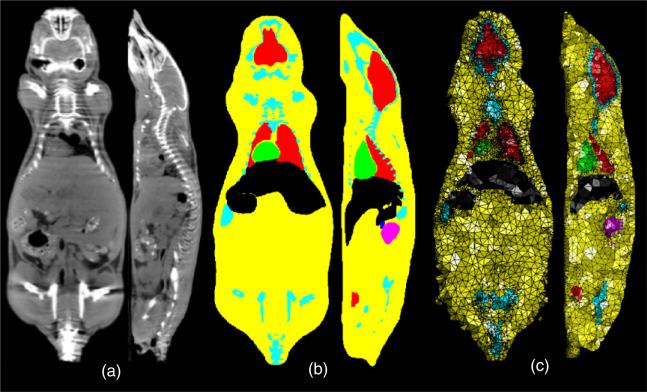
We used the Digimouse atlas http://neuroimage.usc.edu/Digimouse.html as our anatomical template (figure 1). The Digimouse has been generated using co-registered CT and cryosection images of a 28 g nude normal male mouse. Seventeen anatomical structures are labeled in the Digimouse that include the whole brain, external cerebrum, cerebellum, olfactory bulbs, striatum, medulla, massetter muscles, eyes, lachrymal glands, heart, lungs, liver, stomach, spleen, pancreas, adrenal glands, kidneys, testes, bladder, skeleton and the skin. The corresponding volumetric tetrahedral mesh, also available, was generated using the constrained-Delaunay method which conforms to organ boundaries. This mesh contains N = 58 244 vertices and T = 306 773 tetrahedral faces.
Figure 1.
The Digimouse atlas: (a) orthogonal sections from the CT scan, (b) the corresponding sections from the anatomically labeled volume and (c) the same sections from the tetrahedral mesh. The segmented organs of Digimouse are indicated using different colors.
2.2. Deformable elastic modeling of the Digimouse
Mathematical variables and operators used in this and subsequent sections are described in table 1. We model the atlas mouse body as an elastic volume, and therefore displacements to it will be governed by the elastic equilibrium equation (Hughes 1987). We use linearized form of the Piola–Kirchhoff formulation of linear elasticity (Holden 2008). The Piola–Kirchhoff formulation is typically used for finite deformations instead of Cauchy's formulation which is used for infinitesimally small deformations (Hughes 1987).
Table 1.
Mathematical notations used in the paper.
| Variables | Meaning | Dimensions | Mapping |
|---|---|---|---|
| Ω | Mouse atlas volume | ||
| ∂ Ω | Mouse atlas surface (continuous/discretized) | ||
| ∂ P | Set of surface points (continuous/discretized) | ||
| Ŝ | Piola–Kirchhoff strain tensor | ||
| Ĝ | Green–St Venant strain tensor | ||
| S | Linearized Piola–Kirchhoff strain tensor | ||
| G | Linearized Green–St Venant strain tensor | ||
| u | Displacement field over the mouse body | ||
| f | External force field over the mouse body | ||
| U | Vector of displacements at the mesh nodes | ||
| Uk | Displacements at the kth node in the mesh | ||
| L | Elastic energy | ||
| Δ d | Discretized Laplacian operator | ||
| Es | Surface energy | ||
| d | Asymmetric L2 pseudo-distance | ||
| I | Identity operator | ||
| λ | Lameś constant (strain modulus) | ||
| μ | Lameś constant (shear modulus) | ||
| Y | Young's modulus | ||
| ν | Poisson ratio | ||
| N | Number of mesh nodes | ||
| T | Number of tetrahedral faces |
At equilibrium, the elastic energy L(u) corresponding to the displacement u equals the external forces f applied on the body:
| (1) |
where Ŝ denotes the second Piola–Kirchhoff stress tensor defined by Ŝ = λ Tr (Ĝ)I + 2μĜ and Ĝ = ½ (∇uT + ∇u + ∇uT ∇u) represents the Green–St Venant strain tensor. The coefficients λ and μ are the Lamés elastic constants. We use a linearized approximation of (1), given by
| (2) |
where S = λ Tr(G) + 2μG is the linearized stress tensor and G = ½(∇u + ∇uT) is the linearized strain tensor (Postelnicu et al 2009). In our case, the elasticity operator L is discretized using a FEM as described in the appendix. In brief, the equilibrium equation (2) is converted using a variational principle into an energy minimization, that leads to a quadratic form UTKU, where U = [U1, U2, . . . , UN]T is the vector of displacements at N nodes in the tetrahedral mesh. The matrix K discretizes the elastic energy operator and is defined in the appendix.
In the following formulations in sections 2.3 and 2.4, the displacements are first computed at the surface nodes and are then extrapolated for the whole mouse using the elastic deformable model. The surface displacements are modeled as external forces in this formulation and are used to guide the volumetric deformations.
2.3. Posture correction
There are a wide variety of postures in which mice are imaged at various imaging facilities since a standard has not been established. Typically, positions of limbs, the head and the animal orientation vary greatly. As an initial step, the limbs and the head of the Digimouse need to be repositioned to match those of the mouse being imaged. Landmark-based warping cannot be directly employed to warp the elastic atlas because it leads to singularities (Hughes 1987). Instead, we warp the atlas mouse surface first using the surface Laplacian as a regularizer. Then we use the warped atlas mouse surface as a constraint on the volumetric elastic warping. For posture correction we use a two-step procedure: (1) we use a landmark-based method for warping the mouse surface, and (2) we deform the mouse volume with the elastic model from section 2.2.
2.3.1. Surface warping
We select five landmarks from the range data, denoted here by pi ∈ ∂P, i ∈ 1, . . . , 5, namely one each at the ends of four limbs and one at the midpoint between the two ears. The corresponding landmarks ai ∈ ∂Ω, i ∈ 1, . . . , 5, are also selected on the atlas. This gives the five displacement vectors Wi = (pi – ai) required to map the atlas landmarks to the mouse. The displacement vector field is then extrapolated to the whole mouse surface ∂Ω by minimizing the energy
| (3) |
where Δd denotes the discretized Laplacian operator matrix for the atlas surface ∂Ω (Chung and Taylor 2004, Chung et al 2005) and β > 0 is the mismatch penalty parameter. Let the vector be the minimizer of the energy minimization process in (3) and ∂Ωpos = ∂Ω + Upos denote the warped atlas surface.
2.3.2. Volume warping
Having warped the mouse atlas surface to match the posture of the subject mouse, we then warp the internal anatomy of the atlas to fit the new posture, again by elastic energy minimization. We use the elastic model from section 2.2 to warp the internal anatomy of the mouse by minimizing the energy
| (4) |
The solution of this minimization problem is a displacement field Vpos at the volumetric points, which when applied to the atlas Ω leads to a warping of the internal organs consistent with the warped surface. The posture-corrected atlas is now given by Ωpos = Ω + Vpos. Sample postures of the Digimouse atlas that can be obtained using this procedure are shown in figure 2.
Figure 2.
Posture correction for the Digimouse: (a) the Digimouse, (b) a different orientation of the head and (c) a different orientation of the head and the right fore limb.
2.4. Surface fitting and elastic volume warping
Having used the five feature points to adjust the pose of Digimouse to that of the subject mouse, we are now ready to warp the volume atlas to the surface data for that mouse. This is carried out in two steps: (1) surface warping and (2) volumetric warping.
2.4.1. Surface warping
In order to be able to register the complete or incomplete surface topography data recovered from the optical setup to the atlas surface, the matching problem is formulated as an asymmetric L2 pseudo-distance minimization, where the distance is computed from the incomplete surface point-set to the complete surface. We define the asymmetric L2 pseudo-distance metric d between an incomplete acquired surface ∂P and the posture-corrected atlas surface ∂Ωpos by
| (5) |
In this expression, for each location in the surface point-set we find the closest point on the atlas surface. Note that it is a pseudo-distance since it is not symmetric. Our objective, then, is to deform the posture-matched atlas surface ∂Ωpos from section 2.3 such that the distance metric in (5) is minimized. Additionally, we want the displacement field for this operation Upos to be smooth, such that the deformed surface ∂Ωpos + Upos remains smooth. This is achieved by a Laplacian regularizer on the displacement field. The cost function CS is defined as
| (6) |
where Δd denotes the discrete Laplacian (Chung and Taylor 2004) calculated on the triangulated mesh of the mouse atlas surface. The minimization of the pseudo-distance is performed by a searching strategy over the point-set and results in a displacement vector field Us. The searching algorithm is based on the Qhull algorithm (Barber et al 1996) and is implemented in MATLAB®'s dsearchn function. The displacement field Us obtained as a result of this minimization is then applied to the posture-corrected atlas surface ∂Ωpos to get the surface ∂Ωs = ∂Ωpos + Us that matches with the range data ∂P.
2.4.2. Volume warping
Similar to section 2.3, the surface warping field is then extrapolated to the entire mouse volume using the elastic regularizer to further warp the posture adjusted mouse atlas Ωpos to match the surface of the subject mouse. The energy minimization
| (7) |
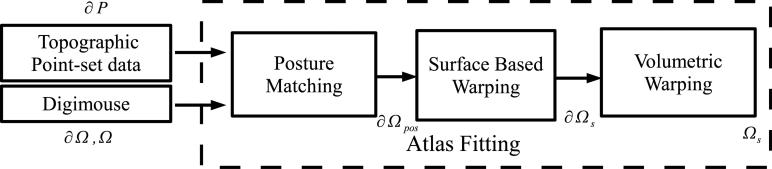
leads to a displacement field Uvol at the volumetric points, which when applied to the posture-corrected atlas Ωpos leads to a warping of the internal organs consistent with the warped surface ∂Ωs. Thus, the posture-corrected and surface-matched atlas is given by Ωs = Ωpos + Uvol. The result is a warped mouse atlas Ωs such that its surface ∂Ωs conforms to the range data ∂P. A flowchart of the complete registration process is shown in figure 3.
Figure 3.
Flowchart for the proposed method of deformable atlas registration to topographic range data.
2.5. Implementation of the warping method
The warping method was implemented in MATLAB®. The conjugate gradient minimization procedure was used for the cost function minimization procedures in sections 2.3 and 2.4. We empirically chose Y = 1, ν = 0.3, α = 3, β = 1. The whole method took approximately 20–30 min of runtime on a Pentium IV 3.6 GHz machine with 4GB RAM.
2.6. Acquisition of surface topographic range data—mouse 1
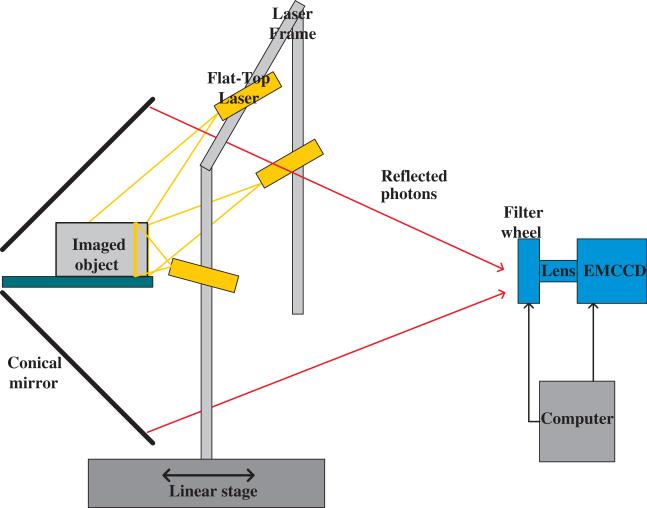
Our surface profiling scheme used a conical mirror with a horizontal stage that held the animal oriented axially inside it. Details of this setup (shown in figure 4) and the associated image acquisition protocol are in Li et al (2009). In brief, to profile the dorsal part of the animal, a laser line was projected on the animal surface. This line traces a planar trajectory from the laser to the surface of the animal. The equation of this plane is estimated by measuring the slope and intercept of the projected line on the stage. The reflection of the line is a bright curve seen on the conical mirror image. Each point on this image curve corresponds to a surface point that lies along the radial line that passes through it. Thus, the corresponding surface point can be determined from the intersection of the radial line and the plane traced by the laser light. By translating the laser source longitudinally we obtain 3D coordinates for a set of points lying on the upper mouse surface. To obtain the full surface, three line lasers are mounted on a frame: one on the top and two on the sides. A normal adult mouse (nu/nu, weight = 24 g) was scanned using this setup. Data acquisition lasted 3 min. The temperature of the animal was monitored throughout the scan. Anesthesia was maintained by means of the delivery of 2% isofluorane via the nose cone.
Figure 4.
Optical setup used for acquisition of optical surface topographic data (Li et al 2009).
2.7. micro-CT acquisition and segmentation for examining skull and skeletal overlap—mouse 1
After optical scanning, the mouse with its stage was transferred to a MicroCAT II CT scanner (Siemens Preclinical Solutions, Knoxville, TN) and a whole body scan was collected. The x-ray voltage and anode current were set to 80 kVp and 200 μA, respectively. The scan lasted 4 min and 2 s. The temperature of the animal was maintained at 37 °C. The anesthesia was maintained by means of the delivery of 2% isofluorane gas via the nose cone. The experiment was performed under a protocol approved by the University of California-Davis Animal Care and Use Committees. Images were reconstructed into 0.097 mm cubic voxels with the Shepp–Logan filter. Using DigiWarp, the Digimouse was warped to the optical range data. Using this computed warping field, the Digimouse CT was transformed. The skull and skeleton of the warped Digimouse CT were extracted by straightforward thresholding using BrainSuite (Shattuck and Leahy 2002). The animal skull and skeleton were also directly extracted from its CT image using BrainSuite.
2.8. MRI acquisition and segmentation for evaluation of soft tissue overlap—mouse 2
Range data were obtained for a second normal adult mouse (nu/nu, weight = 26 g) using the method described in section 2.6. After optical scanning, the mouse with its stage was transferred to the MRI facility and was imaged using the Bruker 7T Biospec small-animal MR scanner equipped with the Bruker B-GA12 gradient coil set. The temperature of the animal was monitored constantly and maintained at 37 °C. 2% isofluorane was used for delivery of gas anesthesia. Whole-body MRI scanning was performed under a protocol approved by the University of California-Davis Animal Care and Use Committees. For data acquisition and image reconstruction, the ParaVision software package (Bruker) was used. The resultant images had a voxel size (0.23×0.23×0.5) mm2. The brain, the heart, the two kidneys and the bladder were manually segmented by an experienced observer using BrainSuite 2.0 (Shattuck and Leahy 2002). This observer was blind to the design of the study. Unique labels were assigned to the segmented organs.
2.9. Assessment of mesh quality
Topologically correct meshes are necessary for solving the forward problems in OBT and FOT. The following mesh quality metrics based on Berzins (1998) were used to evaluate the meshes produced by DigiWarp. Quality measures M1–M5 were computed for each tetrahedron in the meshes based on the following definitions: M1: 3 times the radius of the insphere divided by the radius of the circumsphere; M2: times the radius of the insphere divided by the length of the longest side of the tetrahedron; M3: 12 × (3 × volume)2/3/(sum of squares of edge lengths); M4: the minimum solid angle in a tetrahedron and M5: a uniformity measure, the ratio of the volume of a tetrahedon to maximum tetrahedral volume in the mesh. The maximum and best possible value for each measure is 1, and the minimum and worst value is 0. These measures were calculated using a MATLAB program available from http://people.sc.fsu.edu/~jburkardt/m_src/tet_mesh_quality/tet_mesh_quality.html.
3. Results
3.1. CT-based evaluation of the proposed method—mouse 1
3.1.1. Warping of the Digimouse to range data
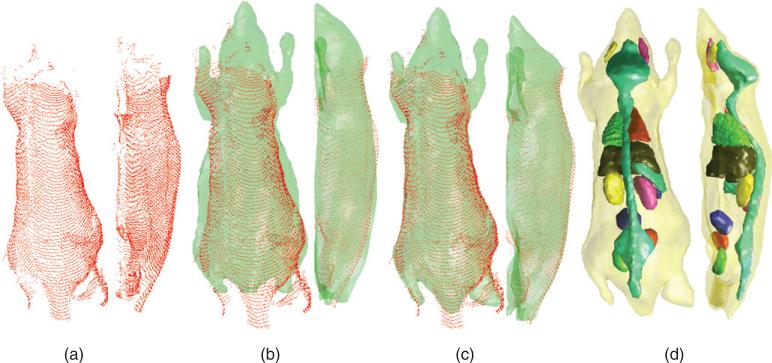
Figure 5(a) shows the point cloud representing the top and side surfaces of mouse 1. The data are incomplete and sparse in many regions. Figure 5(b) shows an overlay of the point cloud on the posture-corrected Digimouse surface (shown in green). This warping was further improved by the asymmetric L2 distance minimization procedure to obtain the results in figure 5(c). The internal organ distribution obtained after volumetric elastic warping using surface deformation as a guide is shown in figure 5(d).
Figure 5.
Estimation of internal anatomy of mouse 1 from optical data: (a) the point cloud obtained from the optical scan, (b) the point cloud and the posture-corrected Digimouse, (c) the point cloud and the final result of surface fitting by the proposed scheme and (d) elastically transformed animal volume whose surface fits the point cloud.
3.1.2. Skull and skeleton overlap comparisons
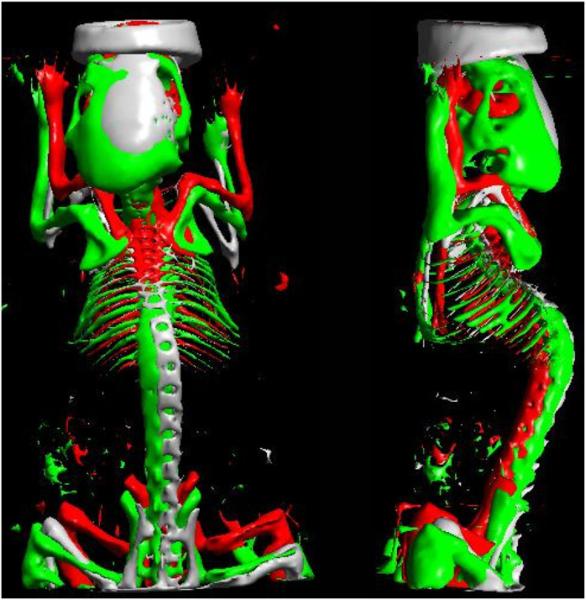
The Digimouse was registered to mouse 1 as shown in figure 5(c). The estimated warping transform was applied to the Digimouse CT data. By applying straightforward thresholding to the warped Digimouse CT, we could segment out the skull and the skeleton. The same procedure of thresholding was followed for mouse 1 and for the rigidly registered Digimouse CT. In figure 6, we show an overlay of the extracted skeletons from the three CT data. The 3D rendering was performed in BrainSuite. In order to quantify the amount of overlap, we use Dice coefficients (defined for volumes A and B as ) (Van Rijsbergen 1979). For skeletons, Dice coefficients were 0.11 for rigid alignment, 0.21 for posture correction and 0.38 for surface-based warping. The low Dice coefficients in this case can be attributed to the fact that skeletons are relatively thin.
Figure 6.
The skeleton extracted from thresholding the CT image of mouse 1 (grey), the rigidly registered Digimouse (red) and the Digimouse registered using DigiWarp (green). We can see that the skeleton of the Digimouse registered by DigiWarp shows better overlap with the original skeleton of the mouse than the one registered rigidly.
3.2. MRI-based evaluation—mouse 2
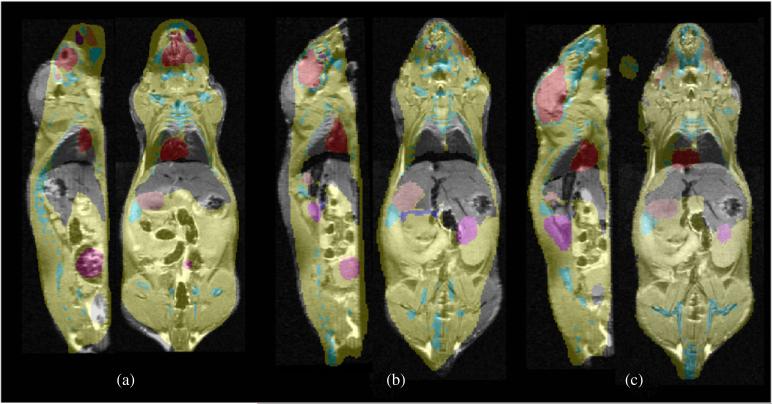
We matched the posture of the Digimouse atlas to that of mouse 2 using the proposed method. The result is a repositioned and posture-matched atlas. The warping field due to posture matching that was generated from the tetrahedral mesh was applied to the labeled volume of the Digimouse. An overlay of warped labels and the mouse MRI is shown in figure 7(b). The asymmetric L2 pseudo-distance-based minimization procedure leads to improved surface and volumetric warping as shown in figure 7(c). For comparison, in figure 7(a), we show an overlay of labels obtained from rigidly registering the Digimouse to the range data for the same sections in figures 7(b) and (c). In figure 8, we show closeup views of the overlay of mouse MRI and the Digimouse warped using DigiWarp.
Figure 7.
Results from the performance evaluation study. In each case, three orthogonal sections from the MRI scan of the subject mouse are overlaid with the corresponding sections of the warped Digimouse labels (a) after only rigid registration, (b) after posture correction and (c) after the asymmetric L2 distance-based method were used.
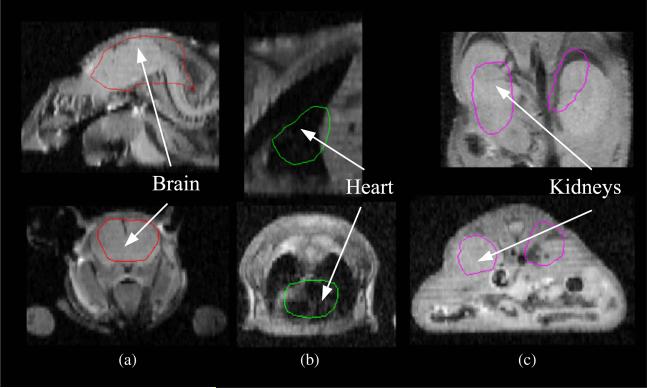
Figure 8.
Results from the performance evaluation study for individual organs. In each case, orthogonal sections from the MRI scan of mouse 2 are overlaid with the corresponding sections of the warped Digimouse labels: (a) head region shows alignment of the brain and skull, (b) mid region shows alignment of the heart and lungs, (c) abdominal region shows alignment of the kidneys.
We compared the accuracy of our method by measuring Dice coefficients of organ overlap between warped atlas labels and labels assigned by manual outlining for the brain, the heart, the two kidneys and the bladder. Our results are tabulated in table 2.
Table 2.
Dice coefficients of organ overlap between warped atlas labels (A) and manually assigned labels (B).
| Organ name | Dice coefficient for rigid registration | Dice coefficient for our method |
|---|---|---|
| Brain | 0.16 | 0.83 |
| Heart | 0.64 | 0.80 |
| Kidneys | 0.11 | 0.32 |
| Bladder | 0.29 | 0.62 |
| Whole body | 0.41 | 0.96 |
3.3. Assessment of mesh quality
We show results from our evaluation (mean and standard deviation of quality measures) of the original Digimouse mesh and the warped meshes for mice 1 and 2 in table 3. Statistically insignificant differences were found in mesh quality measures between the original Digimouse mesh and the deformed meshes generated from DigiWarp. We have employed these meshes for solving OBT and OFT forward models based on Chaudhari et al (2009, 2005), and have not encountered any problems. Detailed validation of the optical forward models using a Monte Carlo-based solver can be performed; however, this is outside the scope of the paper.
Table 3.
Mesh quality measures: mean (standard deviation).
| Measure | Original mesh | Warped mesh (section 3.1) | Warped mesh (section 3.2) |
|---|---|---|---|
| M 1 | 0.617(0.17) | 0.582(0.18) | 0.591(0.17) |
| M 2 | 0.538(0.16) | 0.512(0.16) | 0.524(0.17) |
| M 3 | 0.679(0.16) | 0.652(0.17) | 0.650(0.16) |
| M 4 | 0.349(0.17) | 0.324(0.17) | 0.334(0.17) |
| M 5 | 0.063(0.11) | 0.036(0.18) | 0.053(0.17) |
4. Discussion
We have presented the DigiWarp registration scheme for estimating the internal anatomy of a mouse when only surface topographic information is available. We evaluated DigiWarp against results from anatomical imaging modalities. For the heart and the brain, high Dice coefficients were computed between the animal's MRI image and the warped atlas. This implies that the head and the chest region of the warped atlas have excellent alignment with MRI. The bladder and kidneys show large shape variability across subjects, but a significant improvement over rigid registration for volumetric overlap is achieved using the proposed method. The skeleton also shows improvement as discussed in section 3.1.2. We note that the DigiWarp method can be used with any mouse atlas and is not limited to the Digimouse.
Overall, we found that the organs near the skin and skeleton, especially the brain and heart, were aligned well with our method while the organs in the trunk of the mouse body were aligned less accurately. The limited ability of the DigiWarp method for producing alignment in the trunk may be attributed to the fact that the proposed method is based on surface matching alone and may not be able to model large non-rigid deformations taking place in soft tissue. Additionally, the anatomical variability in the mouse being imaged and in the Digimouse, as well as intra-species variability in mouse anatomy, makes the process of alignment complex (Kovacevic et al 2005). The use of a mouse atlas from the same strain as that of the mouse being imaged, instead of the standard Digimouse atlas, may further improve organ overlap. Creating strain-specific mouse atlases may be challenging, but the proposed approach may provide a potential solution as we describe next.
Owing to reasonable Dice coefficients obtained using the proposed registration scheme, the method can provide an initialization for whole-body mouse segmentation in cases where an intensity-based anatomical image such as CT or MR is available. The organ labels in this case can be initialized using the proposed method, and can further be refined to conform to the anatomical intensity image using deformable shape-based segmentation algorithms (Yushkevich et al 2006, Ghanei et al 1998, Bulpitt and Berry 1998). This technique will need further validation, but if successful it will facilitate the construction of specie-specific atlases. Additionally, in cases where internal anatomical information is available, our proposed method can provide a good initialization for atlas-based registration and facilitate intra-modality comparisons.
The exact alignment of mouse surfaces is not enforced by our method since we wanted the deformation of the atlas surface to be smooth. Thus, we have a Laplacian regularizing term in the matching energy equation (3). This term tends to avoid fold-overs in the deformed surface and leads to topologically correct deformations of the mouse. Quasi-rigid and bending priors can be enforced in our method similar to those proposed by Eckstein et al (2007). Such priors allow rigid movement of the bones and the skeleton while still relaxing the rigidity constraints for other organs. We anticipate that this will lead to further improvement. These constraints will be especially useful in cases where an animal has a superficial tumor that causes an anomaly in the surface profile.
In BLT and FOT, where data are measured on the surface and boundary conditions are applied based on surface topography, the DigiWarp method provides a scheme for estimating internal anatomy. The proposed scheme can allow detailed investigation of anatomical variability on optical source reconstruction and facilitate comparisons with methods that derive anatomical information from all-optical techniques (Tan and Jiang 2008, Hillman and Moore 2007). Additionally, the organ map derived from DigiWarp can serve as an anatomical prior for diffuse optical tomography (Guven et al 2005).
5. Conclusions
We have described a deformable atlas-based registration algorithm for posture correction and surface-based volumetric warping (DigiWarp) with utility for small animal optical tomography studies. A comparison based on Dice coefficients showed that the DigiWarp method resulted in a reasonably accurate alignment of the anatomical organs, even though only surface maps are used. Our method worked well for incomplete data sets and produced topologically correct warps. We conclude that the DigiWarp method potentially may obviate the need of unnecessary anatomical imaging for optical imaging studies. The method also has broader utility for atlas-based small animal registration and image segmentation.
Acknowledgments
The authors would like to thank Dr Ramsey D Badawi, Jennifer Fung and Chris Griesemer of the University of California-Davis Animal Care and Use Committees for helpful discussions and for providing access to the data that were used in this paper. This work was funded by the National Institutes of Health under grants R01CA121783, R01CA129561 and R01RR020060, the Komen Foundation and National Institutes of Health through the NIH Roadmap for Medical Research, grant U54 RR021813 and UL1 RR024146. Information on the National Centers for Biomedical Computing can be obtained from http://nihroadmap.nih.gov/bioinformatics.
Appendix. Finite element method
The solution to the elastic energy minimization problem in section 2.2. is obtained by the FEM. We use tetrahedral elements in order to discretize the elastic energy. Our objective is to find the displacement field at every point in the mouse body. The mouse volume is divided into tetrahedral elements such that every point in the space lies in exactly one tetrahedron. We assume that the displacement field u(x, y, z) is piecewise linear, i.e. if the point is inside tetrahedron, then
| (A.1) |
for coefficients , , , . For the tetrahedron i with nodes , , and , we can write expressions for u in matrix form as
| (A.2) |
Since we assume the function to be piecewise linear over each tetrahedral element, its derivatives are piecewise constants in each tetrahedron, i.e. if the point (x, y, z) is inside the ith tetrahedron, then
| (A.3) |
These coefficients can be obtained by inverting the matrix M in equation (A.2) and therefore the derivative operators for a tetrahedral element el are obtained by arranging the terms of M–1, and are given by
| (A.4) |
| (A.5) |
| (A.6) |
and
| (A.7) |
where r is a resizing function that keeps track of indices of the individual nodes in the whole mesh. This kind of reindexing is commonly performed in FEM techniques (Hughes 1987). Let the matrices L, LW and K be defined as
| (A.8) |
| (A.9) |
| (A.10) |
We arrange the x, y, z components of the displacement (Ux, Uy, Uz) at nodal points in a column vector U = [Ux, Uy, Uz]T. Then the elastic energy, without any external forces, is given by
| (A.11) |
We use this discretization of the energy equation for posture correction as described in section 2.3 and for surface-based volume warping as described in section 2.4.
References
- Alexandrakis G, Rannou F, Chatziioannou AF. Tomographic bioluminescence imaging by use of OPET. Phys. Med. Biol. 2005;50:4225–41. doi: 10.1088/0031-9155/50/17/021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrakis G, Rannou FR, Chatziioannou AF. Effect of optical property estimation accuracy on tomographic bioluminescence imaging: simulation of a combined optical-PET (OPET) system. Phys. Med. Biol. 2006;51:2045–53. doi: 10.1088/0031-9155/51/8/006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arridge S, Dehghani H, Schweiger M, Okada E. The finite element model for the propagation of light in scattering media: a direct method for domains with nonscattering regions. Med. Phys. 2000;27:252. doi: 10.1118/1.598868. [DOI] [PubMed] [Google Scholar]
- Barber C, Dobkin D, Huhdanpaa H. The quickhull algorithm for convex hulls. ACM Trans. Math. Softw. 1996;22:469–83. [Google Scholar]
- Berzins M. A solution-based triangular and tetrahedral mesh quality indicator. SIAM J. Sci. Comput. 1998;19:2051. [Google Scholar]
- Bulpitt A, Berry E. Spiral CT of abdominal aortic aneurysms: comparison of segmentation with an automatic 3D deformable model and interactive segmentation. Proc. SPIE. 1998;3338:938. [Google Scholar]
- Chaudhari AJ, Ahn S, Levenson R, Badawi R D, Cherry SR, Leahy RM. Excitation spectroscopy in multispectral optical fluorescence tomography: methodology, feasibility and computer simulation studies. Phys. Med. Biol. 2009;54:4687. doi: 10.1088/0031-9155/54/15/004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhari AJ, Darvas F, Bading JR, Moats RA, Conti PS, Smith DJ, Cherry SR, Leahy RM. Hyperspectral and multispectral bioluminescence optical tomography for small animal imaging. Phys. Med. Biol. 2005;50:5421–41. doi: 10.1088/0031-9155/50/23/001. [DOI] [PubMed] [Google Scholar]
- Chaudhari AJ, Joshi AA, Darvas F, Leahy RM. A method for atlas-based volumetric registration with surface constraints for optical bioluminescence tomography in small animal imaging. Proc. SPIE. 2007;6510:651024. [Google Scholar]
- Cherry SR. In vivo molecular and genomic imaging: new challenges for imaging physics. Phys. Med. Biol. 2004;49:R13–48. doi: 10.1088/0031-9155/49/3/r01. [DOI] [PubMed] [Google Scholar]
- Chow P, Stout D, Komisopoulou E, Chatziioannou A. A method of image registration for small animal, multi-modality imaging. Phys. Med. Biol. 2006;51:379. doi: 10.1088/0031-9155/51/2/013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung M, Robbins S, Dalton K, Davidson R, Alexander A, Evans A. Cortical thickness analysis in autism with heat kernel smoothing. Neuroimage. 2005;25:1256–65. doi: 10.1016/j.neuroimage.2004.12.052. [DOI] [PubMed] [Google Scholar]
- Chung M, Taylor J. Diffusion smoothing on brain surface via finite element method. IEEE Int. Symp. on Biomedical Imaging: Macro to Nano (2004) 2004:432–35. [Google Scholar]
- Cong W, Durairaj K, Wang L, Wang G. A born-type approximation method for bioluminescence tomography. Med. Phys. 2006;33:679–86. doi: 10.1118/1.2168293. [DOI] [PubMed] [Google Scholar]
- Dogdas B, Stout D, Chatziioannou AF, Leahy RM. Digimouse: a 3D whole body mouse atlas from CT and cryosection data. Phys. Med. Biol. 2007;52:577–87. doi: 10.1088/0031-9155/52/3/003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckstein I, Pons J, Tong Y, Kuo C, Desbrun M. Generalized surface flows for mesh processing. Symp. on Geometry Processing. 2007:183–92. [Google Scholar]
- Ghanei A, Soltanian-Zadeh H, Windham J. A 3D deformable surface model for segmentation of objects from volumetric data in medical images. Comput. Biol. Med. 1998;28:239–53. doi: 10.1016/s0010-4825(98)00007-9. [DOI] [PubMed] [Google Scholar]
- Gibson AP, Hebden JC, Arridge SR. Recent advances in diffuse optical imaging. Phys. Med. Biol. 2005;50:R1–43. doi: 10.1088/0031-9155/50/4/r01. [DOI] [PubMed] [Google Scholar]
- Guven M, Yazici B, Intes X, Chance B. Diffuse optical tomography with a priori anatomical information. Phys. Med. Biol. 2005;50:2837–58. doi: 10.1088/0031-9155/50/12/008. [DOI] [PubMed] [Google Scholar]
- Han W, Kazmi K, Cong W, Wang G. Bioluminescence tomography with optimized optical parameters. Inverse Problems. 2007;23:1215–28. [Google Scholar]
- Hillman E, Moore A. All-optical anatomical co-registration for molecular imaging of small animals using dynamic contrast. Nat. Photonics. 2007;1:526–30. doi: 10.1038/nphoton.2007.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden M. A review of geometric transformations for nonrigid body registration. IEEE Trans. Med. Imaging. 2008;27:111–28. doi: 10.1109/TMI.2007.904691. [DOI] [PubMed] [Google Scholar]
- Hoppe H, DeRose T, Duchamp T, McDonald J, Stuetzle W. Surface reconstruction from unorganized points. Comput. Graph. 1992;26:71–8. [Google Scholar]
- Hughes T. The Finite Element Method. Prentice-Hall; Englewood Cliffs, NJ: 1987. [Google Scholar]
- Jacobs R, Ahrens E, Dickinson M, Laidlaw D. Towards a micro MRI atlas of mouse development. Comput. Med. Imaging Graph. 1999;23:15–24. doi: 10.1016/s0895-6111(98)00059-7. [DOI] [PubMed] [Google Scholar]
- Joshi A, Bangerth W, Sevick-Muraca EM. Adaptive finite element based tomography for fluorescence optical imaging in tissue. Opt. Express. 2004;12:5402–17. doi: 10.1364/opex.12.005402. [DOI] [PubMed] [Google Scholar]
- Joshi A, Rasmussen J, Kwon S, Wareing T, McGhee J, Sevick-Muraca E. Multi-modality CT-PET-NIR fluorescence tomography. 5th IEEE Int. Symp. on Biomedical Imaging: From Nano to Macro. 2008:1601–4. [Google Scholar]
- Joshi A, Shattuck D, Thompson P, Leahy R. Surface-constrained volumetric brain registration using harmonic mappings. IEEE Trans. Med. Imaging. 2007;26:1657–69. doi: 10.1109/tmi.2007.901432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacevic N, Hamarneh G, Henkelman M. Medical Image Computing and Computer-Assisted Intervention—MICCAI 2003. Springer; Berlin: 2003. Anatomically guided registration of whole body mouse MR images; pp. 870–7. Lecture Notes in Computer Science vol 2879. [Google Scholar]
- Kovacevic N, Henderson J, Chan E, Lifshitz N, Bishop J, Evans A, Henkelman R, Chen X. A three-dimensional MRI atlas of the mouse brain with estimates of the average and variability. Cerebral Cortex. 2005;15:639–45. doi: 10.1093/cercor/bhh165. [DOI] [PubMed] [Google Scholar]
- Lasser T, Soubret A, Ripoll J, Ntziachristos V. Surface Reconstruction for free-space 360 degrees fluorescence molecular tomography and the effects of animal motion. IEEE Trans. Med. Imaging. 2008;27:188. doi: 10.1109/TMI.2007.904662. [DOI] [PubMed] [Google Scholar]
- Li C, Mitchell G, Dutta J, Ahn S, Leahy R, Cherry S. A three-dimensional multispectral fluorescence optical tomography imaging system for small animals based on a conical mirror design. Opt. Express. 2009;17:7571–85. doi: 10.1364/oe.17.007571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yankeelov T E, Peterson T E, Gore JC, Dawant BM. Automatic nonrigid registration of whole body CT mice images. Med. Phys. 2008;35:1507–20. doi: 10.1118/1.2889758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Y, Tian J, Cong W, Wang G, Kumar D. MicroCT-guided bioluminescence tomography based on the adaptive finite element tomographic algorithm. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2006;1:381–4. doi: 10.1109/IEMBS.2006.260762. [DOI] [PubMed] [Google Scholar]
- MacKenzie-Graham A, et al. A multimodal, multidimensional atlas of the C57BL/6J mouse brain. J. Anat. 2004;204:93. doi: 10.1111/j.1469-7580.2004.00264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H, Garofalakis A, Zacharakis G, Psycharakis S, Mamalaki C, Kioussis D, Economou EN, Ntziachristos V, Ripoll J. Noncontact optical imaging in mice with full angular coverage and automatic surface extraction. Appl. Opt. 2007;46:3617–27. doi: 10.1364/ao.46.003617. [DOI] [PubMed] [Google Scholar]
- Ntziachristos V, Schellenberger E, Ripoll J, Yessayan D, Graves E, Bogdanov A, Josephson L, Weissleder R. Visualization of antitumor treatment by means of fluorescence molecular tomography with an annexin V-Cy5.5 conjugate. Proc. Natl Acad. Sci. 2004;101:12294–9. doi: 10.1073/pnas.0401137101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntziachristos V, Tung C, Bremer C, Weissleder R. Fluorescence molecular tomography resolves protease activity in vivo. Nat. Med. 2002;8:757–61. doi: 10.1038/nm729. [DOI] [PubMed] [Google Scholar]
- Ntziachristos V, Weissleder R. Charge-coupled-device based scanner for tomography of fluorescent near-infrared probes in turbid media. Med. Phys. 2002;29:803. doi: 10.1118/1.1470209. [DOI] [PubMed] [Google Scholar]
- Pelizzari C, Chen G, Spelbring D, Weichselbaum R, Chen C. Accurate three-dimensional registration of CT, PET, and/or MR images of the brain. J. Comput. Assist. Tomogr. 1989;13:20. doi: 10.1097/00004728-198901000-00004. [DOI] [PubMed] [Google Scholar]
- Postelnicu G, Zollei L, Fischl B. Combined volumetric and surface registration. IEEE Trans. Med. Imaging. 2009;28:508–22. doi: 10.1109/TMI.2008.2004426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice B W, Cable MD, Nelson MB. In vivo imaging of light-emitting probes. J. Biomed. Opt. 2001;6:432–40. doi: 10.1117/1.1413210. [DOI] [PubMed] [Google Scholar]
- Rice B W, Xu H, Kuo C. Surface construction using combined photographic and structured light information. Patent Application. 2006:0268153. [Google Scholar]
- Ripoll J, Schulz RB, Ntziachristos V. Free-space propagation of diffuse light: theory and experiments. Phys. Rev. Lett. 2003;91:103901. doi: 10.1103/PhysRevLett.91.103901. [DOI] [PubMed] [Google Scholar]
- Roy R, Godavarty A, Sevick-Muraca E. Fluorescence-enhanced optical tomography using referenced measurements of heterogeneous media. IEEE Trans. Med. Imaging. 2003;22:824–36. doi: 10.1109/TMI.2003.815072. [DOI] [PubMed] [Google Scholar]
- Schulz R B, Ripoll J, Ntziachristos V. Experimental fluorescence tomography of tissues with noncontact measurements. IEEE Trans. Med. Imaging. 2004;23:492–500. doi: 10.1109/TMI.2004.825633. [DOI] [PubMed] [Google Scholar]
- Schweiger M, Arridge S, Delpy D. Application of the finite-element method for the forward and inverse models in optical tomography. J. Math. Imaging Vis. 1993;3:263–83. [Google Scholar]
- Segars W P, Tsui BM, Frey E C, Johnson GA, Berr SS. Development of a 4-D digital mouse phantom for molecular imaging research. Mol. Imaging Biol. 2004;6:149–59. doi: 10.1016/j.mibio.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Shattuck D, Leahy R. BrainSuite: an automated cortical surface identification tool. Med. Image Anal. 2002;6:129–42. doi: 10.1016/s1361-8415(02)00054-3. [DOI] [PubMed] [Google Scholar]
- Song X, Wang D, Chen N, Bai J, Wang H. Reconstruction for free-space fluorescence tomography using a novel hybrid adaptive finite element algorithm. Opt. Express. 2007;15:18300–17. doi: 10.1364/oe.15.018300. [DOI] [PubMed] [Google Scholar]
- Soubret A, Ripoll J, Ntziachristos V. Accuracy of fluorescent tomography in the presence of heterogeneities: study of the normalized Born ratio. 24. IEEE Trans. Med. Imaging. 2005:1377–86. doi: 10.1109/tmi.2005.857213. [DOI] [PubMed] [Google Scholar]
- Srinivasan S, Pogue BW, Davis S, Leblond F. Improved quantification of fluorescence in 3-d in a realistic mouse phantom. Proc. SPIE. 2007;6434:643405. [Google Scholar]
- Swartling J, Svensson J, Bengtsson D, Terike K, Andersson-Engels S. Fluorescence spectra provide information on the depth of fluorescent lesions in tissue. Appl. Opt. 2005;44:1934–41. doi: 10.1364/ao.44.001934. [DOI] [PubMed] [Google Scholar]
- Tan Y, Jiang H. DOT guided fluorescence molecular tomography of arbitrarily shaped objects. Med. Phys. 2008;35:5703. doi: 10.1118/1.3020594. [DOI] [PubMed] [Google Scholar]
- Thompson P, Toga A. Detection, visualization and animation of abnormal anatomic structure with a deformable probabilistic brain atlas based on random vector field transformations. Med. Image Anal. 1997;1:271–94. doi: 10.1016/s1361-8415(97)85002-5. [DOI] [PubMed] [Google Scholar]
- Van Rijsbergen C. Information Retrieval. Butterworth-Heinemann; Newton, MA: 1979. [Google Scholar]
- Wang G, Cong W, Durairaj K, Qian X, Shen H, Sinn P, Hoffman E, McLennan G, Henry M. In vivo mouse studies with bioluminescence tomography. Opt. Express. 2006;14:7801–9. doi: 10.1364/oe.14.007801. [DOI] [PubMed] [Google Scholar]
- Wang G, Li Y, Jiang M. Uniqueness theorems in bioluminescence tomography. Med. Phys. 2004;31:2289. doi: 10.1118/1.1766420. [DOI] [PubMed] [Google Scholar]
- Yushkevich P, Piven J, Hazlett H, Smith R, Ho S, Gee J, Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31:1116–28. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Zhang Z. Iterative point matching for registration of free-form curves and surfaces. Int. J. Comput. Vis. 1994;13:119–52. [Google Scholar]