Abstract
Several inwardly-rectifying (Kir) potassium channels (Kir1.1, Kir4.1 and Kir4.2) are characterised by their sensitivity to inhibition by intracellular H+ within the physiological range. The mechanism by which these channels are regulated by intracellular pH has been the subject of intense scrutiny for over a decade, yet the molecular identity of the titratable pH-sensor remains elusive. In this study we have taken advantage of the acidic intracellular environment of S. cerevisiae and used a K+-auxotrophic strain to screen for mutants of Kir1.1 with impaired pH-sensitivity. In addition to the previously identified K80M mutation, this unbiased screening approach identified a novel mutation (S172T) in the second transmembrane domain (TM2) that also produces a marked reduction in pH-sensitivity through destabilization of the closed-state. However, despite this extensive mutagenic approach, no mutations could be identified which removed channel pH-sensitivity or which were likely to act as a separate H+-sensor unique to the pH-sensitive Kir channels. In order to explain these results we propose a model in which the pH-sensing mechanism is part of an intrinsic gating mechanism common to all Kir channels, not just the pH-sensitive Kir channels. In this model, mutations which disrupt this pH-sensor would result in an increase, not reduction, in pH-sensitivity. This has major implications for any future studies of Kir channel pH-sensitivity and explains why formal identification of these pH-sensing residues still represents a major challenge.
Key words: pH-sensitivity, Kir channel, pH-sensor, potassium channel, Kir1.1
Introduction
Inwardly-rectifying (Kir) potassium channels are important regulators of cellular electrical activity and K+ transport processes through their ability to couple channel activity to a wide range of metabolic and physiological stimuli.1,2 A subset of the Kir channel superfamily are characterised by their sensitivity to inhibition by intracellular H+ and include Kir1.1, Kir4.1, Kir4.2 and heteromeric Kir4.x/Kir5.1 channels. These ‘pH-sensitive’ Kir channels are abundantly expressed in epithelial and glial cell types where their sensitivity to changes in intracellular pH (pHi) is thought to be important for the regulation of K+ transport and K+ homeostasis.3 Indeed, their importance is highlighted by the discovery that inherited mutations in the Kir1.1 gene (KCNJ1) underlie Type II Bartter's Syndrome, a renal hypokalaemic disorder and mutations in Kir4.1 (KCNJ10) cause SeSAME/EAST syndrome, a complex disorder involving sensorineural deafness, ataxia, mental retardation and electrolyte imbalance.4–6
The molecular mechanism of Kir channel pH-regulation has been the subject of investigation for well over a decade, yet the identity of the titratable pH-sensor remains unknown.7–12 All Kir channels show some sensitivity to intracellular pH and can be inhibited by low pH e.g., pH 5.0. However, only the ‘pH-sensitive’ channels respond to changes in the physiological range; Kir1.1 has an IC50 = 6.5, Kir4.1 IC50 = 6.0, Kir4.2 IC50 = 6.9 and Kir4.1/Kir5.1 IC50 = 7.4.13,14 Initially a lysine residue at the base of TM1 was thought to act as the pH-sensor because mutation of this residue (K80) in Kir1.1 causes a significant reduction in pH-sensitivity.7 Furthermore, a lysine is only found at this TM1 position in the pH-sensitive channels (K67 in Kir4.1 and K66 in Kir4.2) and introduction of a lysine into this position in the non-pH sensitive channels (e.g., Kir2.1-M84K) dramatically increases their pH-sensitivity.7,13 However, several recent studies have now shown that although this lysine is critical for ‘sensitising’ the channel to H+-inhibition, it does not act as the H+-sensor itself.15,16 Instead, this lysine forms an intra-subunit H-bond with the adjacent residue in TM2 and this H-bonding at the helix-bundle crossing enhances Kir1.1 pH-sensitivity by stabilizing the closed state of the channel.
Several previous studies have addressed the identity of the H+-sensor in Kir1.1. Mutation of the histidine residues in Kir1.1 either singly or in multiple combinations, only produces a modest reduction in pH-sensitivity.17 To date the only mutations identified which reduce the IC50 < pH 6.0 are K80M (IC50 = 5.3) and I63R (IC50 = 5.8). However, in both cases the pH-sensitivity is not completely removed, but simply shifted into the acidic range, where their pH-response is similar to the non pH-sensitive channels such as Kir2.1.9,18 By contrast, the vast majority of mutations in Kir1.1 either have no effect on pH-sensitivity or shift it into the alkaline range i.e., they make the channel more sensitive to intracellular pH and this effect is seen with many of the ‘loss of function’ mutations which give rise to Type II Bartter's Syndrome. In these cases, the shift in pH-sensitivity causes the channel to be inhibited within the physiological range, and only activated at highly alkaline pH.4,19
Given the apparent lack of success of such site-directed mutagenesis approaches to identify the pH-sensor, we therefore decided to take a different approach. In a previous study we used a K+-auxotrophic strain of S. cerevisiae to screen for activatory mutations in the prokaryotic potassium channel KcsA.20 This yeast strain (SGY1528) has its primary K+ transport pathways deleted (Δtrk1, Δtrk2) and will not grow on low [K+] media unless it expresses an alternative route for K+ entry. This therefore allows the assay of functional K+ channels and has successfully been used to screen randomly mutated libraries for activatory mutations in Kir2.1 and Kir3.2 as well as many other types of potassium channel.20–24
In this study we also took advantage of the highly acidic intracellular pH of yeast to inhibit Kir1.1 function, thus allowing the screening of randomly mutated libraries of Kir1.1 for mutations which might reduce or abolish pH-sensitivity. We identified several activatory mutations including one novel mutation which produces a marked reduction in pH-sensitivity, but none which completely abolished pH-sensitivity or which would act as a titratable H+-sensor. We therefore propose an alternative model which does not require the pH-sensitive Kir channels to possess a separate independent pH-sensor. Instead we propose that the pH-sensing mechanism is part of a common Kir channel gating mechanism. This model not only explains the results of many different attempts to identify a Kir channel pH-sensor, but also why mutation of the actual H+-sensing residue(s) does not result in the expected reduction in pH-sensitivity and why their identification will be difficult.
Results and Discussion
Expression of pH-sensitive Kir channels in yeast.
Several previous studies have shown that the SGY1528 strain of yeast can be used to identify activatory ‘gain-of-function’ mutations in a wide range of K+ channels, including several different Kir channels.20–22 We therefore decided to adapt this approach to screen for mutations in Kir1.1 which would reduce its sensitivity to intracellular pH. It has been shown that the intracellular pH of S. cerevisiae can drop as low as pH 5.5.25 Such an acidic intracellular pH would cause complete inhibition of Kir1.1 and therefore permit the screening of randomly mutated libraries for mutations which reduce or possibly even abolish its pH-sensitivity.
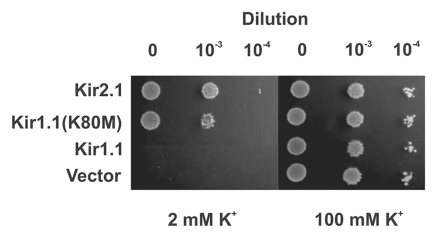
To validate this approach we transformed Kir1.1, Kir2.1 and the Kir1.1 (K80M) mutant channels into SGY1528 and examined their ability to complement growth on low [K+] media. Figure 1 shows that, as expected, the pH-sensitive channel Kir1.1 does not complement growth on 2 mM [K+] media. By contrast, wild-type Kir2.1 and Kir1.1(K80M) which both have a low pH-sensitivity complement growth well, indicating that they are fully functional in this acidic intracellular environment. This demonstrates that channels which are sensitive to pHi within the physiological range are inhibited in this assay and fail to complement. This is also supported by our observation during the development of this assay that two different variants of Kir2.1 behave differently in this assay; the published sequence of mouse Kir2.1 contains a methionine at position 84 and complements growth well (as shown in Fig. 1). However, we found that a variant of mouse Kir2.1,26 with a threonine at position 84 failed to complement growth of SGY1528 yeast. Residue 84 in Kir2.1 is equivalent to lysine 80 in Kir1.1, i.e., the TM1 position which regulates pH-sensitivity. We therefore tested the pH-sensitivity of these two Kir2.1 variants and found that in agreement with previously published reports Kir2.1-84M is fully active at pH5.5. However, Kir2.1-84T shows >70% inhibition at pH5.5 (not shown). Together, these results suggest that this assay can be used to screen for mutations which reduce the sensitivity of Kir1.1 to inhibition by intracellular H+.
Figure 1.
Growth complementation by pH-insensitive Kir channels. The pH-sensitive wild-type Kir1.1 channel does not complement the growth of K+-auxotrophic S. cerevisiae (SGY1528) on low [K+] media (2 mM KCl) presumably due to the low intracellular pH found in yeast. However, the pH-insensitive channel Kir2.1 and the mutant Kir1.1(K80M) channels both complement growth well. In all cases the strains can grow well on high 100 mM [K+] (right hand part). The control vector is the parental pYES-2m.
Random mutagenesis of Kir1.1.
We created a mutant library of Kir1.1 where the entire open reading frame was subjected to random mutagenesis. This library was then transformed into the SGY1528 strain and screened on 2 mM [K+] plates. We recovered approximately 25 positive clones, but upon sequencing found that all of them contained either the K80M or K80I mutation which causes a reduction in pH-sensitivity and permits complementation of SGY1528 (Fig. 1). We next employed a more focused mutagenesis approach in order to avoid mutation of lysine 80; only amino acid residues 84 to 391 were subjected to random mutagenesis by PCR. However, despite multiple rounds of screening, this library failed to generate any positive clones (not shown).
Random mutagenesis of a Kir1.1/Kir4.1 chimera.
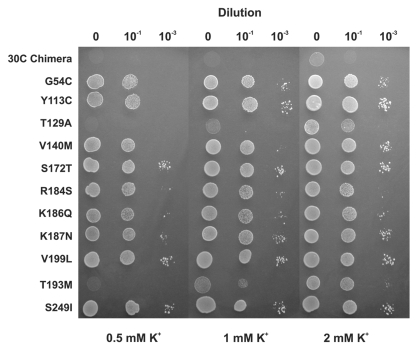
Due to our lack of success with Kir1.1 we decided to screen a mutant library of Kir4.1 which is also a pH-sensitive Kir channel. However, no positive clones could be recovered and it was subsequently found that even mutation of the TM1 lysine residue in Kir4.1 (K67M) failed to permit complementation of SGY1528 yeast. Given that the Kir4.1(K67M) channel has an IC50 of pH 4.3,13 the inability of this channel to complement growth is unlikely to be due to any remaining pH-sensitivity and instead may be because not all types of K+ channels are processed and trafficked correctly in yeast. However, we did not investigate this further. Instead we took advantage of a chimera between Kir1.1 and Kir4.1 which had been created in a previous study.27 This chimera (30C) contains the N-terminus and TM-Pore region of Kir1.1 with the C-terminus of Kir4.1. This chimera did not complement the growth of SGY1528, and neither did the K80M mutant version of this chimera. However, we found that screening of a randomly mutated library of the 30C(K80M) chimera resulted in the recovery of a large number of positive clones. After the exclusion of false positives a total of 42 positive clones were recovered. Several of these clones contained multiple mutations, but either these mutations were represented as single mutants elsewhere or site-directed mutagenesis was used to identify the individual mutation responsible. In total 11 single mutations were identified and confirmed by complementation in SGY1528 (Fig. 2 and Table 1).
Figure 2.
Random mutagenesis identifies activatory mutations. By screening a randomly mutated library of the 30C chimeric channel on low [K+] growth media, 11 unique activatory mutations were identified which complemented growth. All except the S249I mutation were located in the Kir1.1 section of the chimera.
Table 1.
The pH-sensitivity of activatory mutations identified in 30C chimera
| IC50 | |
| WT Kir1.1 | 6.5 ± 0.1 |
| G54C | 6.1 ± 0.1 |
| Y113C | 6.5 ± 0.1 |
| T129A | 6.6 ± 0.1 |
| V140M | N.D. |
| S172T | 5.8 ± 0.1 |
| R184S | 6.1 ± 0.1 |
| K186Q | 6.9 ± 0.1 |
| K187N | 6.3 ± 0.1 |
| T193M | 6.7 ± 0.2 |
| V199L | 6.3 ± 0.1 |
| S249I | N.D. |
All except the S249I mutation were located in the Kir1.1 section of the chimera.
Effects on pH-sensitivity of Kir1.1.
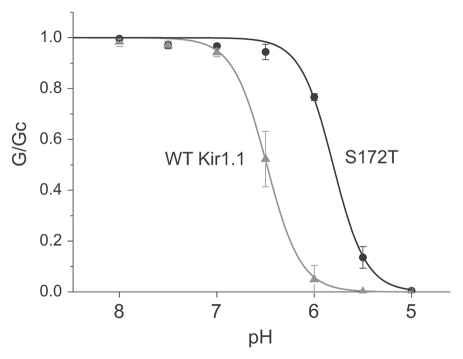
All except one of the these activatory mutations are located in the Kir1.1 portion of the 30C chimera and we therefore examined their effect on pH-sensitivity. The mutations were introduced into wild-type Kir1.1 and expressed in Xenopus oocytes. Giant excised patches were used to measure their sensitivity to changes in intracellular pH and these results are shown in Table 1. The majority of mutations have little effect on pH-sensitivity, whilst mutations G54C and R184S produced only a modest reduction in pH-sensitivity. However, mutation S172T in TM2 produced a significant reduction in pH-sensitivity (IC50 = 5.8) (Fig. 3).
Figure 3.
Reduced pH-sensitivity in the Kir1.1 S172T mutation. Measurement of the pH-sensitivity of the S172T mutation in giant excised patches from Xenopus oocytes. The S172T mutation markedly reduces channel pH-sensitivity (IC50 = 5.8 ± 0.1) compared to wild-type Kir1.1 (IC50 = 6.5 ± 0.1).
Mechanism of activation.
The majority of mutations identified in this assay do not appear to produce a major reduction in pH-sensitivity, though in some cases the modest shifts observed could, in combination with the K80M mutation in the mutant 30C chimera, produce a cumulative effect great enough to permit complementation. However, given their lack of effect on Kir1.1 pH-sensitivity we did not investigate this further. Interestingly, several activatory mutations cluster in the proximal C-terminus. (R184, K186, K187) in an area thought to contribute to the PIP2 binding site.28 Although still controversial, it has been proposed that disruption of channel-PIP2 interactions increases pH-sensitivity through destabilization of the open-state of the channel.28,29 However, if they do weaken the interaction with PIP2 then this has no major effect on pH-sensitivity (Table 1).
Changes in processing and trafficking of the channel could also lead to an increase in channel density in the membrane.30,31 Nevertheless, such explanations need further investigation and we decided not to pursue this further as part of this particular study. However, the ability to express functional Kir1.1 (K80M) channels in yeast now has potential for use of this expression system as a high-throughput screen for Kir1.1 inhibitors for the treatment of hypertension.32
Effects of the S172T mutation.
The S172T mutation produced a significant effect on pH-sensitivity (IC50 = 5.8). We therefore examined whether this mutation would permit complementation of SGY1528 growth by Kir1.1. However, the Kir1.1 (S172T) mutant did not complement (not shown). This would explain why this mutation was not recovered in the original screens of Kir1.1 and only mutations of the K80 residue were found which produce a larger reduction in pH-sensitivity.
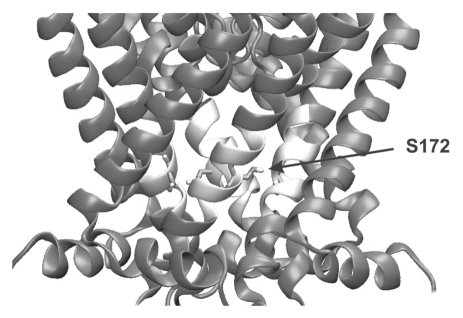
Changing a serine to a threonine at position 172 is a very conservative substitution, yet the functional effect is significant. Closer inspection of the position of S172 in the closed state model shown in Figure 4 shows that this residue sits at the point at which the TM2 helices come closest together and the serine side chain forms several inter-subunit contacts.
Figure 4.
S172 Residue located close to the helix-bundle crossing. The S172 is located close to the bottom of TM2 and to the adjacent TM2 helix in the closed state homology model of Kir1.1.9,16 The TM2 segment containing the S172 residue is shown in white for clarity and the S172 residue is shown as a stick. Opening of the channel involves movement of the TM2 helices and the S172T mutation may reduce pH-sensitivity either by destabilizing the closed state or forming additional interactions in the open state.
We therefore investigated what effect different mutations at this position have on pH-sensitivity. We found that the effect of the threonine mutation was highly specific as mutation to either alanine, valine or isoleucine had very little effect on pH-sensitivity (S172A IC50= 6.6 ± 0.1, S172V IC50= 6.6 ± 0.1 and S172I IC50= 6.5 ± 0.1). The functional effect of the S172T mutation therefore cannot be replicated by a smaller or larger sidechain and although it is a very conservative substitution, the additional methyl group in the presence of the hydroxyl group clearly introduces a very subtle and unique structural change that produces a marked shift in pH-sensitivity. Furthermore, because this threonine side-chain is non-titratable then the effects of this mutation must act indirectly on the pH-sensing mechanism.
This also highlights the usefulness of this random screening approach because such a conservative mutation is unlikely to have been found by systematic scanning mutagenesis or sitedirected mutagenesis approaches which would have naturally favored more radical side-chain substitutions.
Is there a separate titratable pH-sensor?
Mutagenesis studies of pH-sensitivity in other K+ channels have identified specific titratable residues and the existence of modular regulatory domains that confer pH-sensitivity. Furthermore, when these residues are mutated the channels lose their pH-sensitivity, but their other gating mechanisms remain mostly intact.33–35 As a consequence, most previous studies of Kir channel pH-sensitivity have been based upon the idea that the H+-sensor in Kir1.1 is separate and unique to the pH-sensitive Kir channels, and that if this sensor is mutated then inhibitory effect of H+ would be reduced or abolished.
However, in this study (and all previous studies of Kir channel pH-sensitivity) no mutations have been identified which completely remove pH-sensitivity. One simple explanation is therefore that the correct residue has not been mutated. Although it is difficult to prove such a negative result beyond doubt, we believe this not to be the case for two reasons. Firstly, extensive site-directed mutagenesis studies over the last decade have failed to identify any such residues. Secondly, this unbiased random screen of the entire Kir1.1 sequence also failed to identify any such residues even though it was capable of identifying the K80M and S172T mutations.
Alternatively, if no single residue functions as the pH-sensor then perhaps protonation of more than one residue is required to close the channel and no mutagenesis study (including this one) has ever made the correct combination of mutations? Again, we cannot disprove this, but in our previous study of KcsA we were still able to identify many of the individual residues which contribute to a complex pH-sensing mechanism using this random mutagenesis and genetic screening approach.20,36
Therefore we believe that if the pH-sensitive Kir channels possessed a separate, unique and independent pH-sensor consisting of a small number of titratable residues then this approach (and all previous approaches) would have had a high probability of finding such mutations, and that the apparent failure to identify such residues by mutagenesis and functional screening for reduced pH-sensitivity is not just bad luck. Consequently, an alternative explanation for Kir channel pH-sensitivity is required.
A putative model for Kir channel pH-sensitivity.
A relatively simple explanation can account for our results, and those of all previously published studies of Kir channel pH-sensitivity by assuming that the pH-sensitive Kir channels do not possess a unique set of titratable residues which confer H+-sensitivity. Instead, we propose that the pH-sensing mechanism and molecular identity of the H+-sensor is common to all Kir channel sub-classes, and that these pH-sensitive interactions are highly conserved amongst all Kir channels and are involved in stabilization of the channel in the open state conformation. This is consistent with the previously reported sensitivity of all types of Kir channel to H+-inhibition.2 To modulate channel pH-sensitivity therefore only requires subtle variations in channel structure to affect the relative stability of either the open or closed state, e.g., H-bonding between TM1 and TM2 in Kir1.1.16 Such additional interactions would shift the equilibrium of the response to H+ and sensitize the channel to pH changes within the physiological range (i.e., pH 6.0 to pH 8.0) without the requirement for pH-sensitive interactions unique to the pH-sensitive Kir channels alone.
This idea is supported by our previous study on the mechanism by which the K80 residue in Kir1.1 controls pH-sensitivity. H-bonding between K80 and A177 at the helix-bundle crossing influences pH-sensitivity through stabilization of the closed state of the channel i.e., making it easier to close at a given [H+] and thus more ‘pH-sensitive’.9,15,16 Thus although K80 is critical in defining channel pH-sensitivity, it is not the actual H+-sensor and mutation of this residue reduces pH-sensitivity by affecting the stability of the closed state, relative to the open state. This relative destabilization of the closed state is also reflected in a change in gating kinetics because these mutations are able to open at a much faster rate.15,16
The S172T mutation destabilizes the closed-state of Kir1.1.
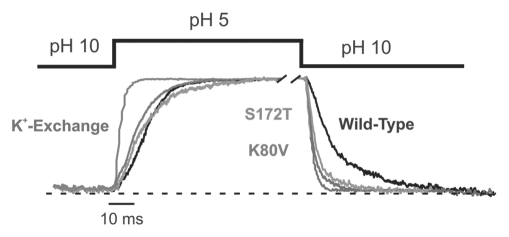
If our hypothesis is correct and the S172T mutation influences pH-sensitivity through a relative destabilization of the closed state then we should also observe an alteration in the gating kinetics. We therefore used a fast piezo-driven solution exchange system to examine the kinetics of pH-gating in the S172T mutant channels.15,16 We found that although the on-rate for pH-inhibition of S172T was slightly increased relative to wild-type Kir1.1, the off rate was dramatically faster and were similar to that seen with the K80V mutation (Table 2 and Fig. 5). Although the on rate increased by greater than 5-fold for the S172T mutant, in reality this increase could be even be greater as it approaches the limits of resolution for the solution exchange system.
Table 2.
Gating kinetics of wild-type and mutant Kir1.1 channels
| On rate | Off rate | |
| K+-Exchange | 5.5 ± 0.5 ms | 16.5 ± 4.5 ms |
| Kir1.1 | 150 ± 7.5 ms | 116 ± 2.6 ms |
| S172T | 104 ± 30 ms | 23 ± 7 ms |
| K80V | 125 ± 8 ms | 20 ± 2 ms |
Time courses were fitted with a monoexponential function. The on rate represents the time course of inhibition from pH 10 to pH 5. The off rate represent the time course of recovery from pH 5 to pH 10 (mean ± SEM, n = 6).
Figure 5.
Destabilization of the closed state by S172T mutation. Time course of pH gating for wild-type and mutant Kir1.1 channels induced by changes in the intracellular pH. Solution exchange was done using a fast piezo-driven application system. Currents for the different channels were equalized for better comparison. The time course of wild-type Kir1.1 currents upon K+ exchange (replacement with Na+ measured at +40 mV) are shown in gray and superimposed on the pH gating time course obtained in the same patch. This demonstrates the maximal temporal resolution of the application system. The pH-inhibition of wild-type Kir1.1 is shown in black, Kir1.1-K80V in red, and the S172T in green. Both the K80V and S172T mutant channels show a faster rate of recovery from inhibition, i.e., faster off rate which reflects a destabilization of the closed state. The values for on and off rates are shown in Table 2. The dotted line represents the zero current level. For color, see online publication.
These changes in gating kinetics clearly demonstrate that although the S172T mutation causes a minor destabilization of the open state it has a far greater effect on the stability of the closed state, and that this destabilization of the closed state could easily account for the change in pH-sensitivity. Furthermore, it demonstrates that differences in titratable side-chains are not necessary to produce relatively large effects on channel pHsensitivity. These concepts are illustrated by the simplified gating model shown in Figure 6.
Figure 6.
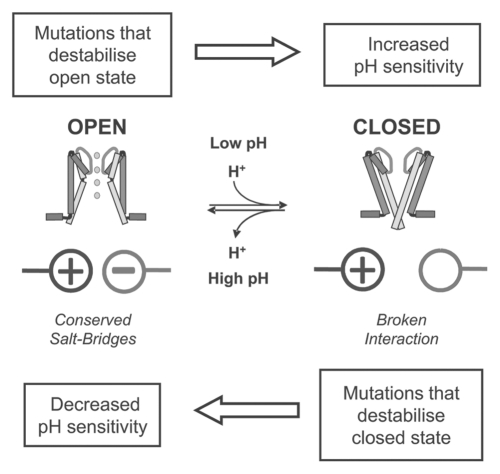
A putative model for Kir channel pH-sensitivity. The model proposes that all Kir channels possess a low intrinsic pH-sensitivity and that this pH-sensitivity is due to titration of one or more salt-bridges in the intracellular domains that stabilize the channel in the open-state. Low pH probably involves titration of carboxylic acid groups to break these stabilizing interactions. Differences in Kir channel pH-sensitivity (i.e., shifting this equilibrium) can therefore arise by altering the relative stability of the open and closed states rather than the presence of additional H+ sensors. The model proposes that direct mutation of the H+-sensing residues will destabilize the open state and therefore result in an increase, not decrease in pH-sensitivity. In some cases this alkaline shift may be so extreme as to produce a non-functional channel. Any other mutations that indirectly destabilize the open state will also result in an increase in pH-sensitivity. In order to decrease channel pH-sensitivity requires mutation of residues which stabilize the closed state or the creation of novel interactions which stabilize the open state.
What is the pH-sensor?
This model also provides a straightforward explanation to why it is so difficult to identify the actual H+-sensor itself; because if it is involved in stabilization of the open state, then its mutagenesis would promote channel closure (i.e., would produce an alkaline shift or a permanently closed channel) instead of producing the expected reduction in pH-sensitivity. It will therefore be difficult, and perhaps impossible, to produce a mutant Kir channel that is completely insensitive to H+-inhibition and explains why so many mutations in Kir channels produce an alkaline shift.19
In the pH-sensitive channels such as Kir1.1 and Kir4.1 or Kir4.2 we propose that additional interactions have evolved which fine-tune this intrinsic pH-sensitivity by their effects on the relative stability of either the open or closed state of the channel. This also explains how heteromultimerisation with Kir5.1 could enhance the pH-sensitivity of Kir4.1, because instead of contributing an additional H+-sensor, the presence of the Kir5.1 subunit simply needs to stabilize the closed state (or destabilize the open state) relative to Kir4.1.14,15
In our model the pH-sensitive Kir channels (Kir1.1, Kir4.1 and Kir4.2) retain a degree of pH-sensitivity and behave like non-pH-sensitive channels such as Kir2.1 even when residues involved in these additional ‘pH-tuning' interactions are mutated (e.g., K80M in Kir1.1). This is because the basic H+-sensing mechanism is common to all Kir channels and absolutely critical for Kir channel gating i.e., maintenance of the open state. Therefore, these actual H+-sensitive interactions cannot be mutated or channel opening will be severely impaired and a non-functional channel result. Likewise, mutations which produce indirect disruptive effects on these interactions would result in an alkaline shift in pH-sensitivity.
So what structural explanation could account for this proposed model? In the KcsA potassium channel a network of saltbridges located at the helix-bundle crossing acts as the H+-sensor, but in this case these interactions hold the channel in the closed state prior to protonation of glutamate residues and activation at low pH.36,37 In TREK1 and TRPM2 channels protonation of intracellular glutamate and aspartate side chains have been shown to act as the H+-sensors.38,39 In Kir channels the pH-sensing residues are most likely located in the intracellular domains and their titration at very low pH values indicates that protonation of carboxylic acid groups are also probably involved. In support of this, we and others have previously shown that mutation of several highly conserved inter-subunit salt-bridges shifts channel pH-sensitivity into the alkaline range.8,9 Therefore we propose that (as summarized in Fig. 6) a series of inter- and/or intra-subunit salt bridges may account for the intrinsic pH-sensitivity of Kir channels, and that protonation of carboxylic acid groups at low pH destabilizes the open state causing inhibition of channel activity. Indeed, a recent study of the pH-sensitivity of the ASIC channel has shown a wide degree of variation in the pKa of aspartic and glutamic acid residues ranging of pH 5.0 to pH 7.4.40 This therefore supports the idea that a carboxylic acid residue could act as a physiological pH-sensor in Kir1.1. However, due to their possible involvement in stabilization of the open state of Kir1.1 their identification will be problematic. Nevertheless, the recent structure of the Kir2.2 channel in the closed state will undoubtedly help with the generation of models of the open state Kir channel,41 and the prediction of residues that may be involved in open state stabilization, as well as those interactions involved in the fine-tuning of channel pH-sensitivity.
In conclusion, we believe that the model we propose now provides a explanation of Kir channel pH-sensitivity that is consistent with all the available functional studies. This model also has major implications for the design and interpretation of any future studies which address Kir channel pH-sensitivity, as well as for other classes of ion channels where the identity of the H+-sensor has so far proven elusive.
Materials and Methods
Molecular biology.
For expression in yeast wild-type and mutant Kir channels were cloned into the methionine regulated pYES2m vector with a modified Kozak sequence of 6 adenine nucleotides for optimal expression (AAA AAA ATG).42 Construction of the 30C chimera was described previously and contains residues 1–197 of Kir1.1 and 185–379 of Kir4.1.27 Randomly mutated libraries were constructed using the GeneMorph-II random mutagenesis kit (Stratagene), which uses non-biased, error-prone PCR. The PCR reactions were quantified according to the manufacturers protocols to produce an error rate of ∼1–3 mutations per open reading frame. PCR products were then subcloned back into their expression vectors. In order to maximise the transformation efficiency, ligations were purified by phenol:chloroform extraction and ethanol precipitation prior to transformation into Library Efficiency DH5α E. coli (Invitrogen) and growth in culture overnight. Analysis of the transformation efficiency prior to overnight growth indicated that the libraries contained approximately >105 independent clones. Randomly selected independent clones were isolated on non-selective media and upon sequencing were found to contain a minimum of 1–3 mutations per open reading frame. All site-directed mutagenesis was performed using the QuikChangeII system (Stratagene).
Yeast transformation and drop-tests.
All media, growth and propagation of this strain is as described previously.20–22 Wild-type and mutant channels were transformed into competent SGY1528 cells using a standard lithium acetate yeast transformation protocol. Transformants were grown on APKOura plates for ∼48 hours before overnight growth in 4 ml APKOura (100 mM KCl). Cultures were washed in APKO-met-ura media (0.5 mM KCl) and 3.5 µl drops of either undiluted, 1:10 or 1:1,000 dilutions made onto APKO-met-ura plates (with [KCl] as specified). Drop test plates were incubated at 30°C for ∼72 hours.
Electrophysiology.
For oocyte expression, constructs were subcloned into the pBF expression vector. mRNAs were synthesized in vitro by using the SP6 mMESSAGE mMACHINE kit (Ambion, Austin, TX, USA) and stored in stock solutions at −80°C. Xenopus oocytes were treated with 0.5 mg/ml collagenase type II (Sigma, Taufkirchen, Germany), defolliculated and incubated at 19°C prior to use. About 50 nl of a solution containing channel specific mRNA was injected into Dumont stage VI oocytes. Giant patch recordings in inside-out configuration under voltage-clamp conditions were made at room temperature 3–7 days after mRNA injection. Pipettes were made from thickwalled borosilicate glass, had resistances of 0.3–0.9 MΩ (tip diameter of 5–15 µm) and filled with (in mM, pH adjusted to 7.2 with KOH) 120 KCl, 10 HEPES and 1.8 CaCl2. K+ free solution contained (in mM, pH adjusted to 7.2 with NaOH) 120 mM N-methylglucamine (NMG+), 10 HEPES and 1.8 CaCl2. Currents were recorded with an EPC9 amplifier (HEKA electronics, Lamprecht, Germany) and sampled at 1 kHz with analog filter set to 3 kHz (−3 dB). Solutions were applied to the cytoplasmic side of excised patches via a multi-barrel pipette. For experiments on the pH gating kinetics a faster solution exchange system was used with double barrel theta-glass capillary attached to a piezo-driven device.
Acknowledgements
This work was supported by grants from the British Heart Foundation and the Deutsche Forschungsgemeinschaft (BA1793\4-2).
Footnotes
Previously published online: www.landesbioscience.com/journals/channels/article/13006
References
- 1.Bichet D, Haass FA, Jan LY. Merging functional studies with structures of inward-rectifier K+ channels. Nat Rev Neurosci. 2003;4:957–967. doi: 10.1038/nrn1244. [DOI] [PubMed] [Google Scholar]
- 2.Hibino H, Inanobe A, Furutani K, Murakami S, Findlay I, Kurachi Y. Inwardly rectifying potassium channels: their structure, function and physiological roles. Physiol Rev. 2010;90:291–366. doi: 10.1152/physrev.00021.2009. [DOI] [PubMed] [Google Scholar]
- 3.Schulte U, Fakler B. Gating of inward-rectifier K+ channels by intracellular pH. Eur J Biochem. 2000;267:5837–5841. doi: 10.1046/j.1432-1327.2000.01671.x. [DOI] [PubMed] [Google Scholar]
- 4.Hebert SC. Bartter syndrome. Curr Opin Nephrol Hypertens. 2003;12:527–532. doi: 10.1097/00041552-200309000-00008. [DOI] [PubMed] [Google Scholar]
- 5.Scholl UI, Choi M, Liu T, Ramaekers VT, Hausler MG, Grimmer J, et al. Seizures, sensorineural deafness, ataxia, mental retardation and electrolyte imbalance (SeSAME syndrome) caused by mutations in KCNJ10. Proc Natl Acad Sci USA. 2009;106:5842–5847. doi: 10.1073/pnas.0901749106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bockenhauer D, Feather S, Stanescu HC, Bandulik S, Zdebik AA, Reichold M, et al. Epilepsy, ataxia, sensorineural deafness, tubulopathy and KCNJ10 mutations. N Engl J Med. 2009;360:1960–1970. doi: 10.1056/NEJMoa0810276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fakler B, Schultz JH, Yang J, Schulte U, Brandle U, Zenner HP, et al. Identification of a titratable lysine residue that determines sensitivity of kidney potassium channels (ROMK) to intracellular pH. EMBO J. 1996;15:4093–4099. [PMC free article] [PubMed] [Google Scholar]
- 8.Leng Q, MacGregor GG, Dong K, Giebisch G, Hebert SC. Subunit-subunit interactions are critical for proton sensitivity of ROMK: evidence in support of an intermolecular gating mechanism. Proc Natl Acad Sci USA. 2006;103:1982–1987. doi: 10.1073/pnas.0510610103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rapedius M, Haider S, Browne KF, Shang L, Sansom MS, Baukrowitz T, et al. Structural and functional analysis of the putative pH sensor in the Kir1.1 (ROMK) potassium channel. EMBO Rep. 2006;7:611–616. doi: 10.1038/sj.embor.7400678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sackin H, Nanazashvili M, Palmer LG, Li H. Role of conserved glycines in pH gating of Kir1.1 (ROMK) Biophys J. 2006;90:3582–3589. doi: 10.1529/biophysj.105.076653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang YY, Sackin H, Palmer LG. Localization of the pH gate in Kir1.1 channels. Biophys J. 2006;91:29019. doi: 10.1529/biophysj.106.087700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choe H, Zhou H, Palmer LG, Sackin H. A conserved cytoplasmic region of ROMK modulates pH sensitivity, conductance and gating. Am J Physiol. 1997;273:516–529. doi: 10.1152/ajprenal.1997.273.4.F516. [DOI] [PubMed] [Google Scholar]
- 13.Pessia M, Imbrici P, D'Adamo MC, Salvatore L, Tucker SJ. Differential pH sensitivity of Kir4.1 and Kir4.2 potassium channels and their modulation by heteropolymerisation with Kir5.1. J Physiol. 2001;532:359–367. doi: 10.1111/j.1469-7793.2001.0359f.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Z, Xu H, Cui N, Qu Z, Chanchevalap S, Shen W, et al. Biophysical and molecular mechanisms underlying the modulation of heteromeric Kir4.1–Kir5.1 channels by CO2 and pH. J Gen Physiol. 2000;116:33–45. doi: 10.1085/jgp.116.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rapedius M, Paynter JJ, Fowler PW, Shang L, Sansom MS, Tucker SJ, et al. Control of pH and PIP2 gating in heteromeric Kir4.1/Kir5.1 channels by H-bonding at the helix-bundle crossing. Channels. 2007;1:327–330. doi: 10.4161/chan.5176. [DOI] [PubMed] [Google Scholar]
- 16.Rapedius M, Fowler PW, Shang L, Sansom MS, Tucker SJ, Baukrowitz T. H bonding at the helix-bundle crossing controls gating in Kir potassium channels. Neuron. 2007;55:602–614. doi: 10.1016/j.neuron.2007.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chanchevalap S, Yang Z, Cui N, Qu Z, Zhu G, Liu C, et al. Involvement of histidine residues in proton sensing of ROMK1 channel. J Biol Chem. 2000;275:78117. doi: 10.1074/jbc.275.11.7811. [DOI] [PubMed] [Google Scholar]
- 18.Schulze D, Krauter T, Fritzenschaft H, Soom M, Baukrowitz T. Phosphatidylinositol 4,5-bisphosphate (PIP2) modulation of ATP and pH sensitivity in Kir channels. A tale of an active and a silent PIP2 site in the N terminus. J Biol Chem. 2003;278:10500–10505. doi: 10.1074/jbc.M208413200. [DOI] [PubMed] [Google Scholar]
- 19.Schulte U, Hahn H, Konrad M, Jeck N, Derst C, Wild K, et al. pH gating of ROMK (Kir1.1) channels: control by an Arg-Lys-Arg triad disrupted in antenatal Bartter syndrome. Proc Natl Acad Sci USA. 1999;96:15298–15303. doi: 10.1073/pnas.96.26.15298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paynter JJ, Sarkies P, Andres-Enguix I, Tucker SJ. Genetic selection of activatory mutations in KcsA. Channels (Austin) 2008;2:413–418. doi: 10.4161/chan.2.6.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yi BA, Lin YF, Jan YN, Jan LY. Yeast screen for constitutively active mutant G protein-activated potassium channels. Neuron. 2001;29:657–667. doi: 10.1016/s0896-6273(01)00241-0. [DOI] [PubMed] [Google Scholar]
- 22.Minor DL, Masseling SJ, Jan YN, Jan LY. Transmembrane structure of an inwardly rectifying potassium channel. Cell. 1999;96:879–891. doi: 10.1016/s0092-8674(00)80597-8. [DOI] [PubMed] [Google Scholar]
- 23.Minor DL. Searching for interesting channels: pairing selection and molecular evolution methods to study ion channel structure and function. Mol Biosyst. 2009;5:802–810. doi: 10.1039/b901708a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chatelain FC, Gazzarrini S, Fujiwara Y, Arrigoni C, Domigan C, Ferrara G, et al. Selection of inhibitor-resistant viral potassium channels identifies a selectivity filter site that affects barium and amantadine block. PLoS ONE. 2009;4:7496. doi: 10.1371/journal.pone.0007496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imai T, Ohno T. Measurement of yeast intracellular pH by image processing and the change it undergoes during growth phase. J Biotechnol. 1995;38:165–172. doi: 10.1016/0168-1656(94)00130-5. [DOI] [PubMed] [Google Scholar]
- 26.Fakler B, Brandle U, Bond C, Glowatzki E, Konig C, Adelman JP, et al. A structural determinant of differential sensitivity of cloned inward rectifier K+ channels to intracellular spermine. FEBS Lett. 1994;356:199–203. doi: 10.1016/0014-5793(94)01258-x. [DOI] [PubMed] [Google Scholar]
- 27.Konstas AA, Korbmacher C, Tucker SJ. Identification of domains that control the heteromeric assembly of Kir5.1/Kir4.0 potassium channels. Am J Physiol Cell Physiol. 2003;284:910–917. doi: 10.1152/ajpcell.00479.2002. [DOI] [PubMed] [Google Scholar]
- 28.Tucker SJ, Baukrowitz T. How highly charged anionic lipids bind and regulate ion channels. J Gen Physiol. 2008;131:431–438. doi: 10.1085/jgp.200709936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leung YM, Zeng WZ, Liou HH, Solaro CR, Huang CL. Phosphatidylinositol 4,5-bisphosphate and intracellular pH regulate the ROMK1 potassium channel via separate but interrelated mechanisms. J Biol Chem. 2000;275:10182–10189. doi: 10.1074/jbc.275.14.10182. [DOI] [PubMed] [Google Scholar]
- 30.Huang CL. Regulation of ROMK trafficking and channel activity. Curr Opin Nephrol Hypertens. 2001;10:693–698. doi: 10.1097/00041552-200109000-00022. [DOI] [PubMed] [Google Scholar]
- 31.Stockklausner C, Klocker N. Surface expression of inward rectifier potassium channels is controlled by selective Golgi export. J Biol Chem. 2003;278:170005. doi: 10.1074/jbc.M212243200. [DOI] [PubMed] [Google Scholar]
- 32.Ramu Y, Xu Y, Lu Z. Engineered specific and high-affinity inhibitor for a subtype of inward-rectifier K+ channels. Proc Natl Acad Sci USA. 2008;105:10774–10778. doi: 10.1073/pnas.0802850105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coulter KL, Perier F, Radeke CM, Vandenberg CA. Identification and molecular localization of a pH-sensing domain for the inward rectifier potassium channel HIR. Neuron. 1995;15:1157–1168. doi: 10.1016/0896-6273(95)90103-5. [DOI] [PubMed] [Google Scholar]
- 34.Rajan S, Wischmeyer E, Xin Liu G, Preisig-Muller R, Daut J, Karschin A, et al. TASK-3, a novel tandem pore domain acid-sensitive K+ channel. An extracellular histidine as pH sensor. J Biol Chem. 2000;275:16650–16657. doi: 10.1074/jbc.M000030200. [DOI] [PubMed] [Google Scholar]
- 35.Sandoz G, Douguet D, Chatelain F, Lazdunski M, Lesage F. Extracellular acidification exerts opposite actions on TREK1 and TREK2 potassium channels via a single conserved histidine residue. Proc Natl Acad Sci USA. 2009;106:14628–14633. doi: 10.1073/pnas.0906267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson AN, Posson DJ, Parsa PV, Nimigean CM. Molecular mechanism of pH sensing in KcsA potassium channels. Proc Natl Acad Sci USA. 2008;105:6900–6905. doi: 10.1073/pnas.0800873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cuello LG, Cortes DM, Jogini V, Sompornpisut A, Perozo E. A molecular mechanism for proton-dependent gating in KcsA. FEBS Lett. 2010;584:1126–1132. doi: 10.1016/j.febslet.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Honore E, Maingret F, Lazdunski M, Patel AJ. An intracellular proton sensor commands lipid- and mechano-gating of the K+ channel TREK-1. EMBO J. 2002;21:2968–2976. doi: 10.1093/emboj/cdf288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du J, Xie J, Yue L. Modulation of TRPM2 by acidic pH and the underlying mechanisms for pH sensitivity. J Gen Physiol. 2009;134:471–488. doi: 10.1085/jgp.200910254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liechti LA, Bernèche S, Bargeton B, Iwaszkiewicz J, Roy S, Michielin O, et al. A combined computational and functional approach identifies new residues involved in pH-dependent gating of ASIC1a. J Biol Chem. 2010;285:16315–16329. doi: 10.1074/jbc.M109.092015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tao X, Avalos JL, Chen J, MacKinnon R. Crystal structure of the eukaryotic strong inward-rectifier K+ channel Kir2.2 at 3.1 A resolution. Science. 2009;326:1668–1674. doi: 10.1126/science.1180310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamilton R, Watanabe CK, de Boer HA. Compilation and comparison of the sequence context around the AUG startcodons in Saccharomyces cerevisiae mRNAs. Nucleic Acids Res. 1987;15:3581–3593. doi: 10.1093/nar/15.8.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]