Abstract
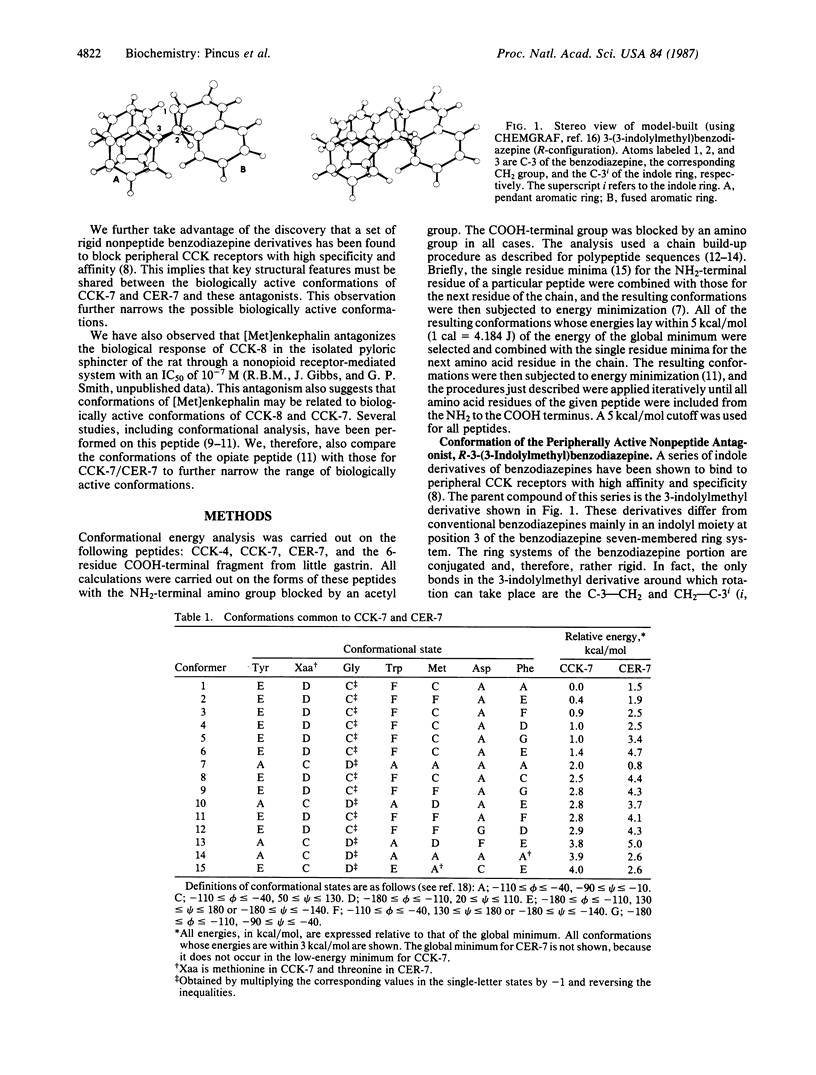
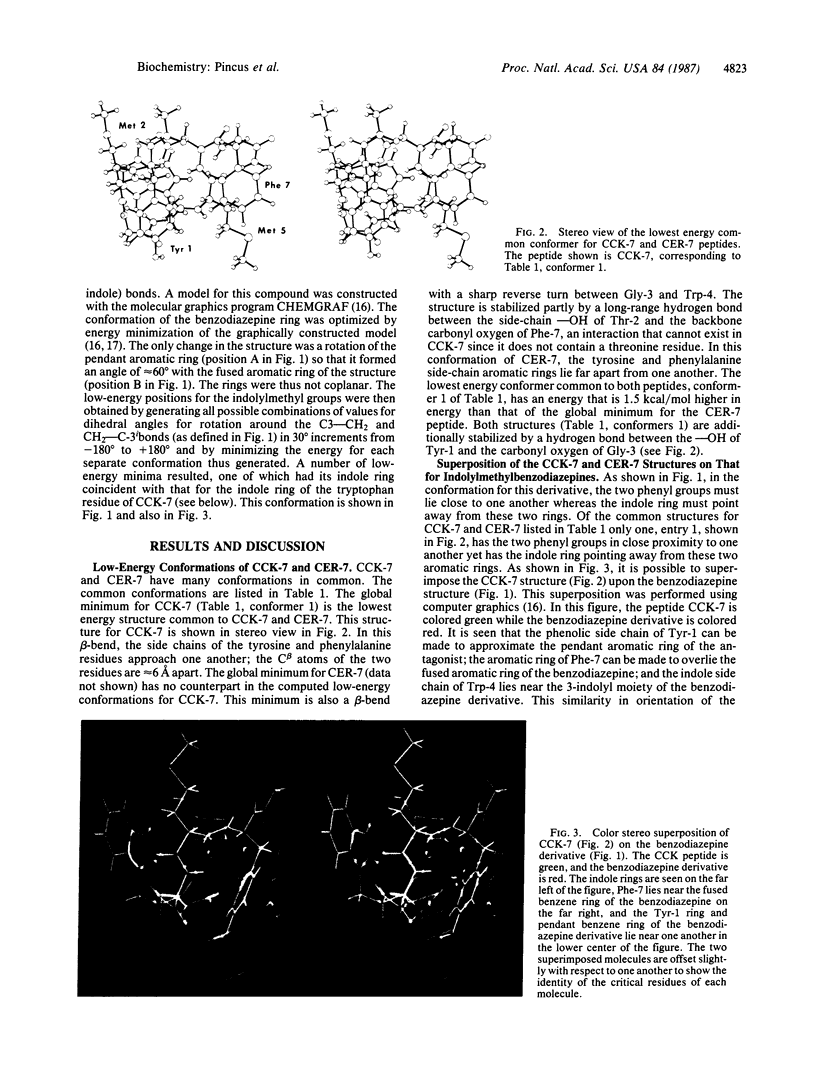
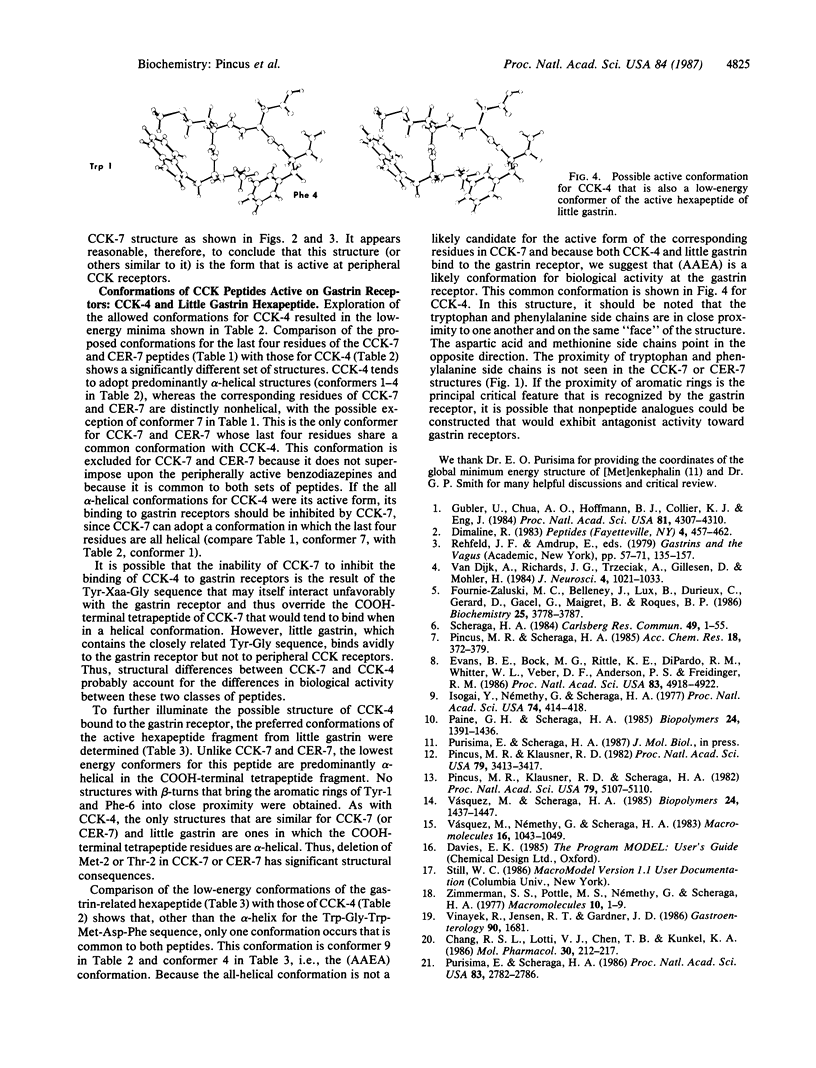
The biologically active conformations of a series of four peptides [four cholecystokinin (CCK)-related peptides and enkephalin] in their interactions with gastrointestinal receptors have been deduced using conformational computational analysis. The two peptides that interact exclusively with peripheral-type CCK receptors are the heptapeptide COOH-terminal fragment from CCK (CCK-7) and the analogous sequence from cerulein (CER-7) in which threonine replaces the methionine proximal to the NH2 terminus. The two peptides that interact exclusively with the gastrin receptor in the stomach are the active COOH-terminal fragment of little gastrin and the COOH-terminal tetrapeptide sequence common to all of these peptides, CCK-4. We find that preferred conformations for the peripherally active peptides CCK-7 and CER-7 are principally beta-bends, whereas little gastrin and CCK-4 are fundamentally helical. In the class of lowest energy structures for both CCK-7 and CER-7, the aromatic rings of the tyrosine and phenylalanine lie close to one another whereas the tryptophan indole ring points in the opposite direction. This structure is superimposable on the structures of a set of rigid indolyl benzodiazepine derivatives that interact with complete specificity and high affinity with peripheral CCK receptors further suggesting that the computed beta-bends are the biologically active conformation. The biologically active conformation for CCK-4 and the little gastrin hexapeptide has also been deduced. By excluding conformations common to CCK-7 and CCK-4, which do not bond to each other's receptors, and then by selecting conformations in common to CCK-4 and the gastrin-related hexapeptide, which do bind to each other's receptors, we deduce that the biologically active conformation at the gastrin receptor is partly helical and one in which the indole of tryptophan and the aromatic ring of phenylalanine are close to one another while the methionine and aspartic acid side chains point in the opposite direction. These major differences in preferred structures between the common CCK-7/CER-7 peptides and the common CCK-4/little gastrin peptides explain the mutually exclusive activities of these two sets of peptides. We have observed that [Met]enkephalin strongly antagonizes the action of the naturally occurring peripherally active CCK-8 (CCK-7 with an NH2-terminal aspartic acid residue added). The computed lowest energy structures for this opiate peptide closely resemble key features of the computed CCK-7/CER-7 structure, further supporting the proposed structure.
Full text
PDF

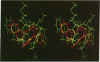
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chang R. S., Lotti V. J., Chen T. B., Kunkel K. A. Characterization of the binding of [3H]-(+/-)-L-364,718: a new potent, nonpeptide cholecystokinin antagonist radioligand selective for peripheral receptors. Mol Pharmacol. 1986 Sep;30(3):212–217. [PubMed] [Google Scholar]
- Dimaline R. Is caerulein amphibian CCK? Peptides. 1983 Jul-Aug;4(4):457–462. doi: 10.1016/0196-9781(83)90049-9. [DOI] [PubMed] [Google Scholar]
- Evans B. E., Bock M. G., Rittle K. E., DiPardo R. M., Whitter W. L., Veber D. F., Anderson P. S., Freidinger R. M. Design of potent, orally effective, nonpeptidal antagonists of the peptide hormone cholecystokinin. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4918–4922. doi: 10.1073/pnas.83.13.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournié-Zaluski M. C., Belleney J., Lux B., Durieux C., Gérard D., Gacel G., Maigret B., Roques B. P. Conformational analysis of cholecystokinin CCK26-33 and related fragments by 1H NMR spectroscopy, fluorescence-transfer measurements, and calculations. Biochemistry. 1986 Jul 1;25(13):3778–3787. doi: 10.1021/bi00361a008. [DOI] [PubMed] [Google Scholar]
- Gubler U., Chua A. O., Hoffman B. J., Collier K. J., Eng J. Cloned cDNA to cholecystokinin mRNA predicts an identical preprocholecystokinin in pig brain and gut. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4307–4310. doi: 10.1073/pnas.81.14.4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai Y., Némethy G., Scheraga H. A. Enkephalin: conformational analysis by means of empirical energy calculations. Proc Natl Acad Sci U S A. 1977 Feb;74(2):414–418. doi: 10.1073/pnas.74.2.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine G. H., Scheraga H. A. Prediction of the native conformation of a polypeptide by a statistical-mechanical procedure. I. Backbone structure of enkephalin. Biopolymers. 1985 Aug;24(8):1391–1436. doi: 10.1002/bip.360240802. [DOI] [PubMed] [Google Scholar]
- Pincus M. R., Klausner R. D. Prediction of the three-dimensional structure of the leader sequence of pre-kappa light chain, a hexadecapeptide. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3413–3417. doi: 10.1073/pnas.79.11.3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincus M. R., Klausner R. D., Scheraga H. A. Calculation of the three-dimensional structure of the membrane-bound portion of melittin from its amino acid sequence. Proc Natl Acad Sci U S A. 1982 Aug;79(16):5107–5110. doi: 10.1073/pnas.79.16.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purisima E. O., Scheraga H. A. An approach to the multiple-minima problem by relaxing dimensionality. Proc Natl Acad Sci U S A. 1986 May;83(9):2782–2786. doi: 10.1073/pnas.83.9.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk A., Richards J. G., Trzeciak A., Gillessen D., Möhler H. Cholecystokinin receptors: biochemical demonstration and autoradiographical localization in rat brain and pancreas using [3H] cholecystokinin8 as radioligand. J Neurosci. 1984 Apr;4(4):1021–1033. doi: 10.1523/JNEUROSCI.04-04-01021.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vásquez M., Scheraga H. A. Use of buildup and energy-minimization procedures to compute low-energy structures of the backbone of enkephalin. Biopolymers. 1985 Aug;24(8):1437–1447. doi: 10.1002/bip.360240803. [DOI] [PubMed] [Google Scholar]
- Zimmerman S. S., Pottle M. S., Némethy G., Scheraga H. A. Conformational analysis of the 20 naturally occurring amino acid residues using ECEPP. Macromolecules. 1977 Jan-Feb;10(1):1–9. doi: 10.1021/ma60055a001. [DOI] [PubMed] [Google Scholar]