Abstract
In the title compound, C15H12O4·H2O, the two benzene rings are not coplanar, making a dihedral angle of 7.24 (16)°. An intramolecular hydroxy–carbonyl O—H⋯O hydrogen bond occurs. In the crystal, four intermolecular O—H⋯O hydrogen bonds involving the hydroxy residues, the carbonyl group and the water molecule lead to the formation of a three-dimensional network. The supramolecular structure is further stabilized by weak C—H⋯O interactions.
Related literature
For the biological activity of the title compound, see: Jang et al. (2008 ▶); Liu et al. (2008 ▶). For a related structure, see: Ma et al. (2005 ▶).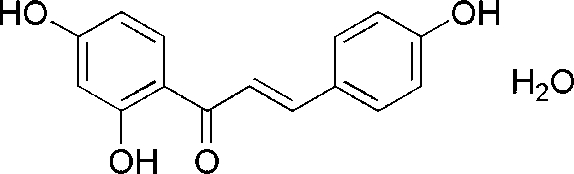
Experimental
Crystal data
C15H12O4·H2O
M r = 274.26
Monoclinic,

a = 11.489 (2) Å
b = 9.5903 (17) Å
c = 12.498 (2) Å
β = 103.649 (3)°
V = 1338.2 (4) Å3
Z = 4
Mo Kα radiation
μ = 0.10 mm−1
T = 298 K
0.12 × 0.10 × 0.10 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.988, T max = 0.990
8297 measured reflections
2625 independent reflections
2115 reflections with I > 2σ(I)
R int = 0.031
Refinement
R[F 2 > 2σ(F 2)] = 0.057
wR(F 2) = 0.144
S = 1.09
2625 reflections
196 parameters
5 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.33 e Å−3
Δρmin = −0.19 e Å−3
Data collection: SMART (Bruker, 1997 ▶); cell refinement: SAINT (Bruker, 1997 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536811006271/go2004sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811006271/go2004Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Enhanced figure: interactive version of Fig. 1
Enhanced figure: interactive version of Fig. 2
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1A⋯O3 | 0.87 (2) | 1.74 (2) | 2.530 (2) | 150 (3) |
| O2—H2A⋯O5i | 0.82 (2) | 1.83 (2) | 2.644 (3) | 175 (4) |
| O4—H4A⋯O3ii | 0.83 (2) | 1.95 (2) | 2.776 (2) | 175 (4) |
| O5—H5A⋯O1iii | 0.84 (2) | 1.99 (2) | 2.802 (2) | 164 (3) |
| O5—H5B⋯O4iv | 0.84 (2) | 1.97 (2) | 2.785 (3) | 165 (3) |
| C9—H9⋯O4v | 0.93 | 2.56 | 3.402 (3) | 151 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  ; (v)
; (v)  .
.
Acknowledgments
This study was funded by the Jiangxi Provincial Department of Education (GJJ08433) and the Jiangxi Provincial Department of Science and Technology (2008ZD06100). The authors thank Professor Xianggao Meng at Hua-Zhong Normal University for the data acquisition.
supplementary crystallographic information
Comment
The title compound exhibits many biological activities such as tracheal relaxation effects (Liu et al., 2008) and suppressing cocaine-induced extracellular dopamine release (Jang et al., 2008).
One (E)-1-(2,4-Dihydroxyphenyl)-3-(4-hydroxyphenyl)prop-2-en-1-one molecule bears one crystalline water molecule (Fig.1). In the molecule, the two benzene rings are not coplanar, the dihedral angle being 7.24 (16)°. The structure displays O—H···O and C—H···O hydrogen bonding (Table 1 and Fig. 2).
Experimental
2, 4-dihydroxyacetophenone (7.6 g, 0.05 mol) and 4-hydroxybenzaldehyde (8.54 g, 0.07 mol) were dissolved in diglycol (25 ml). Then 40% aq. KOH (50 ml) was added, and the reaction mixture was vigorously stirred under nitrogen atmosphere at 333 K for 2 h. The progress of the reaction was monitored by thin- layer chromatography (Si gel, developing solvent V(ethyl acetate)/V(benzene) = 1:2). The mixture was colled to room temperature and 1:1 (v/v) hydrochloric acid was added to acidize the mixture to pH=3 and a solid was obtained. After crystallized by ethanol-water, crystalline yellow needles were obtained, m.p. 472.5–474.2 K.
Refinement
All the carbon-bounded hydrogen atoms were located at their ideal positions with the C—H=0.93Å and Uiso(H)=1.2Ueq(C). All the hydrogen atoms bonded to the oxygen atoms were located from the difference maps and refined with the restraints of O—H=0.82 (1)Å and Uiso(H)=1.5Ueq(O).
Figures
Fig. 1.
View of the molecule of (I) showing the atom-labelling scheme. Displacement ellipsoids are drawn at the 50% probability level.
Fig. 2.
The crystal packing for (I), with O—H···O and C—H···O interactions shown as dashed lines.
Crystal data
| C15H12O4·H2O | F(000) = 576 |
| Mr = 274.26 | Dx = 1.361 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 2288 reflections |
| a = 11.489 (2) Å | θ = 2.7–25.3° |
| b = 9.5903 (17) Å | µ = 0.10 mm−1 |
| c = 12.498 (2) Å | T = 298 K |
| β = 103.649 (3)° | Block, yellow |
| V = 1338.2 (4) Å3 | 0.12 × 0.10 × 0.10 mm |
| Z = 4 |
Data collection
| Bruker SMART CCD area-detector diffractometer | 2625 independent reflections |
| Radiation source: fine-focus sealed tube | 2115 reflections with I > 2σ(I) |
| graphite | Rint = 0.031 |
| phi and ω scans | θmax = 26.0°, θmin = 2.7° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | h = −14→14 |
| Tmin = 0.988, Tmax = 0.990 | k = −11→9 |
| 8297 measured reflections | l = −15→15 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.057 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.144 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.09 | w = 1/[σ2(Fo2) + (0.0569P)2 + 0.499P] where P = (Fo2 + 2Fc2)/3 |
| 2625 reflections | (Δ/σ)max < 0.001 |
| 196 parameters | Δρmax = 0.33 e Å−3 |
| 5 restraints | Δρmin = −0.19 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.19653 (18) | 1.0001 (2) | 0.07068 (17) | 0.0402 (5) | |
| C2 | 0.10947 (18) | 1.1026 (2) | 0.07420 (17) | 0.0412 (5) | |
| C3 | 0.0605 (2) | 1.1156 (2) | 0.16459 (19) | 0.0475 (6) | |
| H3 | 0.0037 | 1.1844 | 0.1655 | 0.057* | |
| C4 | 0.0950 (2) | 1.0275 (2) | 0.25329 (18) | 0.0452 (5) | |
| C5 | 0.1818 (2) | 0.9256 (3) | 0.25306 (19) | 0.0500 (6) | |
| H5 | 0.2061 | 0.8663 | 0.3131 | 0.060* | |
| C6 | 0.2309 (2) | 0.9138 (2) | 0.16359 (19) | 0.0473 (6) | |
| H6 | 0.2892 | 0.8462 | 0.1644 | 0.057* | |
| C7 | 0.24678 (19) | 0.9881 (2) | −0.02538 (18) | 0.0437 (5) | |
| C8 | 0.3403 (2) | 0.8856 (3) | −0.02910 (19) | 0.0494 (6) | |
| H8 | 0.3588 | 0.8183 | 0.0258 | 0.059* | |
| C9 | 0.3990 (2) | 0.8858 (3) | −0.10772 (19) | 0.0493 (6) | |
| H9 | 0.3751 | 0.9540 | −0.1613 | 0.059* | |
| C10 | 0.49523 (19) | 0.7959 (2) | −0.12387 (17) | 0.0431 (5) | |
| C11 | 0.5467 (2) | 0.6925 (3) | −0.05077 (19) | 0.0566 (7) | |
| H11 | 0.5169 | 0.6760 | 0.0111 | 0.068* | |
| C12 | 0.6406 (2) | 0.6134 (3) | −0.0672 (2) | 0.0635 (7) | |
| H12 | 0.6750 | 0.5458 | −0.0161 | 0.076* | |
| C13 | 0.68407 (19) | 0.6350 (3) | −0.16102 (18) | 0.0474 (6) | |
| C14 | 0.6341 (2) | 0.7353 (3) | −0.23509 (18) | 0.0486 (6) | |
| H14 | 0.6628 | 0.7498 | −0.2978 | 0.058* | |
| C15 | 0.5412 (2) | 0.8148 (3) | −0.21662 (19) | 0.0518 (6) | |
| H15 | 0.5080 | 0.8832 | −0.2675 | 0.062* | |
| O1 | 0.07099 (15) | 1.19360 (19) | −0.01001 (14) | 0.0589 (5) | |
| H1A | 0.107 (3) | 1.171 (3) | −0.061 (2) | 0.088* | |
| O2 | 0.04269 (18) | 1.0443 (2) | 0.33756 (14) | 0.0649 (5) | |
| H2A | 0.069 (3) | 0.988 (3) | 0.387 (2) | 0.097* | |
| O3 | 0.21067 (14) | 1.06876 (19) | −0.10709 (13) | 0.0565 (5) | |
| O4 | 0.77775 (17) | 0.5545 (2) | −0.17385 (14) | 0.0668 (6) | |
| H4A | 0.784 (3) | 0.563 (4) | −0.2384 (17) | 0.100* | |
| O5 | 0.12061 (17) | 0.6281 (2) | 0.00416 (14) | 0.0587 (5) | |
| H5A | 0.072 (2) | 0.687 (3) | 0.017 (3) | 0.088* | |
| H5B | 0.149 (3) | 0.586 (3) | 0.0630 (19) | 0.088* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0381 (11) | 0.0444 (12) | 0.0430 (12) | −0.0015 (9) | 0.0192 (9) | −0.0060 (10) |
| C2 | 0.0413 (11) | 0.0446 (12) | 0.0415 (11) | −0.0002 (10) | 0.0174 (9) | 0.0015 (10) |
| C3 | 0.0457 (12) | 0.0512 (14) | 0.0524 (13) | 0.0090 (11) | 0.0251 (10) | −0.0011 (11) |
| C4 | 0.0502 (12) | 0.0514 (13) | 0.0409 (12) | 0.0005 (10) | 0.0247 (10) | −0.0039 (10) |
| C5 | 0.0583 (14) | 0.0530 (14) | 0.0436 (12) | 0.0078 (11) | 0.0217 (11) | 0.0070 (10) |
| C6 | 0.0485 (12) | 0.0485 (13) | 0.0497 (13) | 0.0086 (10) | 0.0213 (10) | −0.0007 (10) |
| C7 | 0.0400 (11) | 0.0520 (14) | 0.0432 (12) | −0.0049 (10) | 0.0183 (9) | −0.0041 (10) |
| C8 | 0.0513 (13) | 0.0551 (14) | 0.0483 (13) | 0.0061 (11) | 0.0249 (10) | −0.0017 (11) |
| C9 | 0.0475 (12) | 0.0576 (15) | 0.0485 (13) | 0.0041 (11) | 0.0229 (10) | 0.0008 (11) |
| C10 | 0.0404 (11) | 0.0506 (13) | 0.0431 (12) | −0.0005 (10) | 0.0193 (10) | −0.0044 (10) |
| C11 | 0.0605 (15) | 0.0757 (18) | 0.0437 (13) | 0.0113 (13) | 0.0323 (12) | 0.0049 (12) |
| C12 | 0.0690 (16) | 0.0791 (19) | 0.0495 (14) | 0.0290 (15) | 0.0285 (12) | 0.0156 (13) |
| C13 | 0.0425 (12) | 0.0596 (15) | 0.0446 (12) | 0.0065 (11) | 0.0193 (10) | −0.0039 (11) |
| C14 | 0.0493 (13) | 0.0617 (15) | 0.0427 (12) | 0.0062 (11) | 0.0267 (10) | 0.0028 (11) |
| C15 | 0.0528 (13) | 0.0586 (15) | 0.0507 (14) | 0.0097 (11) | 0.0254 (11) | 0.0083 (11) |
| O1 | 0.0648 (11) | 0.0665 (12) | 0.0538 (10) | 0.0202 (9) | 0.0307 (8) | 0.0141 (9) |
| O2 | 0.0793 (13) | 0.0756 (13) | 0.0531 (10) | 0.0183 (10) | 0.0424 (10) | 0.0072 (9) |
| O3 | 0.0571 (10) | 0.0710 (12) | 0.0492 (9) | 0.0108 (8) | 0.0284 (8) | 0.0084 (8) |
| O4 | 0.0654 (11) | 0.0920 (14) | 0.0512 (10) | 0.0355 (10) | 0.0302 (9) | 0.0110 (10) |
| O5 | 0.0702 (12) | 0.0605 (12) | 0.0539 (10) | 0.0151 (9) | 0.0315 (9) | 0.0043 (9) |
Geometric parameters (Å, °)
| C1—C6 | 1.405 (3) | C9—H9 | 0.9300 |
| C1—C2 | 1.410 (3) | C10—C11 | 1.383 (3) |
| C1—C7 | 1.454 (3) | C10—C15 | 1.394 (3) |
| C2—O1 | 1.359 (3) | C11—C12 | 1.374 (3) |
| C2—C3 | 1.382 (3) | C11—H11 | 0.9300 |
| C3—C4 | 1.375 (3) | C12—C13 | 1.393 (3) |
| C3—H3 | 0.9300 | C12—H12 | 0.9300 |
| C4—O2 | 1.340 (3) | C13—C14 | 1.364 (3) |
| C4—C5 | 1.397 (3) | C13—O4 | 1.364 (3) |
| C5—C6 | 1.371 (3) | C14—C15 | 1.375 (3) |
| C5—H5 | 0.9300 | C14—H14 | 0.9300 |
| C6—H6 | 0.9300 | C15—H15 | 0.9300 |
| C7—O3 | 1.270 (3) | O1—H1A | 0.867 (18) |
| C7—C8 | 1.465 (3) | O2—H2A | 0.819 (18) |
| C8—C9 | 1.316 (3) | O4—H4A | 0.829 (18) |
| C8—H8 | 0.9300 | O5—H5A | 0.836 (18) |
| C9—C10 | 1.453 (3) | O5—H5B | 0.836 (18) |
| C6—C1—C2 | 116.61 (18) | C8—C9—H9 | 115.0 |
| C6—C1—C7 | 123.2 (2) | C10—C9—H9 | 115.0 |
| C2—C1—C7 | 120.24 (19) | C11—C10—C15 | 117.1 (2) |
| O1—C2—C3 | 117.03 (19) | C11—C10—C9 | 123.66 (19) |
| O1—C2—C1 | 121.81 (18) | C15—C10—C9 | 119.2 (2) |
| C3—C2—C1 | 121.2 (2) | C12—C11—C10 | 121.7 (2) |
| C4—C3—C2 | 120.5 (2) | C12—C11—H11 | 119.1 |
| C4—C3—H3 | 119.7 | C10—C11—H11 | 119.1 |
| C2—C3—H3 | 119.7 | C11—C12—C13 | 119.6 (2) |
| O2—C4—C3 | 117.5 (2) | C11—C12—H12 | 120.2 |
| O2—C4—C5 | 122.6 (2) | C13—C12—H12 | 120.2 |
| C3—C4—C5 | 119.90 (19) | C14—C13—O4 | 122.43 (19) |
| C6—C5—C4 | 119.4 (2) | C14—C13—C12 | 119.9 (2) |
| C6—C5—H5 | 120.3 | O4—C13—C12 | 117.6 (2) |
| C4—C5—H5 | 120.3 | C13—C14—C15 | 119.7 (2) |
| C5—C6—C1 | 122.4 (2) | C13—C14—H14 | 120.1 |
| C5—C6—H6 | 118.8 | C15—C14—H14 | 120.1 |
| C1—C6—H6 | 118.8 | C14—C15—C10 | 121.9 (2) |
| O3—C7—C1 | 119.85 (19) | C14—C15—H15 | 119.0 |
| O3—C7—C8 | 119.07 (19) | C10—C15—H15 | 119.0 |
| C1—C7—C8 | 121.1 (2) | C2—O1—H1A | 107 (2) |
| C9—C8—C7 | 122.1 (2) | C4—O2—H2A | 111 (3) |
| C9—C8—H8 | 119.0 | C13—O4—H4A | 108 (2) |
| C7—C8—H8 | 119.0 | H5A—O5—H5B | 107 (3) |
| C8—C9—C10 | 130.0 (2) | ||
| C6—C1—C2—O1 | 178.7 (2) | C2—C1—C7—C8 | 177.9 (2) |
| C7—C1—C2—O1 | −0.9 (3) | O3—C7—C8—C9 | 8.8 (4) |
| C6—C1—C2—C3 | −0.5 (3) | C1—C7—C8—C9 | −170.2 (2) |
| C7—C1—C2—C3 | 179.9 (2) | C7—C8—C9—C10 | 178.4 (2) |
| O1—C2—C3—C4 | −179.8 (2) | C8—C9—C10—C11 | −3.0 (4) |
| C1—C2—C3—C4 | −0.6 (3) | C8—C9—C10—C15 | 178.3 (3) |
| C2—C3—C4—O2 | −179.0 (2) | C15—C10—C11—C12 | 1.3 (4) |
| C2—C3—C4—C5 | 1.1 (4) | C9—C10—C11—C12 | −177.5 (2) |
| O2—C4—C5—C6 | 179.5 (2) | C10—C11—C12—C13 | −1.5 (4) |
| C3—C4—C5—C6 | −0.6 (4) | C11—C12—C13—C14 | 0.7 (4) |
| C4—C5—C6—C1 | −0.5 (4) | C11—C12—C13—O4 | 179.7 (2) |
| C2—C1—C6—C5 | 1.1 (3) | O4—C13—C14—C15 | −178.9 (2) |
| C7—C1—C6—C5 | −179.4 (2) | C12—C13—C14—C15 | 0.1 (4) |
| C6—C1—C7—O3 | 179.3 (2) | C13—C14—C15—C10 | −0.3 (4) |
| C2—C1—C7—O3 | −1.1 (3) | C11—C10—C15—C14 | −0.4 (4) |
| C6—C1—C7—C8 | −1.7 (3) | C9—C10—C15—C14 | 178.4 (2) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1A···O3 | 0.87 (2) | 1.74 (2) | 2.530 (2) | 150 (3) |
| O2—H2A···O5i | 0.82 (2) | 1.83 (2) | 2.644 (3) | 175 (4) |
| O4—H4A···O3ii | 0.83 (2) | 1.95 (2) | 2.776 (2) | 175 (4) |
| O5—H5A···O1iii | 0.84 (2) | 1.99 (2) | 2.802 (2) | 164 (3) |
| O5—H5B···O4iv | 0.84 (2) | 1.97 (2) | 2.785 (3) | 165 (3) |
| C9—H9···O4v | 0.93 | 2.56 | 3.402 (3) | 151 |
Symmetry codes: (i) x, −y+3/2, z+1/2; (ii) −x+1, y−1/2, −z−1/2; (iii) −x, −y+2, −z; (iv) −x+1, −y+1, −z; (v) −x+1, y+1/2, −z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: GO2004).
References
- Bruker (1997). SMART and SAINT Bruker AXS Inc. Madison, Wisconsin, USA.
- Jang, E. Y., Choe, E. S., Hwang, M., Kim, S. C., Lee, J. R., Kim, S. G., Jeon, J. P., Buono, R. J. & Yang, C. H. (2008). Eur. J. Pharmacol. 587, 124–128. [DOI] [PubMed]
- Liu, B., Yang, J., Wen, Q. S. & Li, Y. (2008). Eur. J. Pharmacol. 587, 257–266 [DOI] [PubMed]
- Ma, C.-J., Li, G.-S., Zhang, D.-L., Liu, K. & Fan, X. (2005). J. Chromatogr. A, 1078, 188–192. [DOI] [PubMed]
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536811006271/go2004sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811006271/go2004Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Enhanced figure: interactive version of Fig. 1
Enhanced figure: interactive version of Fig. 2




