Abstract
In the title molecule, C16H23N3O3, the dihedral angle between the benzimidazole and nitro group planes is 5.34 (9)° and the dihedral angle between the benzimidazole and aliphatic chain mean planes is 73.23 (5)°. The C—C—C—C torsion angles (about 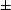 176°) of the nonyl group indicate an all-antiperiplanar conformation. In the crystal, adjacent molecules are linked by pairs of N—H⋯O hydrogen bonds into inversion dimers. These molecules are further connected through C—H⋯O interactions, building tapes parallel to (
176°) of the nonyl group indicate an all-antiperiplanar conformation. In the crystal, adjacent molecules are linked by pairs of N—H⋯O hydrogen bonds into inversion dimers. These molecules are further connected through C—H⋯O interactions, building tapes parallel to (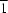 22).
22).
Related literature
For background to the pharmacological and biochemical properties of benzimidazolones, see: Gbadamassi et al. (1988 ▶); Singh et al. (2000 ▶); Derand et al. (2003 ▶); Badarau et al. (2009 ▶). For similar structures, see: Saber et al. (2010 ▶); Ouzidan et al. (2011 ▶).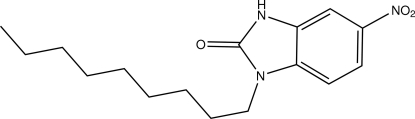
Experimental
Crystal data
C16H23N3O3
M r = 305.37
Triclinic,
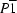
a = 5.483 (1) Å
b = 10.2092 (15) Å
c = 14.746 (3) Å
α = 74.275 (9)°
β = 79.727 (6)°
γ = 83.410 (8)°
V = 779.9 (2) Å3
Z = 2
Mo Kα radiation
μ = 0.09 mm−1
T = 90 K
0.35 × 0.27 × 0.22 mm
Data collection
Nonius KappaCCD diffractometer
21087 measured reflections
6349 independent reflections
5183 reflections with I > 2σ(I)
R int = 0.023
Refinement
R[F 2 > 2σ(F 2)] = 0.040
wR(F 2) = 0.113
S = 1.03
6349 reflections
201 parameters
H-atom parameters constrained
Δρmax = 0.45 e Å−3
Δρmin = −0.28 e Å−3
Data collection: COLLECT (Nonius, 2000 ▶); cell refinement: SCALEPACK (Otwinowski & Minor, 1997 ▶); data reduction: DENZO (Otwinowski & Minor, 1997 ▶) and SCALEPACK; program(s) used to solve structure: SIR97 (Altomare et al., 1999 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶) and PLATON (Spek, 2009 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536811005654/gk2342sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811005654/gk2342Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O1i | 0.88 | 1.89 | 2.7651 (9) | 170 |
| C6—H6⋯O3ii | 0.95 | 2.58 | 3.3139 (11) | 134 |
Symmetry codes: (i) 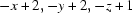 ; (ii)
; (ii) 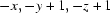 .
.
supplementary crystallographic information
Comment
Benzimidazoles are useful intermediates/subunits for the development of molecules of pharmaceutical or biological interest (Gbadamassi et al., 1988). Benzimidazolone and its derivatives are also an important class of bioactive molecules in the field of drugs and pharmaceuticals (Derand et al., 2003). They found potential applications in diverse therapeutic areas including, anti-hypertensives and anti-virals (Badarau et al., 2009; Singh et al., 2000). The structural studies of benzimidazolone, linked to an isopropenyl and nonyl group respectively, have been published by Saber et al. (2010)and Ouzidan et al. (2011).
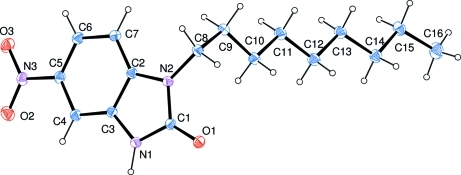
The 5-nitro-1-nonyl-1H-benzimidazol-2(3H)-one molecule structure is built up from two fused six-and five-membered rings linked to C9H19 chain as shown in Fig.1. The aliphatic chain has all-antiperiplanar (all-trans) conformation. Furthermore, the fused-ring system and the nitro group are almost planar, with a maximum deviation of 0.0414 (8) Å and 0.0250 (7) Å for C4 and N1 respectively. The dihedral angle between the two rings and nitro group planes is 5.34 (9)°. The torsion angles C1 N2 C8 C9 and C13 C14 C15 C16 are 113.66 (8)° and 177.26 (7)° respectively.
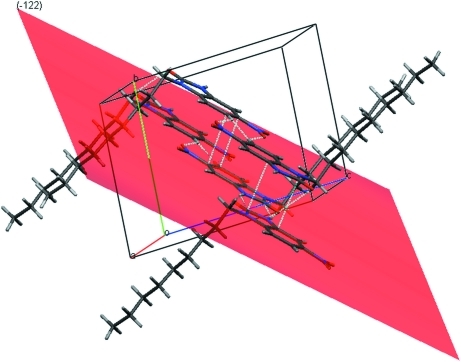
In the crystal, adjacent molecules are linked by pairs of N—H···O hydrogen bonds into inversion dimers. These molecules are further connected through C—H···O hydrogen bonds into a tape parallel to the (-1 2 2) plane, as schown in Fig. 2 and Table 1.
Experimental
To 5-nitro-1H-benzoimidazol-2(3H)-one (0.2 g, 1.1 mmol), potassium carbonate (0.30 g, 2.2 mmol) and tetra-n-butylammonium bromide (0.07 g, 0.2 mmol) in DMF (15 ml) was added 1-bromononane (0.43 ml, 2.2 mmol). Stirring was continued at room temperature for 6 h. The salt was removed by filtration and the filtrate concentrated under reduced pressure. The residue was separated by chromatography on a column of silica gel with ethyl acetate/hexane (1/2) as eluent. Colorless needle-shaped crystals were isolated when the solvent was allowed to evaporate [(m.p. 392–394 K (ethanol)].
Refinement
H atoms were located in a difference map and treated as riding with C—H = 0.99 Å, 0.98, Å, 0.95 Å, and 0.88 Å for –CH2–, –CH3, aromatic CH and NH respectively. All H atoms with Uiso(H) = 1.2 Ueq (aromatic, methylene, N) and Uiso(H) = 1.5 Ueq(methyl).
Figures
Fig. 1.
: Molecular structure of the title compound with displacement ellipsoids drawn at the 50% probability level. H atoms are represented as small circles.
Fig. 2.
Partial packing view of the title compound, showing tapes in the (-1 2 2) plane, built up from molecules linked through N—H···O hydrogen bonds and intermolecular C–H···O contacts (dashed lines).
Crystal data
| C16H23N3O3 | Z = 2 |
| Mr = 305.37 | F(000) = 328 |
| Triclinic, P1 | Dx = 1.300 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 5.483 (1) Å | Cell parameters from 5537 reflections |
| b = 10.2092 (15) Å | θ = 2.5–34.9° |
| c = 14.746 (3) Å | µ = 0.09 mm−1 |
| α = 74.275 (9)° | T = 90 K |
| β = 79.727 (6)° | Needle, colourless |
| γ = 83.410 (8)° | 0.35 × 0.27 × 0.22 mm |
| V = 779.9 (2) Å3 |
Data collection
| Nonius KappaCCD diffractometer | 5183 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.023 |
| graphite | θmax = 34.9°, θmin = 2.8° |
| ω and φ scans | h = −8→8 |
| 21087 measured reflections | k = −15→16 |
| 6349 independent reflections | l = −23→22 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.040 | H-atom parameters constrained |
| wR(F2) = 0.113 | w = 1/[σ2(Fo2) + (0.0578P)2 + 0.1462P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.03 | (Δ/σ)max = 0.001 |
| 6349 reflections | Δρmax = 0.45 e Å−3 |
| 201 parameters | Δρmin = −0.28 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.023 (5) |
Special details
| Geometry. All s.u.'s (except the s.u. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell s.u.'s are taken into account individually in the estimation of s.u.'s in distances, angles and torsion angles; correlations between s.u.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell s.u.'s is used for estimating s.u.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.92795 (11) | 1.07035 (6) | 0.37556 (4) | 0.01618 (12) | |
| O2 | 0.40529 (12) | 0.45052 (6) | 0.71989 (4) | 0.02109 (13) | |
| O3 | 0.12666 (12) | 0.41865 (6) | 0.64166 (5) | 0.02143 (13) | |
| N1 | 0.78463 (12) | 0.88673 (7) | 0.50035 (5) | 0.01369 (12) | |
| H1 | 0.8751 | 0.8904 | 0.5431 | 0.016* | |
| N2 | 0.61923 (12) | 0.93607 (7) | 0.36748 (4) | 0.01292 (12) | |
| N3 | 0.29292 (13) | 0.48382 (7) | 0.65097 (5) | 0.01591 (13) | |
| C1 | 0.79248 (13) | 0.97452 (8) | 0.41127 (5) | 0.01294 (13) | |
| C2 | 0.50448 (13) | 0.82444 (7) | 0.42883 (5) | 0.01255 (13) | |
| C3 | 0.61268 (13) | 0.79137 (8) | 0.51286 (5) | 0.01240 (13) | |
| C4 | 0.54726 (13) | 0.68061 (8) | 0.58770 (5) | 0.01379 (13) | |
| H4 | 0.6229 | 0.6565 | 0.6437 | 0.017* | |
| C5 | 0.36219 (13) | 0.60623 (7) | 0.57568 (5) | 0.01384 (13) | |
| C6 | 0.24677 (14) | 0.63920 (8) | 0.49481 (6) | 0.01500 (13) | |
| H6 | 0.1193 | 0.5860 | 0.4912 | 0.018* | |
| C7 | 0.31799 (14) | 0.75013 (8) | 0.41917 (5) | 0.01431 (13) | |
| H7 | 0.2421 | 0.7740 | 0.3632 | 0.017* | |
| C8 | 0.56136 (14) | 1.00911 (8) | 0.27295 (5) | 0.01453 (13) | |
| H8A | 0.6475 | 1.0948 | 0.2508 | 0.017* | |
| H8B | 0.3804 | 1.0338 | 0.2778 | 0.017* | |
| C9 | 0.63787 (14) | 0.92606 (8) | 0.19926 (5) | 0.01496 (13) | |
| H9A | 0.5714 | 0.9758 | 0.1402 | 0.018* | |
| H9B | 0.5589 | 0.8382 | 0.2237 | 0.018* | |
| C10 | 0.91760 (14) | 0.89612 (8) | 0.17380 (5) | 0.01536 (14) | |
| H10A | 0.9848 | 0.8397 | 0.2311 | 0.018* | |
| H10B | 1.0005 | 0.9829 | 0.1522 | 0.018* | |
| C11 | 0.97314 (15) | 0.82089 (8) | 0.09493 (6) | 0.01648 (14) | |
| H11A | 0.8959 | 0.7325 | 0.1186 | 0.020* | |
| H11B | 0.8936 | 0.8751 | 0.0400 | 0.020* | |
| C12 | 1.24889 (15) | 0.79376 (8) | 0.06003 (6) | 0.01714 (14) | |
| H12A | 1.3281 | 0.7349 | 0.1137 | 0.021* | |
| H12B | 1.3293 | 0.8813 | 0.0386 | 0.021* | |
| C13 | 1.29159 (14) | 0.72462 (8) | −0.02204 (6) | 0.01650 (14) | |
| H13A | 1.2188 | 0.6349 | 0.0010 | 0.020* | |
| H13B | 1.2018 | 0.7809 | −0.0735 | 0.020* | |
| C14 | 1.56428 (15) | 0.70296 (8) | −0.06374 (6) | 0.01707 (14) | |
| H14A | 1.6346 | 0.7928 | −0.0918 | 0.020* | |
| H14B | 1.6575 | 0.6522 | −0.0118 | 0.020* | |
| C15 | 1.59873 (15) | 0.62433 (9) | −0.14015 (6) | 0.01812 (15) | |
| H15A | 1.5350 | 0.5330 | −0.1111 | 0.022* | |
| H15B | 1.4981 | 0.6728 | −0.1903 | 0.022* | |
| C16 | 1.86819 (17) | 0.60708 (10) | −0.18635 (7) | 0.02506 (18) | |
| H16A | 1.9271 | 0.6967 | −0.2215 | 0.038* | |
| H16B | 1.8795 | 0.5492 | −0.2304 | 0.038* | |
| H16C | 1.9712 | 0.5643 | −0.1368 | 0.038* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0171 (2) | 0.0163 (3) | 0.0153 (2) | −0.00631 (19) | −0.00140 (19) | −0.0029 (2) |
| O2 | 0.0265 (3) | 0.0195 (3) | 0.0162 (3) | −0.0041 (2) | −0.0049 (2) | −0.0008 (2) |
| O3 | 0.0224 (3) | 0.0188 (3) | 0.0235 (3) | −0.0096 (2) | −0.0006 (2) | −0.0048 (2) |
| N1 | 0.0147 (3) | 0.0147 (3) | 0.0125 (3) | −0.0040 (2) | −0.0026 (2) | −0.0035 (2) |
| N2 | 0.0138 (3) | 0.0137 (3) | 0.0117 (3) | −0.0029 (2) | −0.0018 (2) | −0.0032 (2) |
| N3 | 0.0177 (3) | 0.0143 (3) | 0.0153 (3) | −0.0031 (2) | 0.0006 (2) | −0.0044 (2) |
| C1 | 0.0130 (3) | 0.0138 (3) | 0.0126 (3) | −0.0014 (2) | −0.0010 (2) | −0.0048 (2) |
| C2 | 0.0120 (3) | 0.0132 (3) | 0.0126 (3) | −0.0014 (2) | −0.0005 (2) | −0.0043 (2) |
| C3 | 0.0119 (3) | 0.0133 (3) | 0.0130 (3) | −0.0020 (2) | −0.0009 (2) | −0.0052 (2) |
| C4 | 0.0143 (3) | 0.0145 (3) | 0.0128 (3) | −0.0020 (2) | −0.0011 (2) | −0.0041 (2) |
| C5 | 0.0147 (3) | 0.0123 (3) | 0.0141 (3) | −0.0024 (2) | 0.0000 (2) | −0.0034 (2) |
| C6 | 0.0140 (3) | 0.0150 (3) | 0.0168 (3) | −0.0028 (2) | −0.0017 (2) | −0.0051 (3) |
| C7 | 0.0132 (3) | 0.0159 (3) | 0.0148 (3) | −0.0020 (2) | −0.0030 (2) | −0.0046 (2) |
| C8 | 0.0158 (3) | 0.0147 (3) | 0.0125 (3) | 0.0003 (2) | −0.0026 (2) | −0.0028 (2) |
| C9 | 0.0157 (3) | 0.0172 (3) | 0.0125 (3) | −0.0017 (2) | −0.0026 (2) | −0.0043 (2) |
| C10 | 0.0163 (3) | 0.0171 (3) | 0.0135 (3) | −0.0011 (2) | −0.0024 (2) | −0.0053 (3) |
| C11 | 0.0173 (3) | 0.0189 (3) | 0.0142 (3) | −0.0006 (3) | −0.0017 (2) | −0.0065 (3) |
| C12 | 0.0179 (3) | 0.0196 (4) | 0.0153 (3) | −0.0003 (3) | −0.0029 (3) | −0.0071 (3) |
| C13 | 0.0168 (3) | 0.0184 (3) | 0.0150 (3) | −0.0001 (3) | −0.0020 (2) | −0.0063 (3) |
| C14 | 0.0173 (3) | 0.0181 (3) | 0.0157 (3) | −0.0004 (3) | −0.0015 (3) | −0.0053 (3) |
| C15 | 0.0200 (3) | 0.0183 (4) | 0.0157 (3) | 0.0012 (3) | −0.0015 (3) | −0.0056 (3) |
| C16 | 0.0217 (4) | 0.0302 (5) | 0.0221 (4) | 0.0022 (3) | 0.0013 (3) | −0.0094 (3) |
Geometric parameters (Å, °)
| O1—C1 | 1.2387 (9) | C9—H9A | 0.9900 |
| O2—N3 | 1.2313 (9) | C9—H9B | 0.9900 |
| O3—N3 | 1.2352 (9) | C10—C11 | 1.5297 (11) |
| N1—C1 | 1.3715 (10) | C10—H10A | 0.9900 |
| N1—C3 | 1.3864 (9) | C10—H10B | 0.9900 |
| N1—H1 | 0.8800 | C11—C12 | 1.5268 (11) |
| N2—C1 | 1.3838 (10) | C11—H11A | 0.9900 |
| N2—C2 | 1.3842 (10) | C11—H11B | 0.9900 |
| N2—C8 | 1.4628 (10) | C12—C13 | 1.5303 (11) |
| N3—C5 | 1.4646 (10) | C12—H12A | 0.9900 |
| C2—C7 | 1.3893 (10) | C12—H12B | 0.9900 |
| C2—C3 | 1.4109 (10) | C13—C14 | 1.5272 (11) |
| C3—C4 | 1.3787 (11) | C13—H13A | 0.9900 |
| C4—C5 | 1.3970 (11) | C13—H13B | 0.9900 |
| C4—H4 | 0.9500 | C14—C15 | 1.5263 (11) |
| C5—C6 | 1.3924 (11) | C14—H14A | 0.9900 |
| C6—C7 | 1.3930 (11) | C14—H14B | 0.9900 |
| C6—H6 | 0.9500 | C15—C16 | 1.5246 (12) |
| C7—H7 | 0.9500 | C15—H15A | 0.9900 |
| C8—C9 | 1.5266 (11) | C15—H15B | 0.9900 |
| C8—H8A | 0.9900 | C16—H16A | 0.9800 |
| C8—H8B | 0.9900 | C16—H16B | 0.9800 |
| C9—C10 | 1.5284 (11) | C16—H16C | 0.9800 |
| C1—N1—C3 | 109.78 (6) | C9—C10—C11 | 110.76 (6) |
| C1—N1—H1 | 125.1 | C9—C10—H10A | 109.5 |
| C3—N1—H1 | 125.1 | C11—C10—H10A | 109.5 |
| C1—N2—C2 | 109.39 (6) | C9—C10—H10B | 109.5 |
| C1—N2—C8 | 124.01 (6) | C11—C10—H10B | 109.5 |
| C2—N2—C8 | 126.52 (6) | H10A—C10—H10B | 108.1 |
| O2—N3—O3 | 123.41 (7) | C12—C11—C10 | 114.85 (6) |
| O2—N3—C5 | 118.20 (7) | C12—C11—H11A | 108.6 |
| O3—N3—C5 | 118.39 (7) | C10—C11—H11A | 108.6 |
| O1—C1—N1 | 127.21 (7) | C12—C11—H11B | 108.6 |
| O1—C1—N2 | 125.74 (7) | C10—C11—H11B | 108.6 |
| N1—C1—N2 | 107.04 (6) | H11A—C11—H11B | 107.5 |
| N2—C2—C7 | 131.59 (7) | C11—C12—C13 | 112.22 (6) |
| N2—C2—C3 | 106.99 (6) | C11—C12—H12A | 109.2 |
| C7—C2—C3 | 121.42 (7) | C13—C12—H12A | 109.2 |
| C4—C3—N1 | 131.24 (7) | C11—C12—H12B | 109.2 |
| C4—C3—C2 | 121.98 (7) | C13—C12—H12B | 109.2 |
| N1—C3—C2 | 106.77 (6) | H12A—C12—H12B | 107.9 |
| C3—C4—C5 | 115.49 (7) | C14—C13—C12 | 114.38 (7) |
| C3—C4—H4 | 122.3 | C14—C13—H13A | 108.7 |
| C5—C4—H4 | 122.3 | C12—C13—H13A | 108.7 |
| C6—C5—C4 | 123.69 (7) | C14—C13—H13B | 108.7 |
| C6—C5—N3 | 118.32 (7) | C12—C13—H13B | 108.7 |
| C4—C5—N3 | 117.95 (7) | H13A—C13—H13B | 107.6 |
| C5—C6—C7 | 120.08 (7) | C15—C14—C13 | 112.45 (7) |
| C5—C6—H6 | 120.0 | C15—C14—H14A | 109.1 |
| C7—C6—H6 | 120.0 | C13—C14—H14A | 109.1 |
| C2—C7—C6 | 117.29 (7) | C15—C14—H14B | 109.1 |
| C2—C7—H7 | 121.4 | C13—C14—H14B | 109.1 |
| C6—C7—H7 | 121.4 | H14A—C14—H14B | 107.8 |
| N2—C8—C9 | 113.04 (6) | C16—C15—C14 | 113.56 (7) |
| N2—C8—H8A | 109.0 | C16—C15—H15A | 108.9 |
| C9—C8—H8A | 109.0 | C14—C15—H15A | 108.9 |
| N2—C8—H8B | 109.0 | C16—C15—H15B | 108.9 |
| C9—C8—H8B | 109.0 | C14—C15—H15B | 108.9 |
| H8A—C8—H8B | 107.8 | H15A—C15—H15B | 107.7 |
| C8—C9—C10 | 115.31 (6) | C15—C16—H16A | 109.5 |
| C8—C9—H9A | 108.4 | C15—C16—H16B | 109.5 |
| C10—C9—H9A | 108.4 | H16A—C16—H16B | 109.5 |
| C8—C9—H9B | 108.4 | C15—C16—H16C | 109.5 |
| C10—C9—H9B | 108.4 | H16A—C16—H16C | 109.5 |
| H9A—C9—H9B | 107.5 | H16B—C16—H16C | 109.5 |
| C3—N1—C1—O1 | −178.59 (7) | C3—C4—C5—N3 | 177.28 (6) |
| C3—N1—C1—N2 | 1.29 (8) | O2—N3—C5—C6 | 175.91 (7) |
| C2—N2—C1—O1 | 179.64 (7) | O3—N3—C5—C6 | −3.41 (11) |
| C8—N2—C1—O1 | −3.45 (12) | O2—N3—C5—C4 | −1.82 (10) |
| C2—N2—C1—N1 | −0.24 (8) | O3—N3—C5—C4 | 178.86 (7) |
| C8—N2—C1—N1 | 176.67 (6) | C4—C5—C6—C7 | 1.40 (12) |
| C1—N2—C2—C7 | 178.96 (8) | N3—C5—C6—C7 | −176.19 (7) |
| C8—N2—C2—C7 | 2.15 (13) | N2—C2—C7—C6 | 178.47 (7) |
| C1—N2—C2—C3 | −0.87 (8) | C3—C2—C7—C6 | −1.72 (11) |
| C8—N2—C2—C3 | −177.68 (6) | C5—C6—C7—C2 | −0.33 (11) |
| C1—N1—C3—C4 | 176.91 (8) | C1—N2—C8—C9 | 113.65 (8) |
| C1—N1—C3—C2 | −1.82 (8) | C2—N2—C8—C9 | −69.97 (9) |
| N2—C2—C3—C4 | −177.25 (7) | N2—C8—C9—C10 | −66.44 (9) |
| C7—C2—C3—C4 | 2.90 (11) | C8—C9—C10—C11 | −176.26 (6) |
| N2—C2—C3—N1 | 1.62 (8) | C9—C10—C11—C12 | 176.47 (7) |
| C7—C2—C3—N1 | −178.23 (7) | C10—C11—C12—C13 | −177.19 (7) |
| N1—C3—C4—C5 | 179.65 (7) | C11—C12—C13—C14 | 176.46 (7) |
| C2—C3—C4—C5 | −1.79 (11) | C12—C13—C14—C15 | 175.71 (7) |
| C3—C4—C5—C6 | −0.32 (11) | C13—C14—C15—C16 | 177.26 (7) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O1i | 0.88 | 1.89 | 2.7651 (9) | 170 |
| C6—H6···O3ii | 0.95 | 2.58 | 3.3139 (11) | 134 |
Symmetry codes: (i) −x+2, −y+2, −z+1; (ii) −x, −y+1, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: GK2342).
References
- Altomare, A., Burla, M. C., Camalli, M., Carrozzini, B., Cascarano, G. L., Giacovazzo, C., Guagliardi, A., Moliterni, A. G. G., Polidori, G. & Rizzi, R. (1999). J. Appl. Cryst. 32, 339–340.
- Badarau, E., Suzenet, F., Bojarski, A. J., Adriana-Luminiţa Fînaru, A. L. & Guillaumet, G. (2009). Bioorg. Med. Chem. Lett. 19, 1600–1603. [DOI] [PubMed]
- Derand, R., Bulteau-Pignoux, L. & Becq, F. (2003). J. Membr. Biol. 194, 109–117. [DOI] [PubMed]
- Farrugia, L. J. (1997). J. Appl. Cryst. 30, 565.
- Gbadamassi, M., Barascut, J. L., Imbach, J. L. & Gayral, P. (1988). Eur. J. Med. Chem. 23, 225–232.
- Nonius (2000). COLLECT Nonius BV, Delft, The Netherlands.
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Vol. 276, Macromolecular Crystallography, Part A, edited by C. W. Carter Jr & R. M. Sweet, pp. 307–326. New York: Academic Press.
- Ouzidan, Y., Kandri Rodi, Y., Butcher, R. J., Essassi, E. M. & El Ammari, L. (2011). Acta Cryst. E67, o283. [DOI] [PMC free article] [PubMed]
- Saber, A., Zouihri, H., Essassi, E. M. & Ng, S. W. (2010). Acta Cryst. E66, o1409. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Singh, S., Syme, C. A., Singh, A. K., Devor, D. C. & Bridges, R. J. (2000). J. Pharmacol. Exp. Ther. 296, 600–611. [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536811005654/gk2342sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536811005654/gk2342Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report