Abstract
Wnt proteins mediate the transduction of at least three major signaling pathways that play central roles in many early and late developmental decisions. They control diverse cellular behaviors, such as cell fate decisions, proliferation, and migration, and are involved in many important embryological events, including axis specification, gastrulation, and limb, heart, or neural development. The three major Wnt pathways are activated by ligands, the Wnts, which clearly belong to the same gene family. However, their signal is then mediated by three separate sets of extracellular, cytoplasmic, and nuclear components that are pathway-specific and that distinguish each of them. Homologs of the Wnt genes and of the Wnt pathways components have been discovered in many eukaryotic model systems and functional investigations have been carried out for most of them. This review extracts available data on the Wnt pathways, from the protist Dictyostelium discoideum to humans, and provides from an evolutionary prospective the overall molecular and functional conservation of the three Wnt pathways and their activators throughout the eukaryotic superkingdom.
Keywords: Wnt pathway, eukaryote, evolution, canonical, PCP, calcium
1. Introduction
The founders of the Wnt gene family were identified and sequenced in the late 1900s independently in Drosophila and mouse. In Drosophila, wingless was identified as a segment polarity gene (1, 2), while in mouse int-1 was cloned as a proto-oncogene, responsible for virally induced mammary tumors (3, 4). The name “Wnt” derived from the combination of both names “wingless” and “int-1” after these two genes were shown to encode homologous proteins (5). Because of the undeniable involvement of these genes in cancer and embryonic development, interest was naturally prompted and led to an exhaustive search for Wnt homologs throughout the eukaryotic superkingdom (for a eukaryotic phylogenic representation, see Fig. 1.1). Combining results from degenerative PCR approaches with bioinformatics surveys using available genomic sequences, more than 100 Wnt genes have now been reported, which comprise the large Wnt multi-gene family. Sequence analyses comparing all of these genes allow the examination of the evolution of the Wnt proteins among eukaryotes. Wnt genes encode secreted ligands that bind to a receptor and activate a signaling pathway. In parallel to the search for Wnt homologs, other studies were thereby carried out to identify the Wnt receptors and the extracellular and intracellular molecules acting alongside or in response to this ligand–receptor interaction. Genetic analyses in Drosophila, Caenorhabditis elegans, and zebrafish, as well as biochemical experiments in mammalian cell culture, and Xenopus embryos, and gene targeting in mouse, authenticated both the receptors and the molecular pathways activated by Wnts. With these studies it became clear that Wnt signals transduce many distinct pathways, three of which have been the most studied these past decades (6–8). Among these three major pathways, the first to be elucidated was the Wnt/β-catenin pathway, often called the “canonical” Wnt pathway. Then, several others called “non-canonical” Wnt pathways were discovered; these involve many of the same components used by the canonical pathway but with molecular relationships between these components that are altered relative to the canonical pathway, or they utilize different transducing molecules. The most studied of these non-canonical Wnt pathways are the planar cell polarity (PCP) pathway and the Wnt-calcium (Wnt/Ca2+) pathway. Like the canonical pathway, these two non-canonical pathways are activated by the same initial events: the interaction of a Wnt ligand with its cognate receptor Frizzled followed by the activation of the cytoplasmic effector Dishevelled (Dsh) (9, 10). However, along with these three molecules, many additional proteins are involved, either to regulate the Wnt/Frizzled interaction or to transduce the signal to the nucleus, and it is the involvement of these various proteins that defines each pathway and provides specificity of function downstream of the individual Wnts. Of these three pathways (canonical, PCP, and calcium), the canonical pathway is the most broadly described throughout living organisms. Equivalent signals have been reported from protists to vertebrates, revealing both a molecular and functional conservation during eukaryotic evolution. In contrast, the PCP and the Wnt-calcium pathways have been less thoroughly studied, reducing the extent of the knowledge on their distribution among eukaryotes; investigations of these pathways have mainly been performed in bilaterian animals. The availability of many genomic sequences for living organisms other than bilaterians permits, however, some speculation on the breadth of the evolutionary distribution of these pathways and the molecular conservation of individual components.
Fig. 1.1.
Schematic eukaryotic phylogenetic representation. All organisms discussed in this volume of Methods in Molecular Biology are represented in this tree. Amoebozoa are protists; however, their relative position on the eukaryotic tree relative to plant and fungi remains controversial.
2. The Wnt Family
Wnt genes are generally defined by sequence homology. They are similar in size (350–400 amino acids in length) and provide characteristic structural features that allow their identification by BLAST sequence analysis. Each contains an amino terminal signal sequence followed by a highly conserved pattern of 22 to 24 cysteine residues essential for the function of the Wnts and the specificity of their responses (13, 14). As mentioned previously more than 100 Wnt genes have now been reported and phylogenetic analyses organize them into 13 subfamilies: Wnt-1 to Wnt-11, Wnt-16, and Wnt-A (Fig. 1.2). So far, no Wnt genes have been found in organisms other than animals (metazoans). Indeed, they appear to be absent from plants (including Arabidopsis thaliana), fungi (such as Saccharomyces cerevisiae), and protists (like Dictyostelium discoideum), although it should be noted that a transducing pathway related to the canonical Wnt pathway has been reported for Dictyostelium (see later). By contrast, in cnidarians, one of the most basal of the animal phyla, 14 Wnt genes have been identified that belong to 12 of the 13 Wnt subfamilies (15, 16); the Wnt-9 subfamily is not represented, while the Wnt-7 and Wnt-8 subfamilies have two paralogous genes (Fig. 1.2). How these Wnt proteins arose is still an outstanding question. One could propose the existence of a single ancestral animal Wnt protein that gave rise to all cnidarian Wnts through multiple rounds of duplication. However, taking into account the diversity of the cnidarian Wnts, the original duplications must have been ancient, perhaps prior to or linked with the emergence of multicellular animals. More ancient than cnidarians is the porifera phyla that includes animals such as sponges. Although no genome data is yet available from this phylum, EST data from the sponge Oscarella carmela revealed that four Wnt genes are expressed in this animal (17), supporting the idea that since the emergence of the metazoans multiple Wnts were present. Wnt genes seem therefore to be animal-specific, identified from the most basal phylum to the most complex, and within each phylum highly diverse. How this family has emerged in the first place is still mysterious, and more genome sequencing would therefore be necessary, especially from basal animals, to determine whether all metazoan Wnts derived from a single ancestral gene or multiple ones.
Fig. 1.2.
Distribution of Wnt proteins throughout the eukaryotic superkingdom. A square represents each gene found in the corresponding genome by protein subfamily. A 0 designates the absence of homologs of that subfamily in the corresponding genome. A question mark indicates orthology uncertainties. Genomes used are: amoebozoa, Dictyostelium discoideum; cnidarians, Nematostella vectensis; protostomes, Drosophila melanogaster and Caenorhabditis elegans (unless stipulated as coming from the mosquito Anopheles gambiae); echinoderms, Strongylocentrotus purpuratus; vertebrates, Homo sapiens (Adapted and Updated from ref. 12).
The distribution of the Wnt proteins within the bilaterian groups, compared to the cnidarian Wnts, reveals another interesting level in the complexity of the Wnt repertoire. First, whereas cnidarians possess 14 Wnts, in Drosophila and C. elegans only 7 and 5 Wnts have been identified, respectively (Fig. 1.2; ref. 18). Thus, if it is assumed that cnidarians reflect the repertoire inherited by the first bilaterians, then the protostomes have secondarily lost Wnt genes. This putative secondary loss is indeed supported by the identification of only 6 and 7 Wnts in mollusks and platyhelminthes, respectively (19), some of which belong to distinct subfamilies than the subfamilies present in Drosophila and C. elegans (Fig. 1.2 and ref. 12). However, full genomes of representatives of the lophotrochozoans (such as mollusks and platyhelminthes) have yet to be sequenced, implying that these conclusions might change once full data sets become available. Second, the Wnt gene family is generally represented by 13 subfamilies; however, an additional subfamily exists (noted Wnt-X in Fig. 1.2) that is ecdysozoan-specific. This subfamily is represented both in Drosophila and C. elegans by a unique member that currently does not have an identified homolog in cnidarian, lophotrochozoan, echinoderm, or vertebrate genomes (18). Third, in cnidarians and vertebrates, the Wnt-7 and Wnt-8 subfamilies contain two members, while other phyla, including arthropods, nematodes, mollusks, echinoderms, and cephalochordates, typically contain only one (Fig. 1.2 and ref. 12). This could be the result of two distinct causes: the individual losses of the Wnts in each of the intervening phyla, or, and most likely, duplication events that took place independently in cnidarians and vertebrates. Indeed, supporting the latter is the proposed model of a whole-genome duplication event that would have occurred at about the time of origin of vertebrates (20); although this notion is still debated (21, 22). Finally, in vertebrates, one of the 13 Wnt subfamilies, Wnt-A, is not represented (Fig. 1.2), and is actually absent from all chordates investigated including vertebrates, cephalochordates, and urochordates (12). In contrast, a Wnt-A homolog has been described in most of the other metazoan phyla from cnidarians to echinoderms (Fig. 1.2 and ref. 12). Neither the role nor the signaling pathway has yet been identified for any of these Wnt-A genes. However, because this subfamily appears to be non-chordate specific, its study might provide interesting insights into the passage from non-chordate to chordate animals.
Another way to classify Wnt genes could be by looking at their expression pattern and function throughout the animal kingdom. In terms of expression, individual and multi-gene studies have highlighted only few conserved patterns. In addition, in many animals several Wnts are expressed at the same time and sometimes in the same tissue during embryogenesis (12, 15). In contrast, experiments performed in Xenopus and mammalian cell culture have established that, based on their ability to induce a secondary body axis and on their oncogenic capacity, Wnt proteins can be grouped into two functional categories: those that induce axis duplication, display transforming activity, and act through the canonical Wnt pathway (Wnt-1, Wnt-3, Wnt-3a, Wnt-7a, Wnt-7b, Wnt-8), and those that do not induce axis duplication, do not have any oncogenic potential, and are activators of the non-canonical Wnt pathways (Wnt-4, Wnt-5a, Wnt-6, Wnt-11) (13, 23). In agreement with this, individual functional analyses performed in other metazoan embryos, such as sea urchin and zebrafish, corroborated these affiliations for some of these Wnts with the canonical or the non-canonical category (e.g., refs. 24 and 25). However, it is important to note that the ability of each of the Wnts to activate either the canonical or the non-canonical signals is not absolute. For example, in Xenopus, Wnt-11, which mediates non-canonical signaling (26, 27), also activates the canonical pathway during dorsal axis specification (28). Thus, while canonical and non-canonical signaling are distinct pathways with distinct roles, how the individual Wnt ligands use them to control their various signaling functions remains an unresolved issue.
3. The Canonical Wnt Pathway
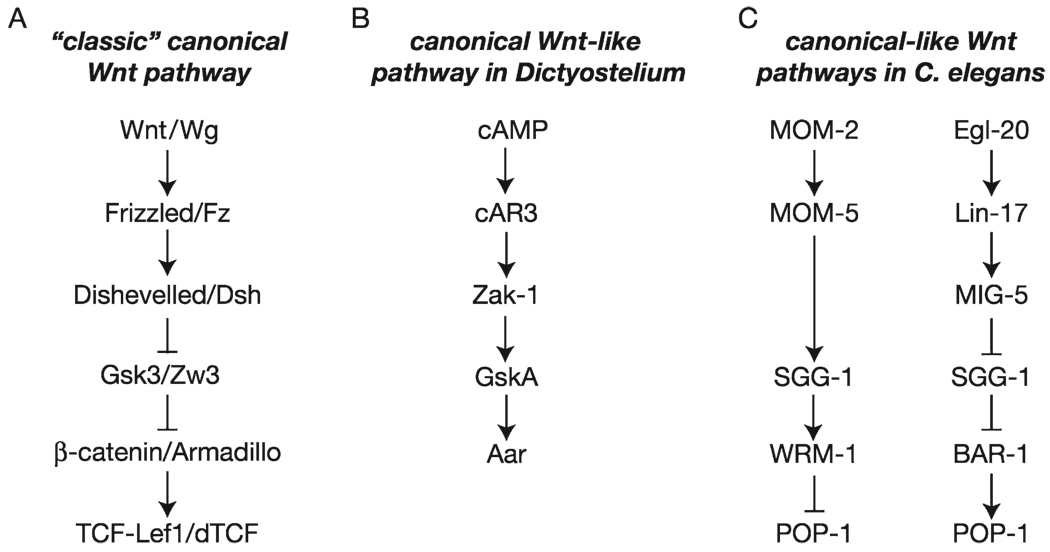
The canonical Wnt pathway is frequently represented by its six key components that include Wnt and Frizzled (the activators of the pathways), Dishevelled (Dsh) and GSK3 (the cytoplasmic transducer), and β-catenin and TCF/Lef (the nuclear factors). As described in many textbooks and observed in all animals studied, the canonical Wnt pathway is activated by the binding of a Wnt ligand to a Frizzled receptor. That interaction triggers the cytoplasmic effector Dsh that ultimately blocks the antagonistic activity of GSK3, which normally phosphorylates β-catenin and targets it for degradation. Preventing the degradation of β-catenin leads to an increase in its cytoplasmic concentration, and its translocation into the nucleus, where it interacts with a TCF/Lef factor and activates transcription of the Wnt target genes (Fig. 1.3A). In the past two decades, searches for homologs of these six main components have identified related canonical Wnt pathways in many organisms ranging from Dictyostelium discoideum to Homo sapiens (Table 1.1).
Fig. 1.3.
Comparison of canonical Wnt signal transduction models among eukaryotes. (A) In most bilaterians, the canonical Wnt pathway is activated by a Wnt/Frizzled interaction, which results in the activation of the cytoplasmic effector Dsh that in turn inhibits the antagonistic GSK3, releasing β-catenin, which now activates TCF/Lef. (B) In the protist Dictyostelium discoideum, GSK3 and β-catenin homologs, GskA and Aar respectively, are activated by the binding of cAMP to its receptor cAR3. This interaction leads to the activation of GskA, which then phosphorylates and activates Aar. (C) In C. elegans, two Wnt pathways involving GSK3 and two distinct β-catenin homologs are observed. While the Wnt/BAR-1 pathway evolved as the classic bilaterians canonical Wnt pathway (right), the Wnt/WRM-1 pathway (left) is similar to the GskA/Aar pathway described in Dictyostelium. Wnt (MOM-2)/Frizzled (MOM-5) interaction induces the activation of the GSK3 homolog SGG-1, which activates WRM-1 by phosphorylation that in turn inhibits the TCF/Lef homolog POP-1 activity. Arrows within this figure do not necessarily represent direct interaction between the molecules.
Table 1.1.
Conservation of the main components of the canonical Wnt pathway within eukaryotes 1
| Amoebozoa | Cnidarians | Protostomes | Deuterostomes | |
|---|---|---|---|---|
| Dsh | o | x | x | x |
| GSK3 | x | x | x | x |
| β-catenin | x | x | x | x |
| TCF | o | x | x | x |
x indicates the presence of a vertebrate gene homolog in the corresponding genomes, while o indicates the absence of such a homolog (amoebozoa, Dictyostelium discoideum; cnidarians, Nematostella vectensis; protostomes, Drosophila melanogaster and Caenorhabditis elegans; deuterostomes, Strongylocentrotus purpuratus and Homo sapiens).
In most plants, fungi, and protists, no Wnts or Frizzled genes have been found (for detail on the evolutionary history of Wnts and Frizzled genes in metazoans see ref. 12), supporting the idea that Wnt pathways per se are animal-specific. In contrast, homologs of GSK3 and β-catenin have been identified in all eukaryotes, including in the protist Dictyostelium discoideum (Table 1.1) where they are both acting in a related canonical Wnt-like pathway (29). In this slime mold, GskA and Aar, the GSK3 and β-catenin homologs, respectively, are not activated by a Wnt ligand but by cyclic AMP (cAMP). cAMP binds to its cognate receptor cAR3, which leads to the activation of the tyrosine kinase Zak-1. Zak-1 then phosphorylates and activates GskA/GSK3, which in turn phosphorylates Aar/β-catenin, leading to gene transcription (Fig. 1.3B). How Aar/β-catenin induces gene transcription is still unknown: whether it works by itself, by interaction with another factor, or by inhibition of another factor has yet to be elucidated; no TCF/Lef homolog has been identified in the Dictyostelium genome (Table 1.1). Nevertheless, this pathway involves the same proteins, GSK3 and β-catenin, for a similar outcome, the control of gene transcription. Therefore, although no Wnt or Fizzled homologs have been identified in the Dictyostelium genome, key components of the canonical pathway are present and functionally linked in this molecular pathway, indicating that whereas the signal per se might have arisen with the emergence of the metazoans, components of the downstream pathway already existed.
Additionally, in contrast to the “classic/textbook” canonical Wnt pathway, activation of this cAMP signal induces activation of GskA, rather than inhibition, and GskA activates Aar/β-catenin, instead of inducing its degradation. These relationships might appear surprising although they are reminiscent of those observed in a “non-canonical” Wnt pathway described in the nematode C. elegans that involves the β-catenin homolog WRM-1. In this pathway, signaling is initiated by homologs of Wnt (MOM-2) and Frizzled (MOM-5). Then, analogous to the situation in Dictyostelium, receptor ligation directly activates the GSK3 homolog (SGG-1) that, independently of Dsh (which is absent from the pathways in both organisms) activates WRM-1/β-catenin by phosphorylation. In turn, WRM-1/β-catenin inhibits the TCF/Lef homolog POP-1, relieving the transcriptional repression of the Wnt/WRM-1 target genes (Fig. 1.3C). Thus, although components of this Wnt pathway are similar to those used by the classic canonical pathway, their relationships to one another are functionally different. Therefore, this raises the interesting possibility that this signal transduction mechanism observed in both Dictyostelium and in C. elegans might actually represent the ancient form of the textbook canonical Wnt pathway observed in all metazoans from cnidarians to vertebrates. Intriguingly, in C. elegans a classic canonical pathway is also present. Unlike most eukaryotes, C. elegans possesses three distinct β-catenin homologs, one of which (HMB-2) is dedicated to cell adhesion, while the other two participate in gene regulation. WRM-1 is involved in the pathway described above, while BAR-1 acts in the classic canonical pathway (Fig. 1.3C; refs. 30 and 31). Therefore, in C. elegans the potentially ancient GSK3- β-catenin functional relationship coexists alongside the modern, textbook relationship.
In a few model organisms, mostly bilaterians, studies on the composition of the canonical Wnt pathway have been explored further to understand the detailed mechanism of this signaling pathway. The motivation of these studies was primarily to identify potential ways to treat cancer and other human diseases associated with the inappropriate activation of this pathway, and secondarily to assess the level of conservation of the pathway between these model systems. Multiple functional analyses performed in vertebrate embryos have led to the identification of a large number of secondary components of the canonical Wnt pathway acting from the extracellular compartment to the nucleus (18). In Drosophila, a high-throughput screen also reported 238 genes involved in regulating the canonical Wnt signal (32). These genes encode for various protein types ranging from membrane-associated proteins to kinases and transcription factors, while a third of them are of unknown function; for many, their involvement has been corroborated by functional analyses (32). In addition, sequence comparisons with human genome databases indicated that 50% of these Drosophila genes have human orthologs (18% of which are associated with human disease) (32). Finally in sea urchin, an invertebrate deuterostome, a large-scale analysis carried out on the genome showed that 88% of the molecules examined (a subset of the known vertebrate components) were conserved between sea urchins and humans (12). Thus, at a molecular level, the canonical Wnt pathway appears to be a highly conserved pathway among metazoans, this conservation extending beyond the simple group of its six key components. Not surprisingly, this level of conservation is lower between protostomes and deuterostomes (e.g. Drosophila-humans) than it is among deuterostome organisms (e.g. sea urchin-humans). However, in both cases this assessment simply measures the conservation of the individual components, rather than the conservation of their genetic relationships or functions. Thus, much more remains to be learned to determine the overall conservation of the complete pathway relative to the simplified version discussed previously.
The canonical Wnt pathway is required in many developmental processes, some of which are particularly well-conserved during animal development, such as blastopore determination and endomesoderm specification. However, the canonical pathway also plays a key role during an earlier embryonic event, axis formation, and this role appears to be the most well-conserved and ancient of its various functions. In Dictyostelium, the canonical Wnt-like pathway (cAMP/GskA) is required for the regulation of the antero-posterior asymmetry of the multicellular slime mold. Active in the posterior region, the cAMP/GskA pathway promotes posterior cell fates leading to the establishment of the longitudinal body axis (33), suggesting that the axis specification function of the canonical pathway may predate the invention of Wnt ligands. Similarly, in all metazoans, from cnidarians to vertebrates, the Wnt/β-catenin pathway controls the establishment of either the antero-posterior, animal-vegetal, or dorso-ventral axis depending on the model system studied (34). Thus, although the protist canonical Wnt-like pathway is molecularly different from the classic canonical Wnt pathway, they are each essential to promote cell fates and establish body axes. Therefore, since this function is observed from protists to vertebrates, it seems to be the most ancient and perhaps the primordial role of the pathway. Interestingly, in Dictyostelium the cAMP/GskA pathway is only required during the multicellular stages of the aggregating slime mold, and not during their unicellular development (35). This exemplifies, therefore, the general assumption that the canonical pathway functions only in the context of multicellularity, suggesting that it may have arisen in particular to facilitate intercellular communication.
4. The Non-canonical Wnt Pathways
The two most studied of the non-canonical Wnt pathways operate through signaling mechanisms that do not involve GSK3 or β-catenin and do not generally regulate cell fate specification in early development, but instead are essential to control morphogenetic cell movements. These pathways are the planar cell polarity (PCP) pathway and the Wnt-calcium (Wnt/Ca2+) pathway. The cell polarity pathway has been named as such because of its function in establishing cell polarity, but it also controls convergent extension movements of cells, which are movements that require polarity guidance for directed displacement. In contrast, the Wnt/Ca2+ pathway mediates cytoskeletal dynamics and cell adhesion, through the regulation of intracellular calcium levels. Although the non-canonical Wnt pathways have been characterized in less detail than the canonical pathway, they nevertheless are both defined by a distinct set of proteins that distinguish the PCP and the Wnt-calcium pathways from one another and from the canonical Wnt pathway.
The PCP pathway involves six core genes aside from Wnt, Fz, and Dsh: the transmembrane Frizzled co-receptor Strabismus (or Van Gogh), the cytoplasmic Strabismus partner Prickle, the cytoplasmic small G proteins RhoA and Rac activated by Dsh, and their respective effectors ROCK (Rho-kinase) and JNK (Jun kinase) (Fig. 1.4). Based on published data and examination of genomes databases, all metazoan model systems analyzed from cnidarians to vertebrates possess a homolog for each of these factors (Table 1.2). Thus, like the canonical Wnt pathway, the PCP pathway appears well-conserved throughout animal evolution. In support, in the sea urchin genome 93% of the 29 molecules presently known to participate in the regulation and the transduction of the PCP pathway are conserved between humans and sea urchins (12). However, in the non-metazoan Dictyostelium genome, Strabismus and Rac were the only PCP components identified (Table 1.2). Similar to GskA and Aar/β-catenin, it is possible that these two molecules act in a pathway that is divergent from the PCP pathway but that has a comparative functional outcome; an hypothesis that has yet to be tested.
Fig. 1.4.
Schematic representation of the two most studied non-canonical pathways. (A) The Planar Cell Polarity pathway. Upon interaction of a Wnt ligand with a Frizzled receptor and activation of the cytoplasmic protein Disheveled, the signal is transduced through two distinct small G proteins Rho and Rac, which convey cell polarity via their respective effectors ROCK and JNK. To establish polarity, however, the membrane receptor Strabismus is also activated but in the neighboring cell (in gray), where via Prickle it inhibits the PCP pathway by sequestering Disheveled. (B) The Wnt/Ca2+ pathway. Activation of the Wnt/calcium pathway leads to the activation of the key component PLC and an increase in intracellular calcium levels, which induces activation of the calcium-sensitive proteins Calcineurin, CamKII, and PKC.
Table 1.2.
Conservation of the main components of the non-canonical Wnt pathways within eukaryotes1
| Amoebozoa | Cnidarians | Protostomes | Deuterostomes | ||
|---|---|---|---|---|---|
| PCP pathway | Strabismus | x | x | x | x |
| Prickle | o | x | x | x | |
| RhoA | o | x | x | x | |
| ROCK | o | x | x | x | |
| Rac | x | x | x | x | |
| JNK | o | x | x | x | |
| Ca2+ pathway | PLC | x | x | x | x |
| PKC | o | x | x | x | |
| CamKII | o | x | x | x | |
| Calcineurin | x | x | x | x | |
x indicates the presence of a vertebrate gene homolog in the corresponding genomes, while o indicates the absence of such a homolog (amoebozoa, Dictyostelium discoideum; cnidarians, Nematostella vectensis; protostomes, Drosophila melanogaster and Caenorhabditis elegans; deuterostomes, Strongylocentrotus purpuratus and Homo sapiens).
The PCP pathway was first discovered in Drosophila where it controls epithelial planar polarity within the eye, wing, and thorax (36, 37). In the eye, this pathway controls the orientation of the ommatidia, while in the wing and thorax it regulates the pattern of setae. In vertebrates, an equivalent genetic pathway was subsequently described that plays an essential role in patterning the fur and the sensory hairs of the inner ear (38, 39). Further, in vertebrates, the PCP pathway also controls convergent extension movements associated with gastrulation and neurulation (40). Convergent extension (CE) movements are polarized rearrangements of cells within a given tissue along an axis (reviewed in ref. 41). In CE movements, cells intercalate and elongate, such that a tissue is narrowed in one direction and elongated in the perpendicular direction. Thus, whether associated with ommatidia, hair patterning, or CE movements, the PCP pathway consistently controls cell polarization, underlying a functional conservation of the pathway between protostomes and vertebrates. Reinforcing this idea, analyses performed on some of the PCP pathway components (Fz5/8, RhoA and Prickle) in sea urchins and ascidians demonstrated the role of this pathway in the control of CE movements in these animals (42–44). Further, in C. elegans the PCP pathway is involved in regulating the asymmetric division of the B cell (which gives rise to the proctodeum of the larvae), thereby conveying cell polarity at another level (45). Thus, all these studies indicate that the PCP pathway is well-conserved, not only at a molecular level, but also at functional level, at least throughout bilaterians. It will now be interesting, therefore, to determine the role of this pathway in more basal animals such as cnidarians, which have all the main PCP components (Table 1.2), although their functions have yet to be investigated.
The Wnt/Ca2+ pathway acts through heterotrimeric G-proteins to activate phospholipase C (PLC), which leads to the release of intracellular calcium. This increase in Ca2+ activates three calcium sensitive components: PKC (protein kinase C), calcineurin, and CamKII (calcium/calmodulin-dependent kinase II) (Table 1.2; for review, see refs. 46 and 47). The Wnt/Ca2+ pathway was discovered in Xenopus and zebrafish (48, 49), but no equivalent pathways have yet been described in any other model system, raising the possibility that the Wnt-calcium signaling is a vertebrate-specific pathway. Alternatively, protein homologs of all key components of the Wnt-calcium pathway have been reported in a number of metazoan model systems (Cnidarians, sea urchin, Drosophila, and C.elegans; Table 1.2), but whether these molecules act together in a related Wnt-activated pathway has yet to be determined. Indeed PLC, PKC, calcineurin, and CamKII are generic proteins involved in many transduction pathways that play important roles in various physiological processes. Their presence in these more basal metazoans could therefore be due to the conservation of these other functions and not their involvement within a particular Wnt pathway, which then could have arisen as a vertebrate-specific acquisition resulting from the co-option of currently available signaling proteins. Supporting this contention, PLC and calcineurin homologs are present in protists, plants, and fungi, although there is no evidence for any Wnt-like pathway in these groups (other than the similarities alluded to earlier in Dictyostelium). In contrast, Cook and his collaborators reported in 1996 the existence of a Wingless–PKC pathway in Drosophila (50), an observation that provides initial support for a potentially broader distribution of the Wnt/Ca2+ pathway within metazoans.
5. Conclusions
The three Wnt pathways discussed previously are involved in many aspects of embryonic developmental events, controlling cell fate, proliferation, polarity, migration, and adhesion. As a result, disruption of their functions causes drastic consequences ranging from tumorigenesis to embryonic lethality. These Wnt pathways have been studied in diverse living organisms allowing us to describe them from an evolutionary prospective. As an overview, all three pathways seem to have emerged with the metazoans, although with regard to canonical signaling, a related pathway is present in the protist Dictyostelium. Furthermore, based on the identification of individual components of the three Wnt pathways, these pathways appear to be well-conserved among all studied metazoans, from cnidarians to vertebrates, perhaps with the exception of the Wnt-calcium pathway for which the data is still limited. Therefore, because of this important molecular and functional conservation of the Wnt pathways throughout the animal kingdom, their studies across divergent animal phyla add to the richness of our knowledge, although additional investigations will be necessary to determine whether this conservation can be extended to all components, as well as to all of their molecular relationships. Nevertheless, reports currently appearing at high frequency concerning the role and the composition of these pathways in non-vertebrate metazoans (cnidarian, Drosophila, C. elegans, and sea urchin) are likely to have functional relevant parallels in vertebrates. The synthesis of all these results will lead therefore to a better understanding of the origin of the pathways and the basis of the diseases associated with their malfunction.
Acknowledgements
The authors thank Dr. Cynthia Bradham for critical evaluation of the manuscript. J. C. thanks Athula Wikramanayake for allowing me the opportunity to contribute to this book. This work was supported by grants NIH 61464 and HD 14483.
References
- 1.Sharma RP, Chopra VL. Effect of the Wingless (wg1) mutation on wing and haltere development in Drosophila melanogaster. Dev Biol. 1976;48:461–465. doi: 10.1016/0012-1606(76)90108-1. [DOI] [PubMed] [Google Scholar]
- 2.Baker NE. Molecular cloning of sequences from wingless, a segment polarity gene in Drosophila: the spatial distribution of a transcript in embryos. Embo J. 1987;6:1765–1773. doi: 10.1002/j.1460-2075.1987.tb02429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nusse R, Varmus HE. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982;31:99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- 4.Van Ooyen A, Nusse R. Structure and nucleotide sequence of the putative mammary oncogene int-1; proviral insertions leave the protein-encoding domain intact. Cell. 1984;39:233–240. doi: 10.1016/0092-8674(84)90209-5. [DOI] [PubMed] [Google Scholar]
- 5.Rijsewijk F, Schuermann M, Wagenaar E, Parren P, Weigel D, Nusse R. The Drosophila homolog of the mouse mammary oncogene int-1 is identical to the segment polarity gene wingless. Cell. 1987;50:649–657. doi: 10.1016/0092-8674(87)90038-9. [DOI] [PubMed] [Google Scholar]
- 6.Dale TC. Signal transduction by the Wnt family of ligands. Biochem J. 1998;329:209–223. doi: 10.1042/bj3290209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huelsken J, Birchmeier W. New aspects of Wnt signaling pathways in higher vertebrates. Curr Opin Genet Dev. 2001;11:547–553. doi: 10.1016/s0959-437x(00)00231-8. [DOI] [PubMed] [Google Scholar]
- 8.Korswagen HC. Canonical and non-canonical Wnt signaling pathways in Caenorhabditis elegans: variations on a common signaling theme. Bioessays. 2002;24:801–810. doi: 10.1002/bies.10145. [DOI] [PubMed] [Google Scholar]
- 9.Wharton KA., Jr Runnin’ with the Dvl: proteins that associate with Dsh/Dvl and their significance to Wnt signal transduction. Dev Biol. 2003;253:1–17. doi: 10.1006/dbio.2002.0869. [DOI] [PubMed] [Google Scholar]
- 10.Sheldahl LC, Slusarski DC, Pandur P, Miller JR, Kuhl M, Moon RT. Dishevelled activates Ca2+ flux, PKC, and CamKII in vertebrate embryos. J Cell Biol. 2003;161:769–777. doi: 10.1083/jcb.200211094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huelsken J, Behrens J. The Wnt signaling pathway. J Cell Sci. 2002;115:3977–3978. doi: 10.1242/jcs.00089. [DOI] [PubMed] [Google Scholar]
- 12.Croce JC, Wu SY, Byrum C, Xu R, Duloquin L, Wikramanayake AH, Gache C, McClay DR. A genome-wide survey of the evolutionarily conserved Wnt pathways in the sea urchin Strongylocentrotus purpuratus. Dev Biol. 2006;300:121–131. doi: 10.1016/j.ydbio.2006.08.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du SJ, Purcell SM, Christian JL, McGrew LL, Moon RT. Identification of distinct classes and functional domains of Wnts through expression of wild-type and chimeric proteins in Xenopus embryos. Mol Cell Biol. 1995;15:2625–2634. doi: 10.1128/mcb.15.5.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Ooyen A, Kwee V, Nusse R. The nucleotide sequence of the human int-1 mammary oncogene; evolutionary conservation of coding and non-coding sequences. Embo J. 1985;4:2905–2909. doi: 10.1002/j.1460-2075.1985.tb04021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kusserow A, Pang K, Sturm C, Hrouda M, Lentfer J, Schmidt HA, Technau U, von Haeseler A, Hobmayer B, Martindale MQ, Holstein TW. Unexpected complexity of the Wnt gene family in a sea anemone. Nature. 2005;433:156–160. doi: 10.1038/nature03158. [DOI] [PubMed] [Google Scholar]
- 16.Lee PN, Pang K, Matus DQ, Martindale MQ. A WNT of things to come: Evolution of Wnt signaling and polarity in cnidarians. Semin Cell Dev Biol. 2006;17:157–167. doi: 10.1016/j.semcdb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Nichols SA, Dirks W, Pearse JS, King N. Early evolution of animal cell signaling and adhesion genes. Proc Natl Acad Sci USA. 2006;103:12451–12456. doi: 10.1073/pnas.0604065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The Wnt homepage. Available at: www.stanford.edu/~rnusse/wntwindow.html.
- 19.Prud’homme B, Lartillot N, Balavoine G, Adoutte A, Vervoort M. Phylogenetic analysis of the Wnt gene family. Insights from lophotrochozoan members. Curr Biol. 2002;12:1395. doi: 10.1016/s0960-9822(02)01068-0. [DOI] [PubMed] [Google Scholar]
- 20.Friedman R, Hughes AL. The temporal distribution of gene duplication events in a set of highly conserved human gene families. Mol Biol Evol. 2003;20:154–161. doi: 10.1093/molbev/msg017. [DOI] [PubMed] [Google Scholar]
- 21.Furlong RF, Holland PW. Polyploidy in vertebrate ancestry: Ohno and beyond. Biological Journal of the Linnean Society. 2004;82:425–430. [Google Scholar]
- 22.Holland PW, Garcia-Fernandez J, Williams NA, Sidow A. Gene duplications and the origins of vertebrate development. Dev Suppl. 1994:125–133. [PubMed] [Google Scholar]
- 23.Wong GT, Gavin BJ, McMahon AP. Differential transformation of mammary epithelial cells by Wnt genes. Mol Cell Biol. 1994;14:6278–6286. doi: 10.1128/mcb.14.9.6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kilian B, Mansukoski H, Barbosa FC, Ulrich F, Tada M, Heisenberg CP. The role of Ppt/Wnt5 in regulating cell shape and movement during zebrafish gastrulation. Mech Dev. 2003;120:467–476. doi: 10.1016/s0925-4773(03)00004-2. [DOI] [PubMed] [Google Scholar]
- 25.Wikramanayake AH, Peterson R, Chen J, Huang L, Bince JM, McClay DR, Klein WH. Nuclear beta-catenin-dependent Wnt8 signaling in vegetal cells of the early sea urchin embryo regulates gastrulation and differentiation of endoderm and mesodermal cell lineages. Genesis. 2004;39:194–205. doi: 10.1002/gene.20045. [DOI] [PubMed] [Google Scholar]
- 26.Tada M, Smith JC. Xwnt11 is a target of Xenopus Brachyury: regulation of gastrulation movements via Dishevelled, but not through the canonical Wnt pathway. Development. 2000;127:2227–2238. doi: 10.1242/dev.127.10.2227. [DOI] [PubMed] [Google Scholar]
- 27.Heisenberg CP, Tada M, Rauch GJ, Saude L, Concha ML, Geisler R, Stemple DL, Smith JC, Wilson SW. Silber-blick/Wnt11 mediates convergent extension movements during zebrafish gastrulation. Nature. 2000;405:76–81. doi: 10.1038/35011068. [DOI] [PubMed] [Google Scholar]
- 28.Tao Q, Yokota C, Puck H, Kofron M, Birsoy B, Yan D, Asashima M, Wylie CC, Lin X, Heasman J. Maternal wnt11 activates the canonical wnt signaling pathway required for axis formation in Xenopus embryos. Cell. 2005;120:857–871. doi: 10.1016/j.cell.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 29.Coates JC, Harwood AJ. Cell-cell adhesion and signal transduction during Dictyostelium development. J Cell Sci. 2001;114:4349–4358. doi: 10.1242/jcs.114.24.4349. [DOI] [PubMed] [Google Scholar]
- 30.Ruvkun G, Hobert O. The taxonomy of developmental control in Caenorhabditis elegans. Science. 1998;282:2033–2041. doi: 10.1126/science.282.5396.2033. [DOI] [PubMed] [Google Scholar]
- 31.Korswagen HC, Herman MA, Clevers HC. Distinct beta-catenins mediate adhesion and signaling functions in C. elegans. Nature. 2000;406:527–532. doi: 10.1038/35020099. [DOI] [PubMed] [Google Scholar]
- 32.DasGupta R, Kaykas A, Moon RT, Perrimon N. Functional genomic analysis of the Wnt-wingless signaling pathway. Science. 2005;308:826–833. doi: 10.1126/science.1109374. [DOI] [PubMed] [Google Scholar]
- 33.Harwood AJ, Plyte SE, Woodgett J, Strutt H, Kay RR. Glycogen synthase kinase 3 regulates cell fate in Dictyostelium. Cell. 1995;80:139–148. doi: 10.1016/0092-8674(95)90458-1. [DOI] [PubMed] [Google Scholar]
- 34.Croce JC, McClay DR. The canonical Wnt pathway in embryonic axis polarity. Semin Cell Dev Biol. 2006;17:168–174. doi: 10.1016/j.semcdb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 35.Plyte SE, O’Donovan E, Woodgett JR, Harwood AJ. Glycogen synthase kinase-3 (GSK-3) is regulated during Dictyostelium development via the serpentine receptor cAR3. Development. 1999;126:325–333. doi: 10.1242/dev.126.2.325. [DOI] [PubMed] [Google Scholar]
- 36.Mlodzik M. Planar polarity in the Drosophila eye: a multifaceted view of signaling specificity and cross-talk. Embo J. 1999;18:6873–6879. doi: 10.1093/emboj/18.24.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adler PN, Lee H. Frizzled signaling and cell-cell interactions in planar polarity. Curr Opin Cell Biol. 2001;13:635–640. doi: 10.1016/s0955-0674(00)00263-5. [DOI] [PubMed] [Google Scholar]
- 38.Guo N, Hawkins C, Nathans J. Frizzled6 controls hair patterning in mice. Proc Natl Acad Sci USA. 2004;101:9277–9281. doi: 10.1073/pnas.0402802101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dabdoub A, Kelley MW. Planar cell polarity and a potential role for a Wnt morphogen gradient in stereociliary bundle orientation in the mammalian inner ear. J Neurobiol. 2005;64:446–457. doi: 10.1002/neu.20171. [DOI] [PubMed] [Google Scholar]
- 40.Solnica-Krezel L. Conserved patterns of cell movements during vertebrate gastrulation. Curr Biol. 2005;15:R213–R228. doi: 10.1016/j.cub.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 41.Mlodzik M. Planar cell polarization: do the same mechanisms regulate Drosophila tissue polarity and vertebrate gastrulation? Trends Genet. 2002;18:564–571. doi: 10.1016/s0168-9525(02)02770-1. [DOI] [PubMed] [Google Scholar]
- 42.Croce J, Duloquin L, Lhomond G, McClay DR, Gache C. Frizzled5/8 is required in secondary mesenchyme cells to initiate archenteron invagination during sea urchin development. Development. 2006;133:547–557. doi: 10.1242/dev.02218. [DOI] [PubMed] [Google Scholar]
- 43.Beane WS, Gross JM, McClay DR. RhoA regulates initiation of invagination, but not convergent extension, during sea urchin gastrulation. Dev Biol. 2006;292:213–225. doi: 10.1016/j.ydbio.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 44.Jiang D, Munro EM, Smith WC. Ascidian prickle regulates both mediolateral and anterior-posterior cell polarity of notochord cells. Curr Biol. 2005;15:79–85. doi: 10.1016/j.cub.2004.12.041. [DOI] [PubMed] [Google Scholar]
- 45.Wu M, Herman MA. A novel noncanonical Wnt pathway is involved in the regulation of the asymmetric B cell division in C. elegans. Dev Biol. 2006;293:316–329. doi: 10.1016/j.ydbio.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 46.Kühl M, Sheldahl LC, Park M, Miller JR, Moon RT. The Wnt/Ca2+ pathway: a new vertebrate Wnt signaling pathway takes shape. Trends Genet. 2000;16:279–283. doi: 10.1016/s0168-9525(00)02028-x. [DOI] [PubMed] [Google Scholar]
- 47.Pandur P, Maurus D, Kuhl M. Increasingly complex: new players enter the Wnt signaling network. Bioessays. 2002;24:881–884. doi: 10.1002/bies.10164. [DOI] [PubMed] [Google Scholar]
- 48.Kühl M, Sheldahl LC, Malbon CC, Moon RT. Ca(2+)/calmodulin-dependent protein kinase II is stimulated by Wnt and Frizzled homologs and promotes ventral cell fates in Xenopus. J Biol Chem. 2000;275:12701–12711. doi: 10.1074/jbc.275.17.12701. [DOI] [PubMed] [Google Scholar]
- 49.Sheldahl LC, Park M, Malbon CC, Moon RT. Protein kinase C is differentially stimulated by Wnt and Frizzled homologs in a G-protein-dependent manner. Curr Biol. 1999;9:695–698. doi: 10.1016/s0960-9822(99)80310-8. [DOI] [PubMed] [Google Scholar]
- 50.Cook D, Fry MJ, Hughes K, Sumathipala R, Woodgett JR, Dale TC. Wingless inactivates glycogen synthase kinase-3 via an intracellular signaling pathway which involves a protein kinase C. Embo J. 1996;15:4526–4536. [PMC free article] [PubMed] [Google Scholar]