Abstract
Background
We examine the potential prognostic and predictive roles of EGFR variant III mutation, EGFR gene copy number (GCN), human papillomavirus (HPV) infection, c-MET and p16INK4A protein expression in recurrent or metastatic squamous cell carcinoma of the head and neck (R/M SCCHN).
Methods
We analyzed the archival tumor specimens of 53 patients who were treated in 4 phase II trials for R/M SCCHN. Two trials involved the EGFR inhibitor erlotinib, and 2 trials involved non-EGFR targeted agents. EGFRvIII mutation was determined by quantitative RT-PCR, HPV DNA by Linear Array Genotyping, p16 and c-MET protein expression by immunohistochemistry, and EGFR GCN by FISH.
Results
EGFRvIII mutation, detected in 22 patients (42%), was associated with better disease control, but no difference was seen between erlotinib-treated versus non-erlotinib treated patients. EGFRvIII was not associated with TTP or OS. The presence of HPV DNA (38%), p16 immunostaining (32%), c-MET high expression (58%) and EGFR amplification (27%), were not associated with response, TTP or OS.
Conclusion
EGFRvIII mutation, present in about 40% of SCCHN, appears to be an unexpected prognostic biomarker associated with better disease control in R/M SCCHN regardless of treatment with erlotinib. Larger prospective studies are required to validate its significance.
Background
The epidermal growth factor receptor (EGFR) is over-expressed in up to 90% of squamous cell carcinoma of the head and neck (SCCHN) and has been postulated to be a key molecular target in this malignancy [1]. EGFR signal transduction leads to cell proliferation, invasion, angiogenesis and metastasis [2]. EGFR overexpression and aberrant EGFR gene copy number (EGFR GCN) have been associated with poorer prognosis and disease-specific survival in SCCHN [1,3,4]. Therapies targeted against EGFR have demonstrated modest activity either alone or in combination with chemotherapy in both locally advanced [5] and recurrent and/or metastatic SCCHN [6-10]. No validated biomarkers exist to predict the response to EGFR inhibitors in SCCHN.
The most common EGFR truncation mutation, EGFR variant III (EGFRvIII), harbors an in-frame deletion of exons 2 to 7 (801 bp), resulting in a truncated extracellular EGF-binding domain that is constitutively activated and ineffectively ubiquinated [11,12]. EGFRvIII is found in many human cancers and is present in ~40% of glioblastomas and 5% of lung squamous cell carcinomas, where it confers tumorigenicity and dose-dependent resistance to gefitinib in pre-clinical models [13,14]. The prevalence of EGFRvIII in SCCHN was first reported as 43% in one study of 33 SCCHN tumors [15]. EGFRvIII-transfected SCCHN cells had decreased apoptosis in response to cisplatin and decreased growth inhibition following treatment with the EGFR monoclonal antibody cetuximab compared with controls [15]. EGFRvIII is an interesting therapeutic target because unlike wild-type EGFR, EGFRvIII is not found in normal tissue. EGFRvIII is proposed to account for limitations in response to current EGFR inhibitors, however in patients with SCCHN tumors harboring EGFRvIII response to EGFR tyrosine kinase inhibition (TKI) is unknown.
HPV infection is a risk factor for the development of SCCHN. HPV DNA is found in 20-30% of SCCHN and up to 40-66% of SCCHN of the oropharynx [16,17]. HPV positive oropharyngeal tumors are clinically and molecularly distinct from HPV negative tumors [18,19] and associated with a more favorable prognosis [20]. HPV positive status prospectively predicts survival and response to induction chemotherapy and chemoradiation in stage III or IV oropharynx cancers [21,22] and better response to radiotherapy alone [23]. The combination of low HPV titers and high EGFR expression was associated with worse overall survival in oropharynx cancer [22]. Inactivation of pRb by HPV E7 protein results in overexpression of p16 protein, thus p16 immunostaining has served as a surrogate marker for HPV-associated SCCHN. Patients with tumors lacking both p16 expression and HPV (p16-/HPV-) had the worst disease-specific survival compared to tumors with p16+/HPV+, p16-/HPV+ or p16+/HPV- types [24]. Despite the importance of HPV in the pathogenesis and prognosis of SCCHN in response to chemotherapy and radiation, the role of HPV DNA and response to EGFR inhibitors in SCCHN is unclear.
c-MET, a proto-oncogene tyrosine kinase receptor, is overexpressed in SCCHN, and its ligand, hepatocyte growth factor (HGF), stimulates cell proliferation, motility and invasion [25]. c-MET overexpression has been associated with disease progression in oral squamous cell carcinoma (OSCC) [26]. Elevated serum HGF is associated with resistance to chemoradiation and reduced survival [27]. c-MET amplification and mutations of MET confer an invasive phenotype associated with metastases in SCCHN [28]. Ligand-independent constitutive activation of c-MET via its heterodimerization with EGFR has been identified as a contributing mechanism of acquired resistance to cetuximab in SCCHN [29]. The role of c-MET in response to EGFR TKI in the clinical setting in SCCHN is unknown.
In this study, we examine the prevalence of EGFRvIII, HPV, p16, c-MET and EGFR GCN in patients with R/M SCCHN and explore the potential prognostic and predictive roles of these biomarkers in patients treated with or without EGFR TKI. We hypothesized that EGFRvIII and c-MET would be associated with poorer prognosis or response to EGFR TKI, while HPV and p16 expression would predict improved clinical outcomes and response to treatment.
Methods
Patients
We obtained approval from the University Health Network Research Ethics Board to evaluate the archival formalin-fixed paraffin embedded (FFPE) tumor specimens of patients with R/M SCCHN who were treated in four phase II trials for R/M SCCHN at Princess Margaret Hospital conducted from 2000-2005. Two of the four trials involved the EGFR TKI erlotinib (phase II trial of erlotinib [8], phase II trial of erlotinib and cisplatin [7]) and the remaining two trials used other non-EGFR targeted agents (phase II trial of the kinesin spindle protein inhibitor ispinesib [30], phase II trial of the multi-kinase antiangiogenic inhibitor sorafenib [31]). The medical records and case report forms were reviewed to obtain patient demographics, primary tumor site, treatment details and clinical outcome (response rate, time to progression and overall survival).
Specimen Characteristics
Archival FFPE tumor specimens were available in 35 of 48 patients (73%) treated with erlotinib and 18 of 37 (49%) patients treated with non-EGFR targeted agents. H&E stained sections were examined by a histopathologist (B.P-O.) to confirm the presence of >80% tumor in the specimens evaluated.
Assay Methods/Molecular Assays
EGFRvIII Mutation Detection
RNA Isolation
RNA was isolated in tumor area on the FFPE slides guided by H&E-stained serial sections. The tissues were deparaffinized by xylene and ethanol. Total RNA from paraffin-embedded tissues was extracted using RecoveryAll™ Total Nucleic Acid isolation Kit (Ambion Diagnostics, Streetsville, Ontario, Canada).
Real-time RT-PCR
Reverse transcription was done using TaqMan Reverse transcription reagent kit (Roche, Branchburg, New Jersey) according to the manufacturer's protocol. Reverse transcription reaction was done in a total volume of 25 mL including RNA template, 1.25 mL random hexamer, reverse transcription buffer, 5.0 mL dNTP, 5.5 mL MgCl, RNase inhibitor, and M-MLV reverse transcriptase.
Real-time PCR was performed in duplicate in 25 mL reaction volumes using Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen, Carlsbad, California) and a 7900HT instrument (Applied Biosystems, Foster City, California). The amplification conditions were: 50°C for 2 min., 95°C for 2 min., 40 cycles of 95°C for 15 sec. and 60°C for 1 min.
Data Analysis of real-time PCR
A mixture of at least eight normal FFPE tissue samples was used as a wildtype, normal control. The EGFRvlll cell line (u373flagEGFRvlll) was used as a positive control.
The relative expression of EGFR exon 4 to EGFR exon 9 was determined using the delta delta Ct (ΔΔCt) method. All samples were run in duplicate, and the mean Ct number was used for data analysis. The difference in Ct values (Δthreshold cycle, ΔCt = Exon 9 Ct - Exon 4 Ct) was calculated for each RNA sample. The ΔCt from the normal tissue mixture was then subtracted from the ΔCt of the test sample to generate a ΔΔCt. A negative result occurs when the fold change (exon 9: exon 4 calculated as 2- ΔΔCt) is less than a value of 5. This value was arbitrarily chosen to ensure that no false positives were called. A positive result occurs when the fold change is the same as or greater than that of the positive control (7). When the fold change of tested samples falls between that of the normal control and the positive control (i.e. between 5 and 7) the results are considered inconclusive.
HPV DNA Detection
The Roche Linear Array HPV Genotyping kit (Roche Molecular Diagnostics, Pleasanton, California) was used to detect 37 low- and high-risk HPV types from FFPE tissues. In brief, FFPE sections were deparaffinized and DNA was extracted using a column based method (QIAamp, Qiagen, Valencia, California). HPV detection was performed using PCR amplification followed by hybridization of the amplified products to oligonucleotide probes and subsequent colorimetric determination. All experiments included an HPV positive control and an HPV negative control.
HPV DNA by in situ hybridization (ISH) using the INFORM HPV III Family 16 probe (Ventana Medical Systems Inc., Tucson, Arizona) which detects genotypes 16, 18, 31, 33, 39, 35, 45, 51, 52, 56, 58 and 66, was performed according to the manufacturer's guidelines using the Ventana Benchmark automated slide staining system. All experiments included an HPV positive control and an HPV negative control. Slides were scored as positive if a punctate or diffuse pattern of signal were observed in the tumor nuclei.
P16 and c-MET Detection
Immunohistochemistry (IHC) for p16 and c-MET using the Ventana Benchmark XT auto-immunostainer (Tucson, Arizona) was performed on FFPE sections cut at 4 mm thick. Standardized staining protocols were provided by Ventana for the CINtec p16 Histology kit (MTM Laboratories Inc, Westborough Massachusetts) and c-MET antibody (SP44, rabbit monoclonal, Ventana Medical Systems Inc., Tucson Arizona). Controls were included in each assay, comprising of positive tissue controls and negative controls. All p16 and c-MET IHC slides were reviewed independently by two observers (B.P.O. and N.G.C.) without knowledge of EGFRvIII, HPV status or clinical outcome. p16 staining in SCCHN is generally observed to be dichotomous and scored as absent (weak or no staining) or present (strong and diffuse staining) [32]. c-MET IHC slides were assigned a semi-quantitative score based on the product of an intensity score (0 = no staining or equal to background, 1 = weak, or 2 = strong) and percent of area stained (0 = 0%, 1 = 1-30%, 2 = 31-60%, 3 = >60%). Sections with an inter-observer variation were reassessed by a double-headed light microscope to achieve consensus.
EGFR Gene Copy Number
Archival tumor specimens were analyzed for EGFR GCN using fluorescent in situ hybridization (FISH) as previously described [33,34]. One hundred non-overlapping interphase nuclei were scored for EGFR and CEP7 copy number and classified into six categories (University of Colorado Scoring system) by a reviewer blinded to clinical outcome (O.L.) [35].
Statistical Methods
Descriptive statistics were used to summarize the study cohort and to estimate the parameters of interest. Ninety-five percent confidence intervals were obtained for estimates of the presence of EGFRvIII, HPV, p16, c-MET and EGFR GCN. Exploratory analyses were performed to characterize the relationships between EGFRvIII, HPV, p16, c-MET and EGFR GCN with baseline patient characteristics and outcomes. Only patients with conclusive EGFRvIII results were included in the correlation analyses. The Kaplan-Meier method was used to estimate the overall survival and time to progression. All biomarkers were examined in univariate analysis of overall survival and time to progression using Cox proportional hazards model. Only those which were significant at 0.10 (two-sided) level in the univariate analysis were entered in the multivariate analysis and markers that remained significant at 0.05 (two-sided) level in the multivariate analysis were considered significant prognostic factors. Statistical analyses were performed using the SAS 9.1 software package (SAS Institute, Cary, North Carolina).
Results
Patients
The clinical characteristics of the 53 patients in our study are described in Table 1. For the entire cohort, the overall response rate (CR+PR) to study treatment was 4/53 (7.5%), median time to progression (TTP) was 1.8 months (95% CI 1.6-2.7) and median overall survival (OS) was 5.9 months (95% CI 4.5-8.7). Patients in the erlotinib group had a higher median OS of 7.9 months (95% CI 4.7-9.8) compared to patients in the non-erlotinib group with median OS of 4.2 months (95% CI 2.9-7.0) (p = 0.011). The erlotinib group had a higher TTP than the non-erlotinib group 2.7 months (95% CI 1.6-3.5) vs 1.5 months (95% CI 1.3-1.8) (p = 0.0009).
Table 1.
Clinical characteristics of the entire study cohort (n = 53)
| Clinical Characteristic | Number | |
|---|---|---|
| Median Age (Range) | 56 (15-78) | |
| Gender | Female:Male | 12:41 |
| ECOG Performance Status | 0:1:2 | 15:34:4 |
| Locoregional Recurrence | Yes:No | 45:8 |
| Distant Metastases | Yes:No | 19:34 |
| Primary Tumor Site | Oropharynx | 20 |
| Larynx | 14 | |
| Oral cavity | 10 | |
| Hypopharynx | 2 | |
| Neck mass unknown | 4 | |
| Paranasal sinus | 3 | |
| Histologic Grade | Well differentiated | 5 |
| Moderately differentiated | 33 | |
| Poorly differentiated | 14 | |
| Infiltrating basaloid | 1 | |
| Prior Therapy | Chemotherapy | 21 |
| Radiation Therapy | 51 | |
| Surgery | 41 | |
| Race | Asian | 8 |
| Black | 42 | |
| Caucasian | 9 | |
| Other | 1 | |
| Smoker | Current | 33 |
| Former | 3 | |
| Never | 15 | |
| Unknown | 2 | |
| Erlotinib | Yes: No | 35:18 |
| Best Response | Partial response | 4 |
| Stable disease | 20 | |
| Progressive disease | 24 | |
| Inevaluable | 5 | |
Expression of EGFRvIII mutation by real-time PCR
As the previously reported immunohistochemistry-suitable antibody [15] against EGFRvIII is no longer available, EGFRvIII expression is analysed using the RT-PCR method. The presence of EGFRvIII mutation was detected in 22 patients (42%) (Table 2), negative in 19 patients and inconclusive in 12 patients (Table 3). The median EGFRvIII fold change was 6.8 (0.56 to 576.36) for all patients, 15.0 (4.1 to 576.36) for patients in the EGFRvIII positive group, 1.8 (0.6 to 4.3) for patients in the EGFRvIII negative group, and 6.5 (6.2 to 6.8) for patients in the inconclusive group.
Table 2.
EGFRvIII mutation positive detected by RT-PCR (n = 22)
| Case | Treatment | Primary Site | Specimen Site | EGFRvIII by RT-PCR | EGFRvIII Fold Changes | HPV DNA by Linear Array | P16 IHC | MET score by IHC | EGFR FISH |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Erlotinib | hypopharynx | Untreated primary | + | 11.33 | - | - | High | Low polysomy |
| 2 | Erlotinib | oral cavity | Untreated primary | + | 7.01 | - | - | High | Disomy |
| 3 | Erlotinib | larynx | Untreated primary | + | 61.77 | - | - | Low | Low polysomy |
| 4 | Erlotinib | larynx | Local recurrence | + | 26.64 | - | - | Low | High polysomy |
| 5 | Erlotinib | neck mass unknown primary | Untreated lymph node | + | 60.04 | 33 + | + | High | Low trisomy |
| 6 | Erlotinib | oral cavity | Untreated lymph node | + | 218.49 | - | - | High | Amplification |
| 7 | Erlotinib | oropharynx | Unknown primary | + | 15.66 | 16 + | + | High | Low trisomy |
| 8 | Erlotinib | oral cavity | Local recurrence | + | 8.94 | - | - | High | Low polysomy |
| 9 | Erlotinib + Cisplatin | neck mass unknown primary | Untreated lymph node | + | 127.84 | 16+ | NE | NE | Low trisomy |
| 10 | Erlotinib + Cisplatin | larynx | Node recurrence | + | 576.36 | 16+ | + | High | Disomy |
| 11 | Erlotinib + Cisplatin | larynx | Untreated primary | + | 8.26 | - | - | Low | Failed |
| 12 | Erlotinib + Cisplatin | larynx | Untreated primary | + | 17.38 | - | - | High | Low polysomy |
| 13 | Erlotinib + Cisplatin | oropharynx | Untreated primary | + | 14.28 | 16+ | + | High | Disomy |
| 14 | Erlotinib + Cisplatin | oral cavity | Untreated primary | + | 69.68 | - | NE | NE | Low polysomy |
| 15 | Erlotinib + Cisplatin | oral cavity | Untreated primary | + | 11.11 | 53+, 58+, 6+, 52+ | - | High | Low polysomy |
| 16 | Sorafenib | oropharynx | Untreated primary | + | 7.71 | 16 + | + | Low | Low polysomy |
| 17 | Sorafenib | oropharynx | Untreated primary | + | 7.93 | 16 + | + | Low | Low trisomy |
| 18 | Ispinesib | neck mass unknown primary | Untreated lymph node | + | 4.12 | 16+, 53+, 51 + | + | High | High polysomy |
| 19 | Ispinesib | oropharynx | Untreated primary | + | 15.61 | 16+, 53+, 33+, 51+, 58 + | - | High | High polysomy |
| 20 | Ispinesib | larynx | Untreated primary | + | 218.26 | - | - | High | Disomy |
| 21 | Ispinesib | oropharynx | Local recurrence | + | 29.25 | 16+, 53+, 51+ | + | Low | Disomy |
| 22 | Ispinesib | hypopharynx | Local recurrence | + | 9.31 | 16+, 53+, 58+, 52+ | - | High | Low polysomy |
Abbreviations: +, positive; -, negative; NE, not evaluable
Table 3.
EGFRvIII mutation negative detected by RT-PCR (n = 19) and inconclusive cases (n = 12)
| Case | Treatment | Primary Site | Specimen Site | EGFRvIII by RT-PCR | EGFRvIII Fold Changes | HPV DNA by Linear Array | P16 IHC | MET score by IHC | EGFR FISH |
|---|---|---|---|---|---|---|---|---|---|
| 23 | Erlotinib | larynx | Local recurrence | - | 1.76 | - | - | High | Low polysomy |
| 24 | Erlotinib | larynx | Untreated primary | - | 0.95 | - | - | NE | NE |
| 25 | Erlotinib | oropharynx | Untreated primary | Incon. | NE | 16 + | - | Low | Failed |
| 26 | Erlotinib | larynx | Untreated | Incon. | NE | - | - | NE | NE |
| 27 | Erlotinib | oropharynx | Local recurrence | - | 2.56 | 16 + | - | High | Failed |
| 28 | Erlotinib | neck mass unknown primary | Local recurrence | - | 1.81 | - | - | High | Low polysomy |
| 29 | Erlotinib | paranasal sinus | Untreated primary | - | 0.56 | - | - | High | High polysomy |
| 30 | Erlotinib | oropharynx | Untreated primary | - | 3.16 | 16 + | + | High | Low trisomy |
| 31 | Erlotinib | larynx | Untreated primary | - | 3.17 | - | - | High | Low polysomy |
| 32 | Erlotinib | oropharynx | Local recurrence | NE | NE | - | + | Low | Low polysomy |
| 33 | Erlotinib | larynx | Untreated primary | - | 3.16 | - | - | High | High trisomy |
| 34 | Erlotinib + Cisplatin | oropharynx | Untreated primary | Incon. | NE | Incon. | + | Low | NE |
| 35 | Erlotinib + Cisplatin | larynx | Untreated primary | - | 4.29 | - | - | Low | Low polysomy |
| 36 | Erlotinib + Cisplatin | oral cavity | Untreated primary | - | 0.9 | - | - | Low | High polysomy |
| 37 | Erlotinib + Cisplatin | paransal sinus | Local recurrence | Incon. | NE | 16+, 53+ | Incon. | Low | Low polysomy |
| 38 | Erlotinib + Cisplatin | larynx | Local recurrence | Incon. | NE | 16+, 53+, 33+, 51+ | + | High | Disomy |
| 39 | Erlotinib + Cisplatin | paranasal sinus | Local recurrence | - | 2.59 | 6+ | - | High | NE |
| 40 | Erlotinib + Cisplatin | oral cavity | Untreated primary | - | 2.01 | - | - | High | Disomy |
| 41 | Erlotinib + Cisplatin | oropharynx | Untreated primary | Incon. | NE | - | + | High | Low trisomy |
| 42 | Erlotinib + Cisplatin | oral cavity | Untreated primary | Incon. | NE | - | - | High | High polysomy |
| 43 | Sorafenib | oral cavity | Untreated primary | Incon. | 6.8 | - | - | Low | Low polysomy |
| 44 | Sorafenib | oropharynx | Untreated primary | - | 0.99 | - | + | High | Amplification/High trisomy |
| 45 | Sorafenib | oral cavity | Local recurrence | Incon. | NE | Incon. | - | Low | Low polysomy |
| 46 | Sorafenib | oropharynx | Untreated primary | Incon. | 6.22 | 16 + | + | Low | Low trisomy |
| 47 | Sorafenib | larynx | Local recurrence | - | 2.28 | - | + | High | High polysomy |
| 48 | Sorafenib | oropharynx | Untreated primary | - | 0.64 | - | - | High | Low polysomy |
| 49 | Sorafenib | oropharynx | Local recurrence | - | 1.4 | - | - | High | High polysomy |
| 50 | Sorafenib | oropharynx | Local recurrence | - | 1.3 | - | - | High | High polysomy |
| 51 | Sorafenib | oropharynx | Local recurrence | - | 1.76 | - | + | Low | Monosomy/Disomy |
| 52 | Ispinesib | oropharynx | Untreated lymph node | Incon. | NE | - | - | High | Low polysomy |
| 53 | Ispinesib | oropharynx | Node recurrence | - | 4.05 | 16 + | - | Low | Amplification |
Abbreviations: +, positive; -, negative; Incon., inconclusive; NE, not evaluable
Patients with tumors harboring the EGFRvIII mutation had similar clinical characteristics to patients without the EGFRvIII mutation (Table 4).
Table 4.
Presence of the EGFRvIII mutation is not significantly associated with any clinical characteristics
| Clinical Characteristic | EGFRvIII absent (n = 19) | EGFRvIII present (n = 22) | p-value |
|---|---|---|---|
| Male - no., (%) | 16 (84%) | 18 (82%) | 0.839 (Fisher's) |
| Age - mean, (+/-SD) | 53.5 (+/- 11.6) | 55.1 (+/- 14.1) | 0.685 (t-test) |
| Oropharynx - no., (%) | 8 (42%) | 6 (27%) | 0.318 (Chi-square) |
| Larynx - no., (%) | 6 (32%) | 6 (27%) | 0.763 (Chi-square) |
| Oral Cavity - no., (%) | 2 (11%) | 5 (23%) | 0.271 (Fisher's) |
| Distant metastasis - no., (%) | 4 (21%) | 12 (55%) | 0.053 (Fisher's) |
| Locoregional recurrence - no., (%) | 18 (95%) | 16 (73%) | 0.099 (Fisher's) |
| Well-moderately differentiated - no., (%) | 15 (79%) | 14 (64%) | 0.325 (Fisher's) |
| Poorly differentiated - no., (%) | 4 (21%) | 8 (36%) | |
| Prior chemotherapy - no., (%) | 9 (47%) | 8 (36%) | 0.476 (Chi-square) |
| Prior radiotherapy - no., (%) | 17 (89%) | 22 (100%) | 0.209 (Fisher's) |
| Prior surgery - no., (%) | 17 (89%) | 17 (77%) | 0.419 (Fisher's) |
| Caucasian - no., (%) | 15 (79%) | 18 (82%) | 0.562 (Fisher's) |
| Erlotinib treatment - no., (%) | 12 (63%) | 15 (68%) | 0.735 (Chi-square) |
| No erlotinib treatment - no., (%) | 7 (38%) | 7 (32%) | |
EGFRvIII is associated with disease control
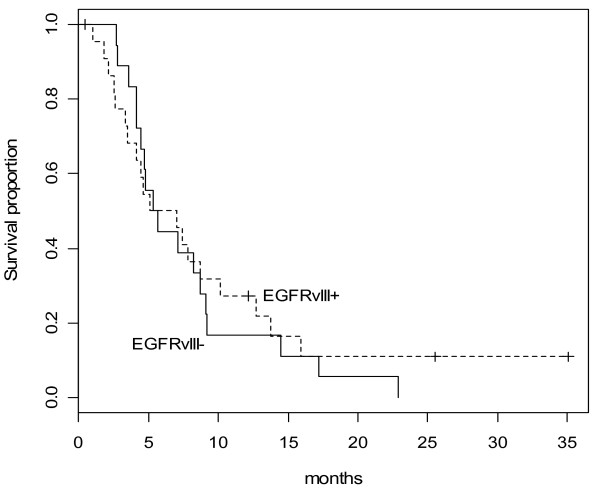
In univariate analysis, the presence of EGFRvIII was associated with better disease control (Table 5). Median EGFRvIII fold changes were higher for patients with disease control than patients with progressive disease (11.11 vs. 3.16, p = 0.04). No significant difference was observed between erlotinib-treated (p = 0.21) versus non-erlotinib (p = 0.10) treated patients due to the small sample size (Table 5). The presence of EGFRvIII mutation was not associated with TTP (HR 0.94 (95% CI 0.33-2.71), p = 0.91) or OS (HR 0.91 (95% CI 0.32-2.60), p = 0.85) (Figure 1).
Table 5.
EGFRvIII mutation is associated with disease control
| EGFRvIII | Best Response | ||
|---|---|---|---|
| Progressive Disease | Disease Control (Partial Response or Stable Disease) | ||
| EGFRvIII absent by ΔΔCt | 12 (67%) | 6 (33%) | P = 0.0099 |
| EGFRvIII present by ΔΔCt | 5 (25%) | 15 (75%) | (Chi-square) |
| EGFRvIII mean fold change | 10.62 | 63.76 | P = 0.04 |
| EGFRvIII median fold change | 3.16 | 11.11 | (Wilcoxon) |
| Erlotinib treated patients | |||
| EGFRvIII absent by ΔΔCt | 6 (55%) | 5 (45%) | P = 0.21 |
| EGFRvIII present by ΔΔCt | 3 (33%) | 10 (77%) | (Fisher's) |
| Non-erlotinib treated patients | |||
| EGFRvIII absent by ΔΔCt | 6 (86%) | 1 (14%) | P = 0.21 |
| EGFRvIII present by ΔΔCt | 2 (29%) | 5 (71%) | (Fisher's) |
Figure 1.
Overall survival by EGFRvIII mutation status (HR = 0.91, 95% CI: 0.32-2.60, p = 0.85).
HPV DNA
HPV DNA testing by PCR was positive in 20 patients (38%), negative in 31 patients and inconclusive in 2 patients (Tables 2 and 3). The most prevalent HPV subtype found in our analysis was the high-risk HPV-16 (18/20 cases). The majority of HPV-16 positive tumors were from the oropharynx (12/18). HPV positive tumor status was not significantly associated with disease control (Table 6), TTP (HR 1.19 (95% CI 0.46-3.11), p = 0.722) or OS (HR 0.88 (95% CI 0.34-2.29), p = 0.788).
Table 6.
HPV by PCR, p16 by IHC, c-MET by IHC are not associated with disease control
| Best Response | |||
|---|---|---|---|
| Progressive Disease | Disease Control (Partial Response or Stable Disease) | ||
| HPV PCR negative | 14 (50%) | 14 (50%) | P = 0.86 |
| HPV PCR positive | 9 (47%) | 10 (53%) | (Chi-square) |
| P16 IHC negative | 14 (47%) | 16 (53%) | P = 0.67 |
| P16 IHC positive | 8 (53%) | 7 (47%) | (Chi-square) |
| c-MET IHC < or = 2 | 9 (56%) | 7 (44%) | P = 0.39 |
| c-MET IHC >2 | 12 (43%) | 16 (57%) | (Chi-square) |
P16
P16 immunoreactivity was detected in 17 patients (32%), absent in 33 patients and inconclusive in 3 patients (Tables 2 and 3). The inter-observer variability rate was 6% and discrepant cases were resolved by consensus review. P16 expression was not associated with disease control (Table 6), TTP (HR 0.50 (95% CI 0.19-1.32), p = 0.16) or OS (HR 0.61 (95%CI 0.24-1.55), p = 0.30).
The discordance between p16 IHC and HPV DNA by PCR was 25% (Table 7). To investigate this further, we performed HPV DNA by ISH. The discordance between p16 IHC and HPV DNA by ISH was lower at 16% (Table 7) and all 7 discordant cases were p16-positive/HPV-ISH-negative. Of these 7 discordant cases, 2 cases were HPV-16 positive by PCR, 4 cases were HPV negative by PCR and 1 case was inconclusive by HPV PCR.
Table 7.
Concordance of p16 IHC status with HPV by PCR and HPV by ISH
| P16 IHC positive | P16 IHC negative | ||
|---|---|---|---|
| HPV PCR positive | 10 | 5 | P = 0.0016 |
| HPV PCR negative | 5 | 25 | (Chi-Square) |
| HPV ISH positive | 6 | 0 | P = 0.0002 |
| HPV ISH negative | 7 | 31 | (Fisher's) |
C-MET
Forty-nine patients had sufficient tumor samples for evaluation of c-MET. Eighteen patients (63%) had low c-MET scores of 0, 1 or 2 and 31 patients had high c-MET (>2) (Tables 2 and 3). Less than 10% inter-observer variability was observed and discrepant cases were resolved by consensus review. High c-MET was not associated with disease control (Table 6), TTP (HR 1.47 (95%CI 0.56-3.85), p = 0.43) or OS (HR 1.72 (95%CI 0.65-4.56), p = 0.27).
EGFR Gene Copy Number
Forty-five patients had sufficient tumor samples for evaluation of EGFR GCN by FISH. High EGFR GCN (amplification and high polysomy) was detected in 13 patients and low EGFR GCN (disomy, low polysomy) was detected in 33 patients (Tables 2 and 3). High EGFR GCN was not predictive for TTP (HR 0.99, p = 0.822) or OS (HR 1.10, p = 0.644). High EGFR GCN was not associated with the presence of EGFRvIII (p = 0.14 Fisher's exact test).
Discussion
To the best of our knowledge, this is the first study to evaluate the role of EGFRvIII in a cohort of patients with R/M SCCHN treated with or without EGFR TKI. This study confirms that EGFRvIII mutation is common in R/M SCCHN, and may play a role in prognosis. We identified EGFRvIII mutation in 42% of 53 R/M SCCHN tumors. This is in keeping with the first description of EGFRvIII expression by IHC and RT-PCR in 42% of 33 SCCHN tumors sampled [15]. In vitro studies suggest that EGFRvIII mutated SCCHN cell lines are resistant to the anti-EGFR monoclonal antibody cetuximab [15]. In this study, EGFRvIII was not associated with an inferior response to erlotinib therapy. Importantly, we observed a significant association between the presence of EGFRvIII (mean fold change and copy number by RT-PCR) with greater disease control, regardless of treatment with erlotinib, suggesting that perhaps EGFRvIII may have a prognostic role.
The prognostic or predictive significance of the EGFRvIII mutation in response to systemic therapy in patients with SCCHN has not been previously described. The potential prognostic role of EGFRvIII appears to be independent of any clinicopathologic characteristics. This is consistent with another study where EGFRvIII detected by IHC in 234 of 681 locally advanced SCCHN tumors (34%) was associated with increased tumor size but not stage or other clinical factors [36]. In our study, EGFRvIII was not associated with overall survival or TTP. To our knowledge, EGFRvIII has not been linked to survival in SCCHN. EGFRvIII has been described more extensively in glioblastoma where it results in enhanced proliferation and reduced apoptosis effects that are mediated through increased levels of activated Ras [37] and activation of the PI3K pathway [38]. However, the role of EGFRvIII as a prognostic or predictive marker of response to EGFR inhibitors in glioblastoma remains controversial. EGFRvIII and PTEN co-expression was associated with response to EGFR TKI in 26 patients out of a cohort of 49 patients with recurrent glioma and a validation set of 33 patients [39]. EGFRvIII has been reported as a prognostic marker for poorer survival in some studies [40,41], but not in others [42,43]. Conflicting results have been attributed to small sample sizes with incomplete clinical data and varying methods to detect EGFRvIII.
The presence of activating mutations conferring a better prognosis has been reported with EGFR mutations in non-small cell lung cancer (NSCLC) [44] and with PIK3CA mutations in breast cancer [45]. Somatic activating mutations (exon 19 deletion and 21 point mutation) in the EGFR tyrosine kinase domain confer sensitivity to EGFR inhibitors in NSCLC. Patients with these mutations also had improved survival and response to chemotherapy alone [46] or placebo [47]. This suggests that EGFR mutations in NSCLC are a good prognostic factor independent of EGFR TKI, hence it may be more difficult to demonstrate the value of EGFR mutations as predictors of benefit to EGFR TKI [44]. The prognostic value of EGFRvIII in SCCHN needs to be verified, and its role as a predictive marker of response to EGFR inhibitor should remain a relevant therapeutic question.
In this study, the prevalence of HPV, p16 and c-MET expression (38%, 32% and 63% respectively) was in keeping with the literature. We did not observe HPV, p16 and c-MET expression to be predictive of disease control, TTP or OS. This may be due to limitations of a small sample size. Consistent with prior reports [21], HPV-16 was the most common HPV subtype in our study. c-MET is a poor prognostic marker in OSCC [48], however the small proportion (11%) of OSCC in our study precludes any meaningful association.
Limitations of this study include its small sample size, potential bias towards patients with available tumor specimens (larger tumor size), potentially variable fixation and quality of the archival tissues and potential variation in marker status of primary tumor compared with recurrent or metastatic tumors (to our knowledge, this is theoretical and has not been described). Due to the absence of an untreated control group in this study ('control' patients received sorafenib or ispinesib), our results cannot conclusively confirm the prognostic versus predictive value of a biomarker. Although our methods did not use an antibody for EGFRvIII detection, we acknowledge that the use of RT-PCR in FFPE samples has demonstrated superior accuracy relative to IHC tests [49] and may allow greater applicability to settings where frozen tissue is unavailable.
Conclusion
Predictors of response to EGFR inhibitors in SCCHN remain elusive. Biomarkers are desperately needed to guide patient selection in SCCHN. EGFRvIII remains an interesting tumor-specific target worthy of further exploration as a prognostic or predictive marker of response to EGFR inhibitor therapy in SCCHN. Larger prospective randomized studies are required to distinguish the prognostic and predictive significance of EGFRvIII, HPV, p16, c-MET and EGFR GCN in SCCHN treated with EGFR inhibitors.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
NC participated in the study design, data acquisition, immunohistochemical interpretation and drafted the manuscript. BP-O participated in the histological examination, immunohistochemical interpretation and manuscript preparation. KZ, NAP, JH, TZ, M-ST, SK-R carried out immunostaining, ISH, PCR analysis, and prepared and reviewed the manuscript. OL performed the FISH analysis and reviewed the manuscript. LW performed the statistical analysis. EC participated in patient management and reviewed the manuscript. LS conceived of the study, participated in its design and coordination, and prepared and revised the manuscript. All authors read and approved the final manuscript.
Contributor Information
Nicole G Chau, Email: nicole.chau@uhn.on.ca.
Bayardo Perez-Ordonez, Email: Bayardo.Perez-Ordonez@uhn.on.ca.
Katherine Zhang, Email: katzhang@uhnres.utoronto.ca.
Nhu-An Pham, Email: npham@uhnres.utoronto.ca.
James Ho, Email: jamesho@uhnres.utoronto.ca.
Tong Zhang, Email: tong.zhang@uhn.on.ca.
Olga Ludkovski, Email: oludkovs@uhnres.utoronto.ca.
Lisa Wang, Email: lisawang@uhnres.utoronto.ca.
Eric X Chen, Email: Eric.Chen@uhn.on.ca.
Ming-Sound Tsao, Email: ming.tsao@uhn.on.ca.
Suzanne Kamel-Reid, Email: Suzanne.Kamel-Reid@uhn.on.ca.
Lillian L Siu, Email: lillian.siu@uhn.on.ca.
Acknowledgements
This work was supported by an unrestricted grant from Pfizer Canada, Inc.
References
- Grandis JR, Tweardy DJ. Elevated levels of transforming growth factor alpha and epidermal growth factor receptor messenger RNA are early markers of carcinogenesis in head and neck cancer. Cancer Res. 1993;53:3579–3584. [PubMed] [Google Scholar]
- Woodburn JR. The epidermal growth factor receptor and its inhibition in cancer therapy. Pharmacol Ther. 1999;82:241–250. doi: 10.1016/S0163-7258(98)00045-X. [DOI] [PubMed] [Google Scholar]
- Temam S, Kawaguchi H, El-Naggar AK, Jelinek J, Tang H, Liu DD, Lang W, Issa JP, Lee JJ, Mao L. Epidermal growth factor receptor copy number alterations correlate with poor clinical outcome in patients with head and neck squamous cancer. J Clin Oncol. 2007;25:2164–2170. doi: 10.1200/JCO.2006.06.6605. [DOI] [PubMed] [Google Scholar]
- Chung CH, Ely K, McGavran L, Varella-Garcia M, Parker J, Parker N, Jarrett C, Carter J, Murphy BA, Netterville J. et al. Increased epidermal growth factor receptor gene copy number is associated with poor prognosis in head and neck squamous cell carcinomas. J Clin Oncol. 2006;24:4170–4176. doi: 10.1200/JCO.2006.07.2587. [DOI] [PubMed] [Google Scholar]
- Bonner JA, Harari PM, Giralt J, Azarnia N, Shin DM, Cohen RB, Jones CU, Sur R, Raben D, Jassem J. et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. NEnglJ Med. 2006;354:567–578. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- Cohen EE, Rosen F, Stadler WM, Recant W, Stenson K, Huo D, Vokes EE. Phase II trial of ZD1839 in recurrent or metastatic squamous cell carcinoma of the head and neck. J Clin Oncol. 2003;21:1980–1987. doi: 10.1200/JCO.2003.10.051. [DOI] [PubMed] [Google Scholar]
- Siu LL, Soulieres D, Chen EX, Pond GR, Chin SF, Francis P, Harvey L, Klein M, Zhang W, Dancey J. et al. Phase I/II trial of erlotinib and cisplatin in patients with recurrent or metastatic squamous cell carcinoma of the head and neck: a Princess Margaret Hospital phase II consortium and National Cancer Institute of Canada Clinical Trials Group Study. J Clin Oncol. 2007;25:2178–2183. doi: 10.1200/JCO.2006.07.6547. [DOI] [PubMed] [Google Scholar]
- Soulieres D, Senzer NN, Vokes EE, Hidalgo M, Agarwala SS, Siu LL. Multicenter phase II study of erlotinib, an oral epidermal growth factor receptor tyrosine kinase inhibitor, in patients with recurrent or metastatic squamous cell cancer of the head and neck. J Clin Oncol. 2004;22:77–85. doi: 10.1200/JCO.2004.06.075. [DOI] [PubMed] [Google Scholar]
- Burtness B, Goldwasser MA, Flood W, Mattar B, Forastiere AA. Phase III randomized trial of cisplatin plus placebo compared with cisplatin plus cetuximab in metastatic/recurrent head and neck cancer: an Eastern Cooperative Oncology Group study. J Clin Oncol. 2005;23:8646–8654. doi: 10.1200/JCO.2005.02.4646. [DOI] [PubMed] [Google Scholar]
- Vermorken JB, Mesia R, Rivera F, Remenar E, Kawecki A, Rottey S, Erfan J, Zabolotnyy D, Kienzer HR, Cupissol D. et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med. 2008;359:1116–1127. doi: 10.1056/NEJMoa0802656. [DOI] [PubMed] [Google Scholar]
- Grandal MV, Zandi R, Pedersen MW, Willumsen BM, van DB, Poulsen HS. EGFRvIII escapes down-regulation due to impaired internalization and sorting to lysosomes. Carcinogenesis. 2007;28:1408–1417. doi: 10.1093/carcin/bgm058. [DOI] [PubMed] [Google Scholar]
- Huang HS, Nagane M, Klingbeil CK, Lin H, Nishikawa R, Ji XD, Huang CM, Gill GN, Wiley HS, Cavenee WK. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J Biol Chem. 1997;272:2927–2935. doi: 10.1074/jbc.272.5.2927. [DOI] [PubMed] [Google Scholar]
- Nishikawa R, Ji XD, Harmon RC, Lazar CS, Gill GN, Cavenee WK, Huang HJ. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci USA. 1994;91:7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Zhao X, Yuza Y, Shimamura T, Li D, Protopopov A, Jung BL, McNamara K, Xia H, Glatt KA. et al. Epidermal growth factor receptor variant III mutations in lung tumorigenesis and sensitivity to tyrosine kinase inhibitors. Proc Natl Acad Sci USA. 2006;103:7817–7822. doi: 10.1073/pnas.0510284103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok JC, Coppelli FM, Thomas SM, Lango MN, Xi S, Hunt JL, Freilino ML, Graner MW, Wikstrand CJ, Bigner DD. et al. Mutant epidermal growth factor receptor (EGFRvIII) contributes to head and neck cancer growth and resistance to EGFR targeting. Clin Cancer Res. 2006;12:5064–5073. doi: 10.1158/1078-0432.CCR-06-0913. [DOI] [PubMed] [Google Scholar]
- Gillison ML, Koch WM, Capone RB, Spafford M, Westra WH, Wu L, Zahurak ML, Daniel RW, Viglione M, Symer DE. et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92:709–720. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- Shi W, Kato H, Perez-Ordonez B, Pintilie M, Huang S, Hui A, O'Sullivan B, Waldron J, Cummings B, Kim J. et al. Comparative prognostic value of HPV16 E6 mRNA compared with in situ hybridization for human oropharyngeal squamous carcinoma. J Clin Oncol. 2009;27:6213–6221. doi: 10.1200/JCO.2009.23.1670. [DOI] [PubMed] [Google Scholar]
- Gillison ML, D'Souza G, Westra W, Sugar E, Xiao W, Begum S, Viscidi R. Distinct risk factor profiles for human papillomavirus type 16-positive and human papillomavirus type 16-negative head and neck cancers. J Natl Cancer Inst. 2008;100:407–420. doi: 10.1093/jnci/djn025. [DOI] [PubMed] [Google Scholar]
- Westra WH, Taube JM, Poeta ML, Begum S, Sidransky D, Koch WM. Inverse relationship between human papillomavirus-16 infection and disruptive p53 gene mutations in squamous cell carcinoma of the head and neck. Clin Cancer Res. 2008;14:366–369. doi: 10.1158/1078-0432.CCR-07-1402. [DOI] [PubMed] [Google Scholar]
- Weinberger PM, Yu Z, Haffty BG, Kowalski D, Harigopal M, Brandsma J, Sasaki C, Joe J, Camp RL, Rimm DL, Psyrri A. Molecular classification identifies a subset of human papillomavirus--associated oropharyngeal cancers with favorable prognosis. J Clin Oncol. 2006;24:736–747. doi: 10.1200/JCO.2004.00.3335. [DOI] [PubMed] [Google Scholar]
- Fakhry C, Westra WH, Li S, Cmelak A, Ridge JA, Pinto H, Forastiere A, Gillison ML. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100:261–269. doi: 10.1093/jnci/djn011. [DOI] [PubMed] [Google Scholar]
- Kumar B, Cordell KG, Lee JS, Worden FP, Prince ME, Tran HH, Wolf GT, Urba SG, Chepeha DB, Teknos TN. et al. EGFR, p16, HPV Titer, Bcl-xL and p53, sex, and smoking as indicators of response to therapy and survival in oropharyngeal cancer. J Clin Oncol. 2008;26:3128–3137. doi: 10.1200/JCO.2007.12.7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassen P, Eriksen JG, Hamilton-Dutoit S, Tramm T, Alsner J, Overgaard J. Effect of HPV-associated p16INK4A expression on response to radiotherapy and survival in squamous cell carcinoma of the head and neck. J Clin Oncol. 2009;27:1992–1998. doi: 10.1200/JCO.2008.20.2853. [DOI] [PubMed] [Google Scholar]
- Smith EM, Wang D, Kim Y, Rubenstein LM, Lee JH, Haugen TH, Turek LP. P16INK4a expression, human papillomavirus, and survival in head and neck cancer. Oral Oncol. 2008;44:133–142. doi: 10.1016/j.oraloncology.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Knowles LM, Stabile LP, Egloff AM, Rothstein ME, Thomas SM, Gubish CT, Lerner EC, Seethala RR, Suzuki S, Quesnelle KM. et al. HGF and c-Met participate in paracrine tumorigenic pathways in head and neck squamous cell cancer. Clinical Cancer Research. 2009;15:3740–3750. doi: 10.1158/1078-0432.CCR-08-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YS, Wang JT, Chang YF, Liu BY, Wang YP, Sun A, Chiang CP. Expression of hepatocyte growth factor and c-met protein is significantly associated with the progression of oral squamous cell carcinoma in Taiwan. Journal of Oral Pathology & Medicine. 2004;33:209–217. doi: 10.1111/j.0904-2512.2004.00118.x. [DOI] [PubMed] [Google Scholar]
- Allen C, Duffy S, Teknos T, Islam M, Chen Z, Albert PS, Wolf G, Van WC. Nuclear factor-kappaB-related serum factors as longitudinal biomarkers of response and survival in advanced oropharyngeal carcinoma. Clin Cancer Res. 2007;13:3182–3190. doi: 10.1158/1078-0432.CCR-06-3047. [DOI] [PubMed] [Google Scholar]
- Di Renzo MF, Olivero M, Martone T, Maffe A, Maggiora P, Stefani AD, Valente G, Giordano S, Cortesina G, Comoglio PM. Somatic mutations of the MET oncogene are selected during metastatic spread of human HNSC carcinomas. Oncogene. 2000;19:1547–1555. doi: 10.1038/sj.onc.1203455. [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Huang S, Kruser TJ, Nechrebecki MM, Armstrong EA, Benavente S, Gondi V, Hsu KT, Harari PM. Mechanisms of acquired resistance to cetuximab: role of HER (ErbB) family members. Oncogene. 2008;27:3944–3956. doi: 10.1038/onc.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang PA, Siu LL, Chen EX, Hotte SJ, Chia S, Schwarz JK, Pond GR, Johnson C, Colevas AD, Synold TW. et al. Phase II study of ispinesib in recurrent or metastatic squamous cell carcinoma of the head and neck. Invest New Drugs. 2008;26:257–264. doi: 10.1007/s10637-007-9098-8. [DOI] [PubMed] [Google Scholar]
- Elser C, Siu LL, Winquist E, Agulnik M, Pond GR, Chin SF, Francis P, Cheiken R, Elting J, McNabola A. et al. Phase II trial of sorafenib in patients with recurrent or metastatic squamous cell carcinoma of the head and neck or nasopharyngeal carcinoma. J Clin Oncol. 2007;25:3766–3773. doi: 10.1200/JCO.2006.10.2871. [DOI] [PubMed] [Google Scholar]
- Weinberger PM, Yu Z, Haffty BG, Kowalski D, Harigopal M, Sasaki C, Rimm DL, Psyrri A. Prognostic significance of p16 protein levels in oropharyngeal squamous cell cancer. Clin Cancer Res. 2004;10:5684–5691. doi: 10.1158/1078-0432.CCR-04-0448. [DOI] [PubMed] [Google Scholar]
- Agulnik M, da Cunha SG, Hedley D, Nicklee T, Dos Reis PP, Ho J, Pond GR, Chen H, Chen S, Shyr Y. et al. Predictive and pharmacodynamic biomarker studies in tumor and skin tissue samples of patients with recurrent or metastatic squamous cell carcinoma of the head and neck treated with erlotinib. Journal of Clinical Oncology. 2007;25:2184–2190. doi: 10.1200/JCO.2006.07.6554. [DOI] [PubMed] [Google Scholar]
- Tsao MS, Sakurada A, Cutz JC, Zhu CQ, Kamel-Reid S, Squire J, Lorimer I, Zhang T, Liu N, Daneshmand M. et al. Erlotinib in lung cancer - molecular and clinical predictors of outcome. N Engl J Med. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- Cappuzzo F, Hirsch FR, Rossi E, Bartolini S, Ceresoli GL, Bemis L, Haney J, Witta S, Danenberg K, Domenichini I. et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97:643–655. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- Eriksen JGLP, Overgaard J. ECCO 15 - 34th ESMO Multidisciplinary Congress. Berlin; 2009. The EGFRvIII variant in squamous cell carcinomas of the head and neck: Expression and correlation with clinico-pathological parameters in 681 patients from the randomised DAHANCA 6/7 study; p. 472. September 2009. [Google Scholar]
- Prigent SA, Nagane M, Lin H, Huvar I, Boss GR, Feramisco JR, Cavenee WK, Huang HS. Enhanced tumorigenic behavior of glioblastoma cells expressing a truncated epidermal growth factor receptor is mediated through the Ras-Shc-Grb2 pathway. J Biol Chem. 1996;271:25639–25645. doi: 10.1074/jbc.271.41.25639. [DOI] [PubMed] [Google Scholar]
- Huang PH, Mukasa A, Bonavia R, Flynn RA, Brewer ZE, Cavenee WK, Furnari FB, White FM. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc Natl Acad Sci USA. 2007;104:12867–12872. doi: 10.1073/pnas.0705158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, Lu KV, Yoshimoto K, Huang JH, Chute DJ. et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- Pelloski CE, Ballman KV, Furth AF, Zhang L, Lin E, Sulman EP, Bhat K, McDonald JM, Yung WK, Colman H. et al. Epidermal growth factor receptor variant III status defines clinically distinct subtypes of glioblastoma. J Clin Oncol. 2007;25:2288–2294. doi: 10.1200/JCO.2006.08.0705. [DOI] [PubMed] [Google Scholar]
- Shinojima N, Tada K, Shiraishi S, Kamiryo T, Kochi M, Nakamura H, Makino K, Saya H, Hirano H, Kuratsu J. et al. Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res. 2003;63:6962–6970. [PubMed] [Google Scholar]
- Heimberger AB, Hlatky R, Suki D, Yang D, Weinberg J, Gilbert M, Sawaya R, Aldape K. Prognostic effect of epidermal growth factor receptor and EGFRvIII in glioblastoma multiforme patients. Clin Cancer Res. 2005;11:1462–1466. doi: 10.1158/1078-0432.CCR-04-1737. [DOI] [PubMed] [Google Scholar]
- Liu L, Backlund LM, Nilsson BR, Grander D, Ichimura K, Goike HM, Collins VP. Clinical significance of EGFR amplification and the aberrant EGFRvIII transcript in conventionally treated astrocytic gliomas. J Mol Med. 2005;83:917–926. doi: 10.1007/s00109-005-0700-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard DA, Giaccone G, Johnson BE. Biomarkers of response to epidermal growth factor receptor inhibitors in Non-Small-Cell Lung Cancer Working Group: standardization for use in the clinical trial setting. J Clin Oncol. 2008;26:983–994. doi: 10.1200/JCO.2007.12.9858. [DOI] [PubMed] [Google Scholar]
- Kalinsky K, Jacks LM, Heguy A, Patil S, Drobnjak M, Bhanot UK, Hedvat CV, Traina TA, Solit D, Gerald W, Moynahan ME. PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res. 2009;15:5049–5059. doi: 10.1158/1078-0432.CCR-09-0632. [DOI] [PubMed] [Google Scholar]
- Eberhard DA, Johnson BE, Amler LC, Goddard AD, Heldens SL, Herbst RS, Ince WL, Janne PA, Januario T, Johnson DH. et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–5909. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- Shepherd FA, Tsao MS. Unraveling the mystery of prognostic and predictive factors in epidermal growth factor receptor therapy. J Clin Oncol. 2006;24:1219–1220. doi: 10.1200/JCO.2005.04.4420. author reply 1220-1211. [DOI] [PubMed] [Google Scholar]
- Lo ML, Leonardi R, Mignogna MD, Pannone G, Rubini C, Pieramici T, Trevisiol L, Ferrari F, Serpico R, Testa N. et al. Scatter factor receptor (c-Met) as possible prognostic factor in patients with oral squamous cell carcinoma. Anticancer Research. 2004;24:1063–1069. [PubMed] [Google Scholar]
- Yoshimoto K, Dang J, Zhu S, Nathanson D, Huang T, Dumont R, Seligson DB, Yong WH, Xiong Z, Rao N. et al. Development of a real-time RT-PCR assay for detecting EGFRvIII in glioblastoma samples. Clin Cancer Res. 2008;14:488–493. doi: 10.1158/1078-0432.CCR-07-1966. [DOI] [PubMed] [Google Scholar]