Abstract
Background
Xuesaitong (XST) injection, consisting of total saponins from Panax notoginseng, was widely used for the treatment of cardio- and cerebro-vascular diseases in China. This study develops a simple and global quality evaluation method for the quality control of XST.
Methods
High performance liquid chromatography-ultraviolet detection (HPLC-UV) was used to identify and quantify the chromatographic fingerprints of the XST injection. Characteristic common peaks were identified using HPLC with photo diode array detection/electrospray ionization tandem mass spectrometry (HPLC-PDA/ESI-MSn).
Results
Representative fingerprints from ten batches of samples showed 27 'common saponins' all of which were identified and quantified using ten reference saponins.
Conclusion
Chemical fingerprinting and quantitative analysis identified most of the common saponins for the quality control of P. notoginseng products such as the XST injection.
Background
Xuesaitong (XST) injection, consisting of total saponins from Panax notoginseng (Sanqi), was widely used for the treatment of cardiovascular and cerebrovascular diseases in China. As total saponins (including ginsenosides and notoginsenosides) in the XST injection are its active ingredients, quality control of total saponins in the XST injection is critical for its safety, efficacy and stability. Single or simultaneous determination of main components of the total saponin extracts from P. notoginseng using high performance liquid chromatography-ultraviolet detection (HPLC-UV) [1-5], high performance liquid chromatography-evaporative light scattering detection (HPLC-ELSD) [6], high performance liquid chromatography-mass spectroscopy (HPLC-MS) [7-13] have been reported but over half of the total saponins were not quantified in these studies due to the lack of saponin references or poor chromatographic resolution. A comprehensive and systematic quality control of saponin extracts is much needed.
Fingerprint analysis is currently developed for quality control in Chinese medicine [14-26] and has been accepted by the WHO for the assessment of herbal medicines [27]. The State Food and Drug Administration (SFDA) of China requires all herbal medicine-derived injections and related materials to use chromatographic fingerprints [28] in standardization.
This article reports a novel fingerprint analytical method for quality control of the XST injection, which may be applicable to other herbal products. Over the previous studies [1-13], the new method features the following advantages. (1) The representative fingerprints show good chromatographic separation for most of visible peaks in the chromatographic profiles at 203 nm; (2) All main saponins (27 visible peaks in chromatographic profiles) are identifiable using high performance liquid chromatography-photo diode array detection/electrospray ionization tandem mass spectrometry (HPLC-PDA/ESI-MSn) technique, ten saponin references or data from literature [8-14].
Methods
Materials and reagents
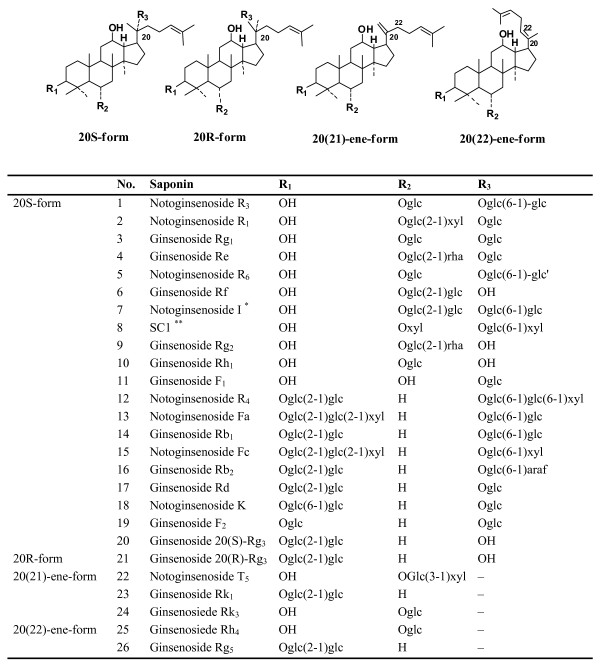
Acetonitrile and methanol (HPLC grade) were purchased from Merck (Darmstadt, Germany). Acetic acid glacial (HPLC grade) was from Tedia (Fairfield, OH, USA). The water used was purified by Milli-Q system (Millipore, USA). Reference compounds, namely notoginsenoside R1, ginsenoside Rg1, Rg2, Rh1, Rb1, Rb2, Rd, Re, 20(S)-Rg3 and 20(R)-Rg3 were purchased from Jilin University (Shenyang, China). The structures of these compounds are shown in Figure 1. Mixed standard stock solution containing accurately weighed reference compounds was directly prepared in 80% aqueous methanol (v/v). Working standard solutions were prepared by diluting the stock solution with 80% aqueous methanol (v/v) to obtain a series of concentrations for the calibration curves.
Figure 1.
Structures of the investigated saponins in P. notoginseng. glc, β-D-glucose; glc', α-D-glucosexyl, β-D-xylose; rha, α-L-rhamnose; araf, α-L-arabinose (furanose). Notoginsenoside I *, H is instead of OH (C12) in 20S-form. SC1 **, 6-O-β-D-xylopyranosyl -20-β-D-xylopyranosyl-(1→6)-β-D-glucopyranosyl dammar-24-ene-3β, 6α, 12β, 20(S)tetraol.
HPLC instrumentationadditional 1 and chromatographic conditions
An Agilent 1100 HPLC system (Agilent Technologies, USA) consisted of a quaternary solvent delivery system, an on-line degasser, an auto-sampler, a column temperature controller and ultraviolet detector coupled with an analytical workstation and an Ultimate™ XB-C18 column, 5 μm, 250 mm × 4.6 mm i.d. (Welch Materials, USA) were used in the HPLC-UV experiments. Flow rate was 1.0 ml/min and sample injection volume was 10 μl. Detection wavelength was set at 203 nm and the column temperature was at 30°C. Mobile phase contained deionized water-acetic acid (A; 100:0.01, v/v) and acetonitrile-acetic acid (B; 100:0.01, v/v). The gradient elution was as follows: 19-21.2% B at 0-30 min; 21.2-26% B at 30-35 min; 26-28% B at 35-40 min; 28-38% B at 40-50 min; 38-55% B at 50-60 min; 55% B at 60-65 min; 55-80% B at 65-70 min; 80-95% B at 70-75 min. Re-equilibrium was 10 min; the total run time was 85 min.
HPLC-MSn instrumentation and chromatographic conditions
Analysis was performed on an Agilent 1100 series LC system equipped with a binary solvent delivery system, an auto-sampler, a column temperature controller, a photo diode array detector and a Finnigan LCQ Deca XPplus ion trap mass spectrometer (Thermo Finnigan, USA) via an ESI interface. The chromatographic conditions were the same for HPLC-UV as described in the previous section. The operating parameters for MS in the negative mode were as follows: collision gas, ultrahigh-purity helium (He); nebulizing gas, high purity nitrogen (N2); ion spray voltage, -4.5 kV; sheath gas (N2) at a flow rate of 60 arbitrary units; auxiliary gas (N2) at a flow rate of 20 arbitrary units; capillary temperature, 350°C; capillary voltage, -15 V; tube lens offset voltage, -30 V. Full scan data acquisition was performed from m/z 80 to 1800 in MS scan mode. The MSn spectra were obtained with the collision energy for collision-induced dissociation adjusted to 30%-40% of maximum and the isolation width of precursor ions was 2.0Th.
Sample preparation
Ten samples of the XST injection (Batch No. 20090307, 20090510, 20090310, 20081018, 9042213, 20090312, 20090421, 20090512, 20090504, 20090203), manufactured by three Chinese pharmaceutical companies, were obtained either from pharmacies or factories. For HPLC-PDA-MSn analysis, a certain volume of the injection, according to its nominal content of total saponins, was transferred to a 50 ml volumetric flask and was diluted with 80% aqueous methanol (v/v) to obtain total saponins at a concentration of about 1 mg/ml. For HPLC-UV analysis, the injection was diluted with 80% aqueous methanol (v/v) to obtain total saponins at a concentration of about 0.5 mg/ml. Prior to analysis, the sample solutions were filtered through a 0.45 μm nylon membrane (Whatman, Britain). Spiked injection was produced by mixing sample solutions with the reference solutions at the ratio of 1:1.
Data analysis
Data analysis was carried out with Similarity Evaluation System for Chromatographic Fingerprint of Traditional Chinese Medicine (version 2004A, National Committee of Pharmacopoeia, China) recommended by the SFDA.
Results and discussion
Optimization of HPLC separation
We optimized the separation conditions including the column, mobile phase, detection wavelength, elution gradient and column temperature in this study. Four reversed-phase columns, Agilent Zorbax Eclipse SB-C18 columns (250 mm × 4.6 mm, 5 μm; 150 mm × 4.6 mm, 3.5 μm; 100 mm × 2.1 mm, 1.8 μm) and Ultimate™ XB-C18 column (250 mm × 4.6 mm, 5 μm) were tested. The results showed that all four columns obtained good peak resolutions in 75 min, 75 min, 45 min and 75 min respectively; however, only two columns with the length of 250 mm (Zorbax Eclipse SB-C18 and Ultimate™ XB-C18) produced more peaks in chromatograms. Ultimate™ XB-C18 column (250 mm × 4.6 mm, 5 μm) was selected in the fingerprint analysis due to its lower cost than Zorbax Eclipse SB-C18 column.
The effects of mobile phase composition on chromatographic separation were also studied. The cetonitrile/water system produced more sharp peaks than the methanol/water system; the addition of 0.01% acetic acid in the acetonitrile/water system further improved the peak shape. Moreover, as the retention time of some components such as ginsenoside 20(S)-Rg3 and 20(R)-Rg3 was long, gradient elution was used in HPLC analysis. Satisfactory separation was achieved in 75 min.
There was no strong absorption for most of saponins in the region of ultraviolet and visible spectra due to their structural characteristics, eg lack of conjugation groups in the molecular structures. As the end adsorption wavelength 203 nm is suitable for the assay of ginsenosides and notoginsenosides [1-5], it was selected as the detection wavelength in the experiment. Furthermore, the effects of column temperature on chromatographic separation were also examined. Four column temperatures, namely 20, 25, 30 and 35°C were tested. We found that at 30°C most peaks in chromatography had good resolutions; therefore, 30°C was chosen as the column temperature for the fingerprint analysis.
HPLC-UV fingerprinting of the XST injection
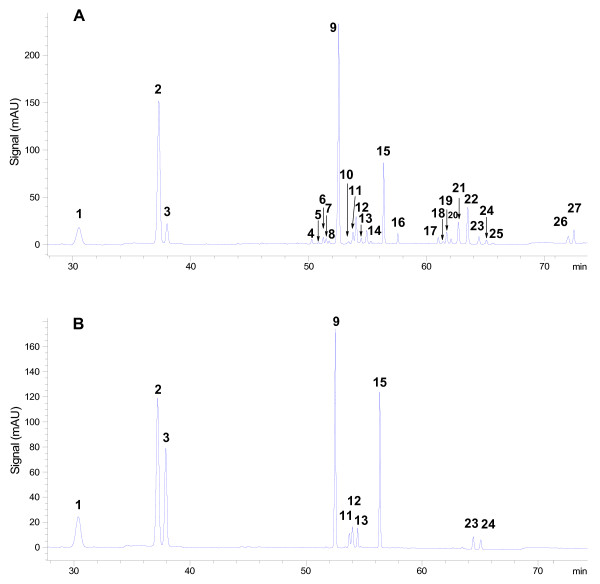
To standardize the fingerprints, we analyzed ten samples using the optimized HPLC-UV method. Peaks found in all ten samples with good resolution were assigned as 'characteristic peaks' and there were 27 characteristic peaks in the fingerprint chromatograms (Figure 2A). The software of Similarity Evaluation System for Chromatographic Fingerprint of Traditional Chinese Medicine was used to evaluate these chromatograms. To exclude the effects of the solvent and baseline fluctuation, we selected the chromatographic data of these ten samples and treated them within the time frame of 28 min to 75 min. The similarities of chromatograms for the ten samples to the reference fingerprints were established using the means of all chromatograms (Additional file 1). The results showed that the ten samples possessed similarities to the reference fingerprints (Additional file 2). While the HPLC-UV fingerprints from different batches and companies varied, the 27 characteristic peaks were common in all samples. Therefore, the detection of these common peaks in HPLC fingerprints is useful in assessing the quality of the XST injection.
Figure 2.
Chromatograms of (A) the representative fingerprint, (B) mixture standard compounds including (1) notoginsenoside R1, (2) ginsenoside Rg1, (3) ginsenoside Re, (9) ginsenoside Rb1, (11) ginsenoside Rg2, (12) ginsenoside Rh1, (13) ginsenoside Rb2, (15), ginsenoside Rd, (23) ginsenoside 20 (S)-Rg3 and (24) ginsenoside 20 (R)-Rg3.
Identification of characteristic peaks
HPLC-PDA/ESI-MSn was used for the components analysis and all 27 characteristic peaks were identified. In the ESI-MS experiment, the molecular weight of each peak was also obtained. By comparing with the ESI-MSn data and HPLC retention time of standard sanponins (Figure 2B and Additional file 3), we identified 10 peaks as notogisenoside R1, ginsenoside Rg1, Re, Rb1, Rg2, Rh1, Rb2, Rd and 20(S)-Rg3, 20(R)-Rg3. A total of 17 peaks were identified tentatively with the aid of the ESI-MSn data and HPLC retention time of some saponins from previous reports [1-13]. All the identification results are shown in Table 1. In addition, The UV spectra of all peaks in the XST injection were obtained from the PDA chromatogram (Additional file 3). The results showed that among all the peaks in the chromatogram of the XST injection no strong UV absorption within the wavelength range from 210 nm to 400 nm was obtained, suggesting that the XST injection consisted of saponins with few other natural components possessing strong UV absorption, such as flavonoids, lignins, anthraquinones and alkaloids.
Table 1.
The identification results of saponins in the XST injection by LC/MSn
| Peak No. | Identification | Retention time (min) | MS[M-H]- | MS data (m/z) |
|---|---|---|---|---|
| 1 | Notoginsenoside R1 | 34.89 | 932 | 799 [M-H-Xyl]-; 637 [M-H-Xyl-Glc]-; 475 Agl |
| 2 | Ginsenoside Rg1 | 39.32 | 800 | 637 [M-H-Glc]-; 619 [M-H-H2O-Glc]-; 475 Agl |
| 3 | Ginsenoside Re | 39.72 | 945 | 783 [M-H-Glc]-; 637 [M-H-Glc-Rha]-; 475 Agl |
| 4 | Notoginsenoside R4 | 51.24 | 1240 | 1107 [M-H-Xyl]-; 1077 [M-H-Glc]]-; 945 [M-H-Xyl-Glc; 783 [M-H-Xyl-2Glc]- |
| 5 | Ginsenoside Rf | 51.89 | 800 | 637 [M-H-Glc]]-; 475 Agl |
| 6 | Notoginsenoside Fa | 52.17 | 1240 | 1107 [M-H-Xyl]-; 1077 [M-H-Glc]]-; 945 [M-H-Xyl-Glc; 783 [M-H-Xyl-2Glc]- |
| 7 | Notoginsenoside I | 52.39 | 1092 | 929[M-H-Glc]-; 767 [M-H-2Glc]-; 605[M-H-3Glc]- |
| 8 | SC1 | 52.56 | 901 | 769 [M-H-Xyl]-; 637 [M-H-2Xyl]-; 475 Agl |
| 9 | Ginsenoside Rb1 | 53.48 | 1107 | 945 [M-H-Glc]-; 783 [M-H-2Glc]-; 621 [M-H-3Glc]-; 459 Agl |
| 10 | Notoginsenoside Fc | 54.32 | 1209 | 1077 [M-H-Xyl]-; 945 [M-H-2Xyl]-; 783 [M-H-2Xyl-Glc]-; 621 [M-H-2Xyl-2Glc]-; 459 Agl |
| 11 | Ginsenoside Rg2 | 54.75 | 783 | 637 [M-H-Rha]-; 621 [M-H-Glc]-; 475 Agl |
| 12 | Ginsenoside Rh1 | 55.04 | 637 | 475 [M-H-Glc]- |
| 13 | Ginsenoside Rb2 | 55.30 | 1077 | 945[M-H-Arap]-; 915[M-H-Glc]-; 783[M-HArap-Glc]-; 621[M-H-Arap-2Glc]-; 459 Agl |
| 14 | Ginsenoside F1 | 55.84 | 637 | 475 [M-H-Glc]- |
| 15 | Ginsenoside Rd | 57.16 | 945 | 783 [M-H-Glc]-; 621[M-H-2Glc]-; 459Agl |
| 16 | Notoginsenoside K | 58.32 | 945 | 783 [M-H-Glc]-; 621[M-H-2Glc]-; 459Agl |
| 17 | Notoginsenoside T5/Unkown | 61.70 | 752 | 619[M-H-Xyl]-; 457 Agl |
| 18 | Unkown | 62.09 | 765 | 603[M-H-Glc]- |
| 19 | Notoginsenoside T5/Unkown | 62.42 | 752 | 619[M-H-Xyl]-; 457 Agl |
| 20 | Unkown | 62.81 | 765 | 603[M-H-Glc]- |
| 21 | Ginsenoside Rk3 | 63.42 | 619 | 551 [M-H-C5H10]- |
| 22 | Ginsenoside Rh4 | 64.18 | 619 | 551 [M-H-C5H10]- |
| 23 | 20(S)-ginsenoside Rg3 | 65.14 | 783 | 621 [M-H-Glc]-; 459 Agl |
| 24 | 20(R)-ginsenoside Rg3 | 65.86 | 783 | 621 [M-H-Glc]-; 459 Agl |
| 25 | Ginsenoside F2 | 66.05 | 783 | 621 [M-H-Glc]-; 459 Agl |
| 26 | Ginsenoside Rk1 | 72.47 | 765 | 603[M-H-Glc]- |
| 27 | Ginsenoside Rg5 | 72.89 | 765 | 603[M-H-Glc]- |
Determination of the main saponins in the XST injection
As shown in Figure 2A, 27 saponins were well separated, of which 25 were potentially identified (Table 1). The ratio of total saponin peak area to all peaks (except for solvent peaks and baseline fluctuation in 0-28 min) in the chromatogram of each sample was beyond 95%. Thus, a method for quantification of the 27 saponins should provide a global and systematical evaluation for the quality control of the XST injection. However, it was difficult to obtain the reference compounds for all 27 saponins; we were only able to obtain ten, including notoginsenoside R1, ginsenoside Rg1, Re, Rb1, Rg2, Rh1, Rb2, Rd, 20(S)-Rg3 and 20(R)-Rg3. Some reports [1-3] found that the slopes of regression equations for most of the determined saponins, such as notoginsenoside R2, R4, Fa, ginsenoside Rg1, Re, Rf, Rb1, Rg2, Rh1 and Rd were approximately negatively correlated to their molecular weights by HPLC-UV at 203 nm (Additional file 4) and that the regression equations of some saponins with similar molecular weights were also close to each other under the same chromatographic condition (Additional file 5, 6, 7, 8 and 9).
Ten saponins, namely R1, ginsenoside Rg1, Re, Rb1, Rg2, Rh1, Rb2, Rd, 20(S)-Rg3 and 20(R)-Rg3 were quantitatively determined and the rest 17 saponins without standard references were semi-quantified using substitutive standard substances. The calibration curves for the quantification of each saponin were selected and listed in Table 2. The developed analytical method was successfully applied to analysis of ten batches of the XST injection. All of the 27 characteristic peaks were determined simultaneously and the results are in Table 3. In the XST injection, the content of ginsenoside Rb1 was the most (26.17%-29.60%), followed by ginsenoside Rg1 (20.50%-25.43%), Rd (6.82%-8.10%), notoginsenoside R1 (5.29%-6.89%) and ginsenoside Re (2.91%-4.92%). The total content of the five saponins made up 61.69%-71.39% of the total saponins in the XST injection (total saponins nominal: 50 mg/ml). The ten saponins with available standard substances were quantitatively determined and made up 68.46%-75.85% of the total saponins nominal. Thus, combined with the semi-quantification data, 81.81%-95.71% components in the XST injection could be examined.
Table 2.
Calibration curves, detection limits and quantification limits of the saponins by HPLC-UV
| Peak No. | Saponins | M.W. | Calibration curve a | Linear range (μg/ml) | R2 | LOD (μg/ml) |
|---|---|---|---|---|---|---|
| 21 | Ginsenoside Rk3 | 619 | y = 6.7519x - 7.6085 | |||
| 22 | Ginsenoside Rh4 | 619 | y = 6.7519x - 7.6085 | |||
| 12 | Ginsenoside Rh1 | 637 | y = 6.7519x - 7.6085 | 4.28-68.5 | 0.9993 | 2.14 |
| 14 | Ginsenoside F1 | 637 | y = 6.7519x - 7.6085 | |||
| 17 | Notoginsenoside T5/Unkown | 752 | y = 5.4845x - 4.8387 | |||
| 19 | Notoginsenoside T5/Unkown | 752 | y = 5.4845x - 4.8387 | |||
| 18 | Unkown | 765 | y = 5.4845x - 4.8387 | |||
| 20 | Unkown | 765 | y = 5.4845x - 4.8387 | |||
| 26 | Ginsenoside Rk1 | 765 | y = 5.4845x - 4.8387 | |||
| 27 | Ginsenoside Rg5 | 765 | y = 5.4845x - 4.8387 | |||
| 11 | Ginsenoside Rg2 | 783 | y = 5.6715x - 5.6679 | 3.34-53.5 | 0.9993 | 1.67 |
| 23 | 20(S)-Rg3 | 783 | y = 5.4845x - 4.8387 | 2.95-47.3 | 0.9990 | 1.48 |
| 24 | 20(R)-Rg3 | 783 | y = 5.0923x - 2.8995 | 2.63-42.0 | 0.9994 | 1.75 |
| 25 | Ginsenoside F2 | 783 | y = 5.4845x - 4.8387 | |||
| 2 | Ginsenoside Rg1 | 800 | y = 5.1367x - 76.471 | 16.64-1065 | 0.9990 | 10.29 |
| 5 | Ginsenoside Rf | 800 | y = 5.1367x - 76.471 | |||
| 8 | SC1 | 901 | y = 4.3254x - 5.0843 | |||
| 1 | Notoginsenoside R1 | 932 | y = 4.3254x - 5.0843 | 10.26-492.5 | 0.9997 | 7.42 |
| 3 | Ginsenoside Re | 945 | y = 4.4123x - 29.465 | 43.28-692.5 | 0.9993 | 4.73 |
| 15 | Ginsenoside Rd | 945 | y = 4.1199x - 5.5681 | 16.64-532.5 | 0.9993 | 4.43 |
| 16 | Notoginsenoside K | 945 | y = 4.1199x - 5.5681 | |||
| 13 | Ginsenoside Rb2 | 1077 | y = 3.8757x + 2.4182 | 4.84-77.5 | 0.9995 | 1.95 |
| 7 | Notoginsenoside I | 1092 | y = 3.8757x + 2.4182 | |||
| 9 | Ginsenoside Rb1 | 1107 | y = 3.5815x - 29.548 | 15.98-1022.5 | 0.9992 | 7.91 |
| 10 | Notoginsenoside Fc | 1209 | y = 3.5815x - 29.548 | |||
| 4 | Notoginsenoside R4 | 1240 | y = 3.5815x - 29.548 | |||
| 6 | Notoginsenoside Fa | 1240 | y = 3.5815x - 29.548 | |||
a y: peak area of analyte; x: concentration of analyte (μg/ml)
Table 3.
Contents (%) of the 27 saponins in the XST injection (total saponins nominal: 50 mg/ml) a
| Peak No. | Saponins | S1 | S2 | S3 | S4 | S5 | S6 | S7 | S8 | S9 | S10 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Notoginsenoside R1 (%) | 6.64 | 5.29 | 6.89 | 6.47 | 6.27 | 5.86 | 5.33 | 6.41 | 6.07 | 6.35 |
| 2 | Ginsenoside Rg1 (%) | 25.43 | 20.50 | 24.53 | 23.99 | 23.76 | 20.29 | 21.15 | 22.23 | 22.31 | 23.33 |
| 3 | Ginsenoside Re (%) | 3.43 | 2.91 | 4.92 | 3.61 | 3.55 | 3.56 | 3.35 | 3.04 | 3.03 | 3.69 |
| 4 | Notoginsenoside R4 (%) | 1.52 | 1.19 | 1.24 | 1.33 | 1.28 | 1.33 | 1.31 | 1.11 | 1.15 | 1.38 |
| 5 | Ginsenoside Rf (%) | 1.24 | 0.95 | 0.98 | 1.15 | 1.15 | 0.97 | 1.03 | 1.03 | 1.03 | 1.00 |
| 6 | Notoginsenoside Fa (%) | 1.45 | 1.21 | 1.90 | 1.35 | 1.44 | 1.43 | 1.35 | 1.29 | 1.29 | 1.34 |
| 7 | Notoginsenoside I (%) | 0.89 | 0.62 | 0.17 | 0.80 | 0.80 | 0.76 | 0.81 | 0.73 | 0.66 | 0.83 |
| 8 | SC1 (%) | 0.65 | 0.51 | 2.28 | 0.56 | 0.62 | 0.46 | 0.54 | 0.52 | 0.49 | 0.54 |
| 9 | Ginsenoside Rb1 (%) | 28.39 | 26.17 | 26.34 | 28.30 | 28.78 | 29.58 | 29.60 | 28.00 | 28.14 | 27.78 |
| 10 | Notoginsenoside Fc (%) | 1.30 | 0.94 | 0.99 | 1.13 | 1.12 | 1.06 | 0.98 | 1.05 | 1.05 | 1.15 |
| 11 | Ginsenoside Rg2 (%) | 1.02 | 1.31 | 1.08 | 1.18 | 0.98 | 0.78 | 1.44 | 1.38 | 1.38 | 1.17 |
| 12 | Ginsenoside Rh1 (%) | 1.77 | 3.06 | 2.25 | 2.22 | 1.65 | 1.06 | 2.90 | 3.19 | 3.22 | 2.17 |
| 13 | Ginsenoside Rb2 (%) | 1.09 | 0.69 | 2.18 | 1.07 | 1.06 | 1.00 | 0.90 | 0.81 | 1.11 | 1.04 |
| 14 | Ginsenoside F1 (%) | 0.76 | 1.77 | 0.29 | 1.14 | 0.85 | 0.50 | 1.59 | 1.90 | 1.88 | 1.24 |
| 15 | Ginsenoside Rd (%) | 7.50 | 6.82 | 7.25 | 7.22 | 7.24 | 7.27 | 8.10 | 7.41 | 7.48 | 7.18 |
| 16 | Notoginsenoside K (%) | 1.01 | 0.72 | 1.05 | 1.18 | 1.24 | 1.33 | 1.36 | 0.96 | 1.04 | 1.43 |
| 17 | Notoginsenoside T5/Unkown (%) | 0.39 | 0.69 | 0.58 | 0.69 | 0.47 | 0.39 | 0.79 | 0.87 | 0.86 | 0.83 |
| 18 | Unkown (%) | 0.30 | 0.37 | 1.11 | 0.45 | 0.36 | 0.23 | 0.56 | 0.50 | 0.50 | 0.46 |
| 19 | Notoginsenoside T5/Unkown (%) | 0.72 | 1.31 | 0.41 | 1.19 | 0.82 | 0.63 | 1.51 | 1.51 | 1.54 | 1.20 |
| 20 | Unkown (%) | 0.39 | 0.55 | 0.31 | 0.55 | 0.37 | 0.39 | 0.70 | 0.66 | 0.67 | 0.55 |
| 21 | Ginsenoside Rk3 (%) | 0.90 | 2.30 | 1.59 | 1.78 | 1.10 | 0.80 | 2.35 | 2.52 | 2.57 | 1.77 |
| 22 | Ginsenoside Rh4 (%) | 1.27 | 3.66 | 2.47 | 2.69 | 1.49 | 0.91 | 3.70 | 3.87 | 3.88 | 2.65 |
| 23 | 20S-Rg3 (%) | 0.37 | 1.01 | 0.75 | 0.81 | 0.44 | 0.43 | 1.21 | 1.09 | 1.14 | 0.83 |
| 24 | 20R-Rg3 (%) | 0.21 | 0.70 | 0.52 | 0.51 | 0.25 | 0.22 | 0.78 | 0.76 | 0.82 | 0.56 |
| 25 | Ginsenoside F2 (%) | 0.36 | 0.38 | 0.23 | 0.28 | 0.14 | 0.10 | 0.78 | 0.42 | 0.43 | 0.25 |
| 26 | Ginsenoside Rk1 (%) | 0.41 | 1.13 | 1.22 | 0.81 | 0.66 | 0.47 | 1.62 | 1.02 | 1.28 | 0.80 |
| 27 | Ginsenoside Rg5 (%) | 0.32 | 1.30 | 1.17 | 1.05 | 0.65 | 0.46 | 1.95 | 1.31 | 1.50 | 1.03 |
| Total (%) b | 89.41 | 86.78 | 93.54 | 92.47 | 87.90 | 81.81 | 95.71 | 94.27 | 95.02 | 91.50 |
a Mean values of samples (n = 3)
b Total content of the 27 saponins in samples
Conclusion
The fingerprint profiles of ten batches of samples showed 27 characteristic peaks. Ten of these 27 saponins in the XST injections were quantitatively determined with their standard references; the rest 17 saponins were semi-quantified with the substitutive standard references.
Abbreviations
XST: Xuesaitong; HPLC-UV: high performance liquid chromatography-ultraviolet detection; HPLC-PDA/ESI-MSn: HPLC with photo diode array detection/electrospray ionization tandem mass spectrometry; HPLC-ELSD: high performance liquid chromatography-evaporative light scattering detection; HPLC-MS: high performance liquid chromatography-mass spectroscopy; SFDA: State Food and Drug Administration (China)
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
XHF designed the study. HY performed the fingerprint and quantitative analysis and wrote the manuscript. PYS and QS assisted HY to identify the characteristic peaks using HPLC-PDA/ESI-MSn. All authors read and approved the final version of the manuscript.
Supplementary Material
The chromatogram of similarity analysis of the fingerprints of 10 samples.
The similarities of chromatograms of 10 samples (n = 3).
PDA Chromatograms. standard compounds (A) and a XST injection (C), and total ion current chromatograms of standard compounds (B) and a XST injection (D). 1-27 were the characteristic peaks, listed in Table 2
Plots of slopes of calibration curves vs. molecular weights (MW) of saponins. From literatures (A) [Journal of Pharmaceutical and Biomedical Analysis 41 (2006) 274-279], (B) [Journal of Pharmaceutical and Biomedical Analysis 48 (2008) 1361-1367], (C) [Journal of Pharmaceutical and Biomedical Analysis 38 (2005) 45-51], (D) [Journal of Chromatography A 1011 (2003) 77-87], (E) [Journal of Shenyang Pharmaceutical University Vol. 20, No.1 (2003) 27-31], and (F) [Chinese Pharmaceutical Journal Vol. 38, No.9 (2003) 698-699]
The method validation for simultaneous determination of the twenty-seven saponins in XST injection. The quantitative and semi-quantitative methods were validated and the semi-quantitative principle were discussed in detail.
Precisions and repeatability. The results of precision and repeatability for simultaneous determination of the twenty-seven saponins
Recovery. The results of recovery for simultaneous determination of the twenty-seven saponins
Plots of slopes of calibration curves vs molecular weights (MW) with different chromatography columns. (A) Ultimate™ XB-C18 (250 mm × 4.6 mm, 5 μm), (B) Zorbax Eclipse SB-C18 (250 mm × 4.6 mm, 5 μm) and (C) Zorbax Eclipse SB-C18 (100 mm × 2.1 mm, 1.8 μm)
Regression equation using different columns. Columns: Zorbax Eclipse SB-C18 (250 mm × 4.6 mm, 5 μm) and Zorbax Eclipse SB-C18 (100 mm × 2.1 mm, 1.8 μm)
Contributor Information
Hong Yao, Email: yauhung@126.com.
Peiying Shi, Email: peiying_shi1983@163.com.
Qing Shao, Email: shaoq@zju.edu.cn.
Xiaohui Fan, Email: fanxh@zju.edu.cn.
Acknowledgements
This work was supported by the National S&T Major Project (No. 2009ZX09502-005 & 2009ZX09311-002) and Zhejiang Provincial Natural Science Foundation, China (R2080693).
References
- Lau AJ, Woo SO, Koh HL. Analysis of saponins in raw and steamed Panax notoginseng using high-performance liquid chromatography with diode array detection. J Chromatogr A. 2003;1011:77–87. doi: 10.1016/S0021-9673(03)01135-X. [DOI] [PubMed] [Google Scholar]
- Lau AJ, Seo BH, Woo SO, Koh HL. High-performance liquid chromatographic method with quantitative comparisons of whole chromatograms of raw and steamed Panax notoginseng. J Chromatogr A. 2004;1057:141–149. doi: 10.1016/j.chroma.2004.09.069. [DOI] [PubMed] [Google Scholar]
- Li L, Zhang JL, Sheng YX, Guo DA, Wang Q, Guo HZ. Simultaneous quantification of six major active saponins of Panax notoginseng by high-performance liquid chromatography-UV method. J Pharm Biomed Anal. 2005;38:45–51. doi: 10.1016/j.jpba.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Guan J, Lai CM, Li SP. A rapid method for the simultaneous determination of 11 saponins in Panax notoginseng using ultra performance liquid chromatography. J Pharm Biomed Anal. 2007;44:996–1000. doi: 10.1016/j.jpba.2007.03.032. [DOI] [PubMed] [Google Scholar]
- Qian ZM, Wan JB, Zhang QW, Li SP. Simultaneous determination of nucleobases, nucleosides and saponins in Panax notoginseng using multiple columns high performance liquid chromatography. J Pharm Biomed Anal. 2008;48:1361–1367. doi: 10.1016/j.jpba.2008.09.038. [DOI] [PubMed] [Google Scholar]
- Wan JB, Yang FQ, Li SP, Wang YT, Cui XM. Chemical characteristics for different parts of Panax notoginseng using pressurized liquid extraction and HPLC-ELSD. J Pharm Biomed Anal. 2006;41:1596–1601. doi: 10.1016/j.jpba.2006.01.058. [DOI] [PubMed] [Google Scholar]
- Wang XY, Zhao T, Gao XF, Dan M, Zhou MM, Jia W. Simultaneous determination of 17 ginsenosides in rat urine by ultra performance liquid chromatography-mass spectrometry with solid-phase extraction. Anal Chim Acta. 2007;594:265–273. doi: 10.1016/j.aca.2007.05.032. [DOI] [PubMed] [Google Scholar]
- Lai CM, Li SP, Yu H, Wan JB, Kan KW, Wang YT. A rapid HPLC-ESI-MS/MS for qualitative and quantitative analysis of saponins in "XUESETONG" injection. J Pharm Biomed Anal. 2006;40:669–678. doi: 10.1016/j.jpba.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Li L, Tsao R, Dou JP, Song FR, Liu ZQ, Liu SY. Detection of saponins in extract of Panax notoginseng by liquid chromatography-electrospray ionisation-mass spectrometry. Anal Chim Acta. 2005;536:21–28. doi: 10.1016/j.aca.2004.12.064. [DOI] [Google Scholar]
- Li XY, Sun JG, Wang GJ, Hao HP, Liang Y, Zheng YT, Yan B, Sheng LS. Simultaneous determination of panax notoginsenoside R1, ginsenoside Rg1, Rd, Re and Rb1 in rat plasma by HPLC/ESI/MS: platform for the pharmacokinetic evaluation of total panax notoginsenoside, a typical kind of multiple constituent traditional Chinese medicine. Biomed Chromatogr. 2007;21:735–746. doi: 10.1002/bmc.813. [DOI] [PubMed] [Google Scholar]
- Liu HL, Xia L, Cao J, Li P, Qi LW. Simultaneous determination of twelve saponins in Radix et Rhizoma Notoginseng by rapid resolution LC-ESI-TOF-MS. Chromatographia. 2008;68:1033–1038. doi: 10.1365/s10337-008-0796-y. [DOI] [Google Scholar]
- Chan ECY, Yap SL, Lau AJ, Leow PC, Toh DF, Koh HL. Ultra-performance liquid chromatography/time-of-flight mass spectrometry based metabolomics of raw and steamed Panax notoginseng. Rapid Commun Mass Spectrom. 2007;21:519–528. doi: 10.1002/rcm.2864. [DOI] [PubMed] [Google Scholar]
- Dan M, Su MM, Gao XF, Zhao T, Zhao AH, Xie GX, Qiu YP, Zhou MM, Liu Z, Jia W. Metabolite profiling of Panax notoginseng using UPLC-ESI-MS. Phytochemistry. 2008;69:2237–2244. doi: 10.1016/j.phytochem.2008.04.015. [DOI] [PubMed] [Google Scholar]
- Wang XJ, Lv HT, Sun H, Jiang XG, Wu ZM, Sun WJ, Wang P, Liu L, Bi KS. Quality evaluation of Yin Chen Hao Tang extract based on fingerprint chromatogram and simultaneous determination of five bioactive constituents. J Sep Sci. 2008;31:9–15. doi: 10.1002/jssc.200700376. [DOI] [PubMed] [Google Scholar]
- Liu AH, Lin YH, Yang M, Guo H, Guan SH, Sun JH, Guo DA. Development of the fingerprints for the quality of the roots of Salvia miltiorrhiza and its related preparations by HPLC-DAD and LC-MSn. J Chromatogr B. 2007;846:32–41. doi: 10.1016/j.jchromb.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Han C, Shen Y, Chen JH, Lee FSC, Wang XR. HPLC fingerprinting and LC-TOF-MS analysis of the extract of Pseudostellaria heterophylla (Miq.) Pax root. J Chromatogr B. 2008;862:125–131. doi: 10.1016/j.jchromb.2007.11.041. [DOI] [PubMed] [Google Scholar]
- Qiao CF, Han QB, Song JZ, Mo SF, Kong LD, Kung HF, Xu HX. Chemical fingerprint and quantitative analysis of Fructus Psoraleae by high-performance liquid chromatography. J Sep Sci. 2007;30:813–818. doi: 10.1002/jssc.200600339. [DOI] [PubMed] [Google Scholar]
- Ding S, Dudley E, Plummer S, Tang J, Newton RP, Brenton AG. Fingerprint profile of Ginkgo biloba nutritional supplements by LC/ESI-MS/MS. Phytochemistry. 2008;69:1555–1564. doi: 10.1016/j.phytochem.2008.01.026. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Li SP, Wang YT, Chen XJ, Tu PF. Differentiation of Herba Cistanches by fingerprint with high-performance liquid chromatography-diode array detection-mass spectrometry. J Chromatogr A. 2009;1216:2156–2162. doi: 10.1016/j.chroma.2008.04.040. [DOI] [PubMed] [Google Scholar]
- Jin XF, Lu YH, Wei DZ, Wang ZT. Chemical fingerprint and quantitative analysis of Salvia plebeia R.Br. by high-performance liquid chromatography. J Pharm Biomed Anal. 2008;48:100–104. doi: 10.1016/j.jpba.2008.05.027. [DOI] [PubMed] [Google Scholar]
- Kong WJ, Zhao YL, Xiao XH, Jin C, Li ZL. Quantitative and chemical fingerprint analysis for quality control of Rhizoma Coptidischinensis based on UPLC-PAD combined with chemometrics methods. Phytomedicine. 2009;16:950–959. doi: 10.1016/j.phymed.2009.03.016. [DOI] [PubMed] [Google Scholar]
- Li W, Deng YL, Dai RJ, Yu YH, Saeed MK, Li L, Meng WW, Zhang XS. Chromatographic fingerprint analysis of Cephalotaxus sinensis from various sources by high-performance liquid chromatography-diodearray detection-electrospray ionization-tandem mass spectrometry. J Pharm Biomed Anal. 2007;45:38–46. doi: 10.1016/j.jpba.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Dumarey M, van Nederkassel AM, Deconinck E, Vander Heyden Y. Exploration of linear multivariate calibration techniques to predict the total antioxidant capacity of green tea from chromatographic fingerprints. J Chromatogr A. 2008;1192:81–88. doi: 10.1016/j.chroma.2008.03.052. [DOI] [PubMed] [Google Scholar]
- Teo CC, Tan SN, Yong JWH, Hew CS, Ong ES. Validation of green-solvent extraction combined with chromatographic chemical fingerprint to evaluate quality of Stevia rebaudiana Bertoni. J Sep Sci. 2009;32:613–622. doi: 10.1002/jssc.200800552. [DOI] [PubMed] [Google Scholar]
- Ni YN, Lai YH, Brandes S, Kokot S. Multi-wavelength HPLC fingerprints from complex substances: An exploratory chemometrics study of the Cassia seed example. Anal Chim Acta. 2009;647:149–158. doi: 10.1016/j.aca.2009.06.021. [DOI] [PubMed] [Google Scholar]
- Li J, Li WZM, Huang W, Cheung AWH, Bi CWC, Duan R, Guo AJY, Dong TTX, Tsim KWK. Quality evaluation of Rhizoma Belamcandae (Belamcanda chinensis (L.) DC.) by using high-performance liquid chromatography coupled with diode array detector and mass spectrometry. J Chromatogr A. 2009;1216:2071–2078. doi: 10.1016/j.chroma.2008.05.082. [DOI] [PubMed] [Google Scholar]
- World Health Organization. Guidelines for the Assessment of Herbal Medicines. WHO, Munich, Geneva; 1991. [Google Scholar]
- State Food and Drug Administration of China. Technical Requirements for the Development of Fingerprints of TCM Injections. SFDA, Beijing; 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The chromatogram of similarity analysis of the fingerprints of 10 samples.
The similarities of chromatograms of 10 samples (n = 3).
PDA Chromatograms. standard compounds (A) and a XST injection (C), and total ion current chromatograms of standard compounds (B) and a XST injection (D). 1-27 were the characteristic peaks, listed in Table 2
Plots of slopes of calibration curves vs. molecular weights (MW) of saponins. From literatures (A) [Journal of Pharmaceutical and Biomedical Analysis 41 (2006) 274-279], (B) [Journal of Pharmaceutical and Biomedical Analysis 48 (2008) 1361-1367], (C) [Journal of Pharmaceutical and Biomedical Analysis 38 (2005) 45-51], (D) [Journal of Chromatography A 1011 (2003) 77-87], (E) [Journal of Shenyang Pharmaceutical University Vol. 20, No.1 (2003) 27-31], and (F) [Chinese Pharmaceutical Journal Vol. 38, No.9 (2003) 698-699]
The method validation for simultaneous determination of the twenty-seven saponins in XST injection. The quantitative and semi-quantitative methods were validated and the semi-quantitative principle were discussed in detail.
Precisions and repeatability. The results of precision and repeatability for simultaneous determination of the twenty-seven saponins
Recovery. The results of recovery for simultaneous determination of the twenty-seven saponins
Plots of slopes of calibration curves vs molecular weights (MW) with different chromatography columns. (A) Ultimate™ XB-C18 (250 mm × 4.6 mm, 5 μm), (B) Zorbax Eclipse SB-C18 (250 mm × 4.6 mm, 5 μm) and (C) Zorbax Eclipse SB-C18 (100 mm × 2.1 mm, 1.8 μm)
Regression equation using different columns. Columns: Zorbax Eclipse SB-C18 (250 mm × 4.6 mm, 5 μm) and Zorbax Eclipse SB-C18 (100 mm × 2.1 mm, 1.8 μm)