Abstract
Cofilin is a major cytoskeletal protein that binds to both monomeric (G-) and polymeric (F-) actin and is involved in microfilament dynamics. Although an atomic structure of the G-actin-cofilin complex does not exist, models of the complex have been built using molecular dynamics simulations, structural homology considerations, and synchrotron radiolytic footprinting data. The hydrophobic cleft between actin subdomains 1 and 3 and, alternatively, the cleft between actin subdomains 1 and 2 have been proposed as possible high affinity cofilin binding sites. In this study, the proposed binding of cofilin to the subdomain 1/3 region on G-actin has been probed using site-directed mutagenesis, fluorescence labeling, and chemical cross-linking with yeast actin mutants containing single reactive cysteines in the actin hydrophobic cleft and cofilin mutants carrying reactive cysteines in the regions predicted to bind to G-actin. Mass spectrometric analysis of the cross-linked complex revealed that cysteine 345 in subdomain 1 of mutant G-actin was cross-linked to native cysteine 62 on cofilin. A cofilin mutant that carried a cysteine substitution in the α3 helix (residue 95) formed a cross-link with residue 144 in actin subdomain 3. Distance constraints imposed by these cross-links provide experimental evidence for cofilin binding between actin subdomains 1 and 3 and fit a corresponding, docking-based structure of the complex. The cross-linking of the N-terminal region of recombinant yeast cofilin to actin residues 346 and 374 with dithio-bis-maleimidoethane (DTME, 12.4 Å) and via disulfide bond formation was also documented. This set of cross-linking data confirms the important role the N-terminal segment of cofilin in the interactions with G-actin.
Keywords: actin, cofilin, cross-linking, molecular docking
INTRODUCTION
Rapid remodeling of the actin cytoskeleton is critical to the development and function of eukaryotic organisms. The ADF/cofilin family proteins are key regulators of actin dynamics in vivo and are essential for cytokinesis, phagocytosis, endocytosis and other cellular processes that depend on actin dynamics (1 ;2). On the other hand, ADF/cofilin family members are also involved in the development of pathologies such as Hirano bodies formation, Williams syndrome, ischemic kidney disease, and ADF/cofilin rod formation associated with Alzheimer disease (2). Altered cofilin expression levels have been detected in some cancer tissues, and recently cofilin has been shown to play a role in the progression of apoptosis (2 ;3).
Several lines of evidence support the assumption that ADF/cofilin effects in vivo depend on its local concentration. It was shown that at optimal low concentrations, cofilin severs actin filaments (4 ;5), promoting dissociation of actin protomers from the pointed ends (6). However, the ability of cofilin to accelerate pointed-end depolymerization of actin filaments is still debated (6 ;7). Surprisingly, at high cofilin concentrations filament severing appears inhibited, filaments are stabilized (5 ;6), and cofilin even favors the nucleation of new filaments probably by stabilizing the longitudinal dimer, which is the first intermediate in the spontaneous assembly of actin (6 ;8). Electron microscopy reconstruction and solution studies revealed the unique ability of ADF/cofilins to change the filament twist by ~5° per protomer and weaken the longitudinal contacts in F-actin in a cooperative manner (9–12). This destabilizing effect appears to affect cooperatively more than 100 protomers upon binding of one cofilin molecule (11 ;13 ;14), but non-cooperative changes in the tilt of actin filament have been observed as well (10).
Such different modes of cofilin interaction with actin make it essential to develop a structural understanding of the actin-cofilin complex. To date, an atomic structure of the complex has not been reported. The nucleation activity of ADF/cofilin (at high concentrations) and its ability to rescue the polymerization of some assembly-incompetent actin derivatives complicates the crystallization efforts (6 ;15). In the absence of X-ray crystallographic data, site directed mutagenesis, NMR and synchrotron radiolytic footprinting have been employed successfully to probe the actin-binding interface on cofilin (16–18). However, data related to the cofilin interface on actin have led to different models of cofilin binding to F- and G-actin.
Electron microscopy (EM) studies revealed two binding modes for cofilin on F-actin. One cofilin molecule appears to interact with two actin protomers within the filament: with an upper protomer at the hydrophobic cleft between subdomains 1 and 3, and with a lower protomer at the interface formed by subdomains 1 and 2 (10). The earlier observation that ADF can bind to F-actin with a stoichiometry ~1.3:1 was subsequently confirmed by EM reconstruction of actin filaments decorated with cofilin, which suggested the existence of a second (weak) interaction site within subdomain 1 (11 ;19). Alternative regions on F-actin have been implicated in cofilin binding based on competition experiments with gelsolin segment 2 and peptide mimetic assays (20–23).
The exact location of a high affinity binding site for cofilin on G-actin is still disputed. On the basis of structural homology between ADF/cofilins and gelsolin segment 1, it has been suggested that these proteins share the same binding site on G-actin, at the hydrophobic cleft between subdomains 1 and 3 (24–26). However, systematic mutagenesis study has shown that in comparison with gelsolin segment 1 (GS1) the residues important for cofilin-actin interaction are distributed more widely on the surface of cofilin molecule and are located in regions other than those at the corresponding positions in the GS1 structure (16). Moreover, recent synchrotron radiolysis footprinting data appeared to position the main cofilin binding site at the cleft between subdomains 1 and 2 on G-actin (27). The presence of a weak second (cryptic) binding site for cofilin on G-actin in the subdomain 2 region was shown by peptide overlapping array and chemical cross-linking methods (28 ;29).
In the absence of experimental evidence for the proposed binding of cofilin to the hydrophobic cleft between subdomains 1 and 3, the goal of this study was to test for such a binding and to obtain distance-based experimental constraints for independent molecular docking of cofilin to actin. This has been done by mapping the distances between selected residues on actin and cofilin using chemical cross-linking and mass spectrometric analysis of the cross-linked peptides. To ensure a matching system of interacting proteins, and to avoid possible artifacts of a heterologous system, both actin and cofilin were of yeast origin.
On the basis of molecular docking of cofilin to G-actin, as constrained by the results of the cross-linking data and in agreement with earlier models of the complex (24–26), we confirm binding of cofilin within the hydrophobic cleft between subdomains 1 and 3 on G-actin. The fluorescence experiments are consistent with the cross-linking data and show that the cofilin-binding interface on actin is different in detail from that for GS1 and KabC.
RESULTS
Choice of mutation sites
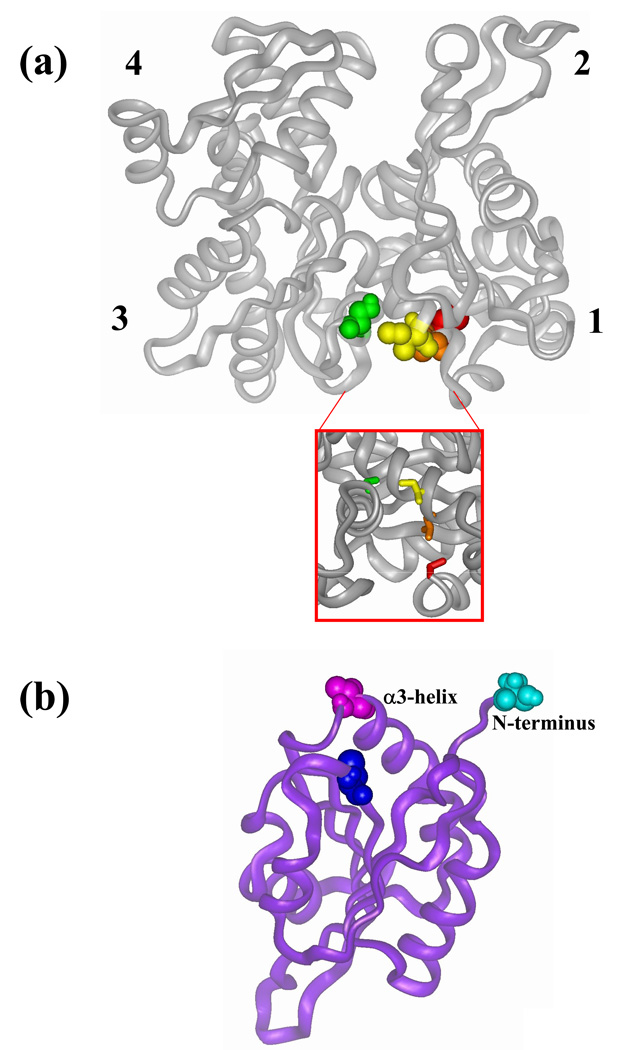
To assess the possibility of cofilin binding in the hydrophobic cleft between subdomains 1 and 3 on G-actin, we employed actin and cofilin mutants that could test the predictions of this model. We used three site-directed actin mutants containing a single reactive cysteine in that cleft and a native cysteine 374 mutated to alanine. These mutants allowed for the cross-linking and attachment of fluorescent probes to specific sites on actin (Figure 1(a)). The mutants included C144 (S144C/C374A), which has a reactive cysteine within helix 137–145 in subdomain 3, and mutants C345 (I345C/C374A) and C346 (L346C/C374A) on the opposite side of the hydrophobic cleft in which isoleucine 345 and leucine 346 in helix 335–348 in subdomain 1 were replaced with cysteine. Yeast actin mutant C345 was previously employed to study the actomyosin interface. It was shown to polymerize into F-actin and bind to skeletal S1 (30). Actin mutants C144 and C346 are characterized for the first time in this work.
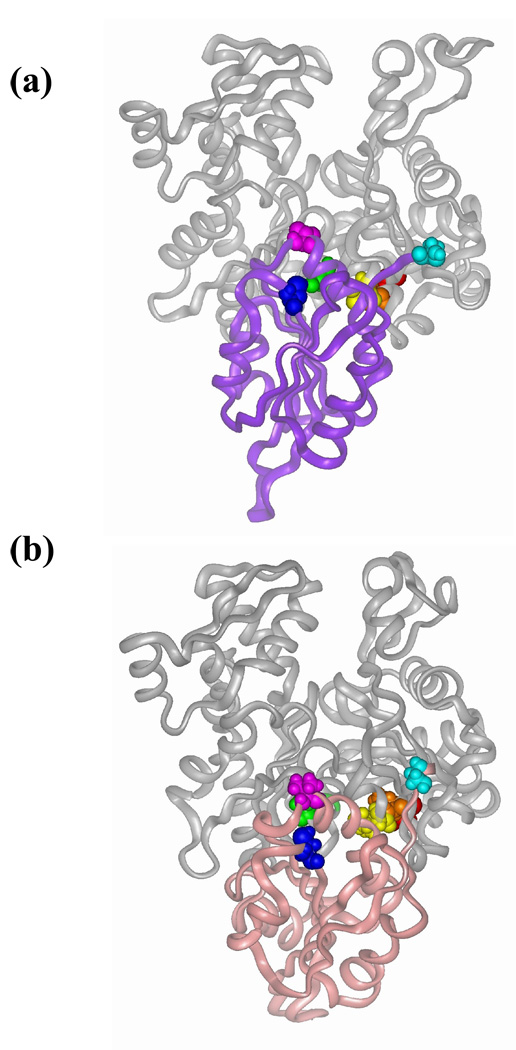
Figure 1.
Location of mutated residues on yeast (Saccharomyces cerevisieae) actin (a) and cofilin (b). Wild-type residues 374 (on actin) and 62 (on cofilin) are highlighted in red and blue, respectively. (a). Residues mutated on actin to cysteine are color coded as follows: Ile 345 in yellow, Leu 346 in orange and Ser 144 in green. Actin subdomains are numbered in the figure. Inset: the hydrophobic cleft of actin. (b) Valine 95 on cofilin is colored in purple. Serine 2 on cofilin N-terminus is colored in cyan. The last five residues at the N-terminus of cofilin were added by homology modeling to cofilin crystal structure (1COF) from Schizosaccharomyces pombe (2I2Q) and to human cofilin NMR structures (1Q8G and 1Q8X) using InsightII (Accelrys Inc.).
The cofilin residues targeted for mutation to cysteine were chosen on the basis of prior indications that the N-terminal segment, the α3 helix, and the region connecting strand β6 and helix α4 are critical for actin-cofilin interactions (16–18). The mutations included the addition of an extra cysteine residue at the N-terminus (cofC1*) and the substitution V95C in the α3 helix (cofC95) with the native cysteine 62 replaced by serine in both cases. The replacement of cysteine 62 with serine was made to achieve specific labeling and/or cross-linking of the engineered cysteine residues. This was found to be necessary because the native Cys 62 was thiol-reactive, especially in the actin-cofilin complex (see Materials and Methods).
Functional properties of actin and cofilin mutants
The rates of nucleotide (ε-ATP) exchange in the C346 and C144 mutants were similar to the exchange rate for WT actin (Table 1). In the C345 actin the nucleotide exchange was slowed by a factor of 2 compared to that in WT (Table I). These data show that single cysteine substitutions in the hydrophobic cleft, together with the replacement of cysteine 374 by alanine, had only a small, if any, effect on the nucleotide exchange rate (within a factor of 2).
Table 1.
Functional properties of actin and cofilin mutants
| Actin mutants | |||
|---|---|---|---|
| mutant | kobs × 103 | Kd, µM | |
| actin | cofilin | (without cofilin), s−1 | |
| WT | WT | 16.21 | 0.162±0.052 |
| C345 | WT | 8.67 | 0.205±0.021 |
| C346 | WT | 19.60 | 7.750±1.770 |
| C144 | WT | 16.83 | 0.116±0.047 |
| Cofilin mutants | |||
| WT | cofC95 | 0.443±0.043 | |
| WT | cofC1* | 0.459±0.049 | |
Kd values were determined from plots of kobs, rate constants of nucleotide exchange in G-actin, vs cofilin concentration (Figure 2).
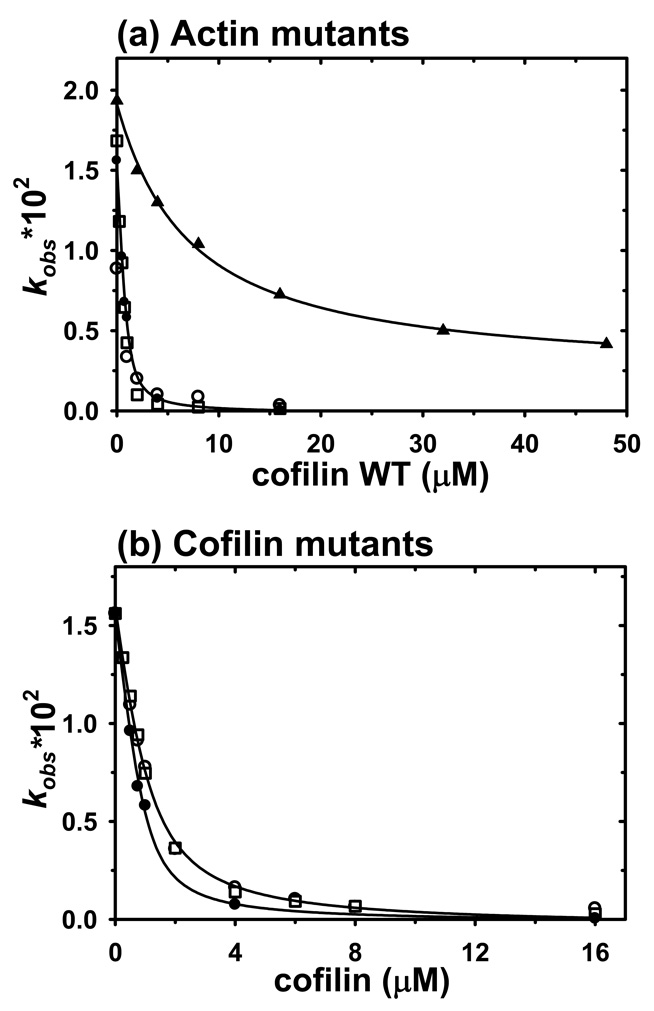
The nucleotide exchange assay was used also to determine the Kd values for cofilin binding to actin mutants (19 ;31) (Figure 2(a)). Kd values of 0.16, 0.12 and 0.21 µM were calculated for WT actin, C144 and C345, respectively, using nonlinear least squares fitting of the observed exchange rates over a range of cofilin concentrations (Table 1). These Kd values are comparable with previously published data (19). However, the inhibition of ε-ATP exchange by cofilin was much weaker for the C346 mutant than for C345, C144 and WT, and yielded a Kd = 7.75 µM, which indicated much lower affinity of cofilin to this actin. Because of the reduced affinity, the formation of the C346-cofilin complexes under our experimental conditions was confirmed by native PAGE for each preparation.
Figure 2.
Inhibition of nucleotide exchange in G-actin by WT-cofilin and cofilin mutants. Kd values for actin-cofilin complexes were determined by nonlinear least-squares fitting of rate constant data (solid lines) (a) Effect of WT-cofilin on rate constants of ε-ATP exchange on G-actin (1–2 µM) upon addition of a 100-fold excess of ATP. WT actin control (filled circles), C345 (open circles), C144 (open squares), C346 (filled triangles). (b) Effect of WT cofilin and mutants on rate constants of ε-ATP release from WT-G-actin (1 µM) upon addition of 100 µM ATP. WT cofilin control (filled circles), cofilin-95C (open circles) and cofilin-C1* (open squares)
The polymerization ability of actin mutants, as tested by light scattering, high-speed pelleting experiments and EM, was similar to that of WT yeast actin, except that the C346 mutant appeared to form shorter filaments compared to WT (data not shown).
Inhibition of nucleotide exchange on WT actin was used also as the main assay for the functional properties of cofilin mutants. For cofC1* and cofC95 the Kd-s for WT G-actin increased approximately by a factor of 2 compared to WT cofilin (~0.45 µM vs 0.16 µM for WT) (Figure 2(b), Table 1).
Fluorescence experiments
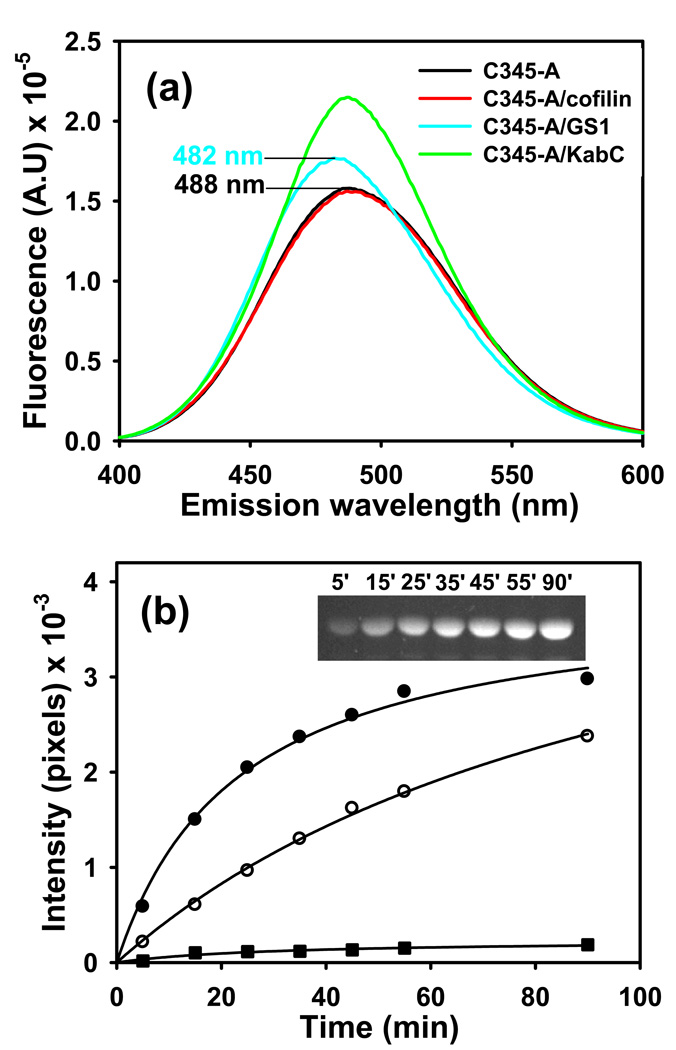
To compare the effects of cofilin, GS1 and KabC on the environment of actin residue C345, we used acrylodan as a thiol probe. The fluorescence of acrylodan-labeled C345 G-actin increased by 12 and 36% upon GS1 and KabC binding, respectively. A small (~6 nm) blue shift appeared upon GS1 binding to actin, indicating that the label was protected from solvent, which is consistent with the atomic structure of the G-actin-GS1 complex (32). In contrast, no spectral changes were detected upon binding of cofilin to acrylodan-labeled C345 actin (Figure 3(a)). Similarly, cofilin had no significant effect on the spectra of acrylodan labeled C144 and C346 G-actin. Native gel analysis confirmed the binding of cofilin to the acrylodan labeled actins, but indicated also a decreased affinity of cofilin for the labeled C345 mutant (data not shown).
Figure 3.
Fluorescence emission spectra of acrylodan-labeled C345 yeast G-actin and its complexes with actin-binding factors. (a) C345 actin mutant labeled with acrylodan (5 µM) in the absence (black) and presence of 5 µM yeast cofilin (red), gelsolin segment 1 (blue) and KabC (green). (b) Time course of acrylodan modification of C345 yeast G-actin (5 µM) in the absence (filled circles) and presence 7.5 µM of yeast cofilin (open circles) and gelsolin segment 1 (filled squares). In the presence of KabC the labeling by acrylodan was below our detection limit. The modification was started with the addition of 5 µM acrylodan to the sample, and was carried out at 24 °C. Aliquots of the samples were withdrawn at selected time points and the labeling was stopped by adding 2 mM NEM. Reaction mixtures were examined by 12% SDS PAGE and the labeled actin was visualized under UV light as shown for G-actin C345, control (inset).
To test more directly the environment of Cys 345 on actin, unperturbed by the attached label, we also monitored the modification of this residue in mutant G-actin in the absence and presence of cofilin, GS1 and KabC (Figure 3(b)). The data (Figure 3(b)) shows that cofilin protected actin cysteine 345 from labeling by acrylodan, but not to the same extent found for GS1 and KabC. The interaction of actin residue 345 with GS1 and KabC is known from the crystal structures of these complexes (18 ;32 ;33). The much smaller effect of cofilin on C345 labeling provides additional evidence for some differences in the binding of cofilin to actin compared to that of GS1 and KabC.
Cross-linking of cofilin to C345, C346 and WT G-actin with dithio-bis-maleimidoethane (DTME)
In general, the cross-linking efficiency depends on the reactivity of the modified residues, stability of the cross-linking reagent in the solution, and experimental conditions (pH, temperature, reaction time). The competition for the thiol specific reagent between free proteins and protein complex in the reaction mixture may affect the cross-linking yield as well. For purification of the cross-linked species the conditions were optimized to achieve maximum yield of actin-cofilin heterodimers.
Preliminary experiments showed that Cys 345 and Cys 346 on G-actin can be efficiently cross-linked to WT cofilin using N`N`-p-phenylenedimaleimide (p-PDM, 9.2 - 12.3 Å (34)). Similar experiments showed that N`N`-o-phenylenedimaleimide (o-PDM, 7.7 – 10.5 Å (34)) can cross-link WT cofilin to yeast WT G-actin. Initial attempts to identify the cross-linking sites on cofilin using combined liquid chromatography-electrospray mass spectrometry and tandem mass spectrometry (LC-MS/MS) of trypsin cleaved peptides were unsuccessful, probably because of the hydrophobicity of the cross-linked peptides (unpublished results).
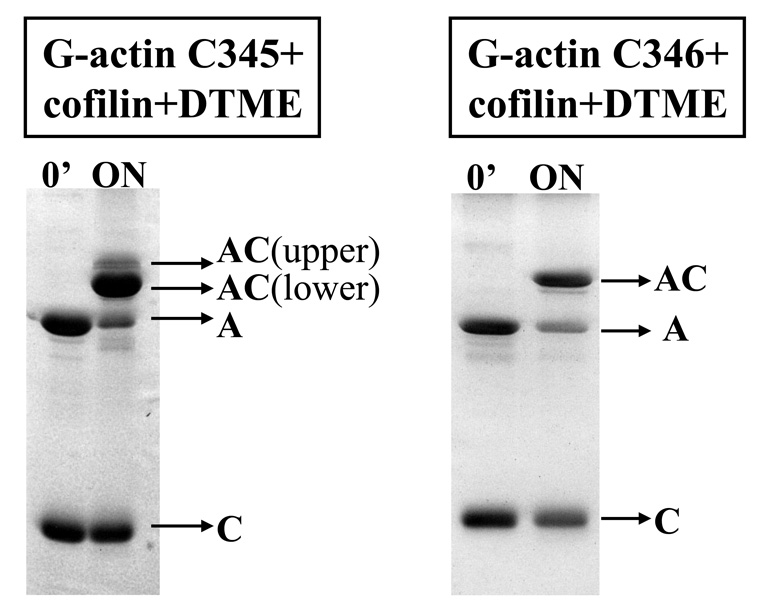
To identify the cross-linking sites, we employed a cleavable reagent, dithio-bis-maleimidoethane (DTME, 6.7 – 16.1 Å (34)), which is similar in length to o- and p-PDM. After DTME reduction, the thio-bis-maleimidoethane (TME) labeled tryptic peptides were traced by the mass increase of 157 Da. The cross-linking patterns of WT cofilin and actin (mutants and WT) with DTME were the same as those previously observed with o- and p-PDM. However, the advantage of the cleavable reagent is that it reduces the complexity of mapping relatively large cross-linked species to the task of identifying much smaller TME tagged peptides, after reagent cleavage and protein separation. In the case of the C345 actin mutant, two populations of actin-cofilin heterodimers were detected by SDS PAGE: the higher (major population) and lower mobility (minor population) complexes. The cross-linking of WT cofilin to WT yeast G-actin (data not shown) and to the C346 mutant yielded a single band with migration consistent with actin-cofilin heterodimer (Figure 4). After DTME cleavage, TME-modified cofilin was separated from actin and purified for mass spectrometry analysis (see Materials and Methods).
Figure 4.
Cross-linking of WT cofilin to C345 and C346 actin mutants with dithio-bis-maleimidoethane (DTME). A, actin; C, cofilin, AC, actin-cofilin heterodimer. The cross-linking was carried out at 4 °C, overnight.
Actin residue 345 (subdomain 1, 338-348 helix)
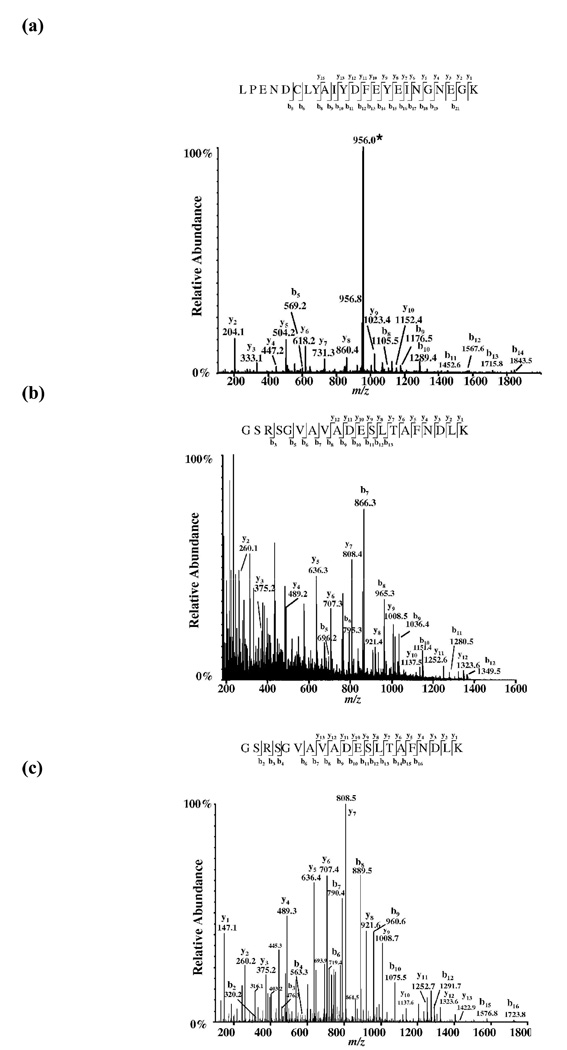
To localize the cross-linking site on cofilin by mass spectrometry, the TME-modified cofilin was purified and digested with Lys-C and trypsin. Cleavage products were identified and sequenced by ESI-MS/MS. A unique peptide that did not appear in the control (WT cofilin digest) corresponded to the mass of a cofilin peptide spanning through residues 57 to 79 (monoisotopic mass 2708.19 Da) plus the mass of the TME fragment derived from DTME reduction (monoisotopic mass 157.02 Da). Collisionally-activated dissociation of the [M+3H]3+ precursor ion at m/z 956.0, and mass measurement of the resulting fragments (tandem mass spectrometry, MS/MS) yielded an extensive y- and b- ions series that assigned cysteine 62 on cofilin as the residue cross-linked to Cys 345 on yeast actin mutant (Figure 5(a)).
Figure 5.
Mass spectrometry peptide sequencing of the TME-modified yeast cofilin peptides. (a) Identification of the G-actin C345-cofilin cross-link by mass spectrometry sequencing of the TME modified cofilin peptide (57–79). Cysteine 62 carries the TME moiety and therefore is the residue cross-linked to cysteine 345 on mutant actin. The triply-charged precursor ion is denoted with an asterisk (*). (b) Identification of the G-actin C346-cofilin cross-link. ESI-MS/MS spectrum of the [M+3H]3+ parent ion at m/z 763.4 from yeast cofilin peptide 1–20 is modified with TME on glycine 1. Top down MS/MS performed on the (M+8H)8+ ion at m/z 2008.34 of the intact TME-cofilin yielded a diagnostic b3 product ion. In both cases b and y fragment ions were consistent with TME-cofilin sequence. (c) Identification of WT yeast G-actin-cofilin cross-link. ESI-MS/MS product ion mass spectrum of the modified yeast cofilin peptide precursor ion [M+3H]3+ at m/z 738.1. Glycine 1 is TME modified and cross-links residue Cysteine 374 on WT actin.
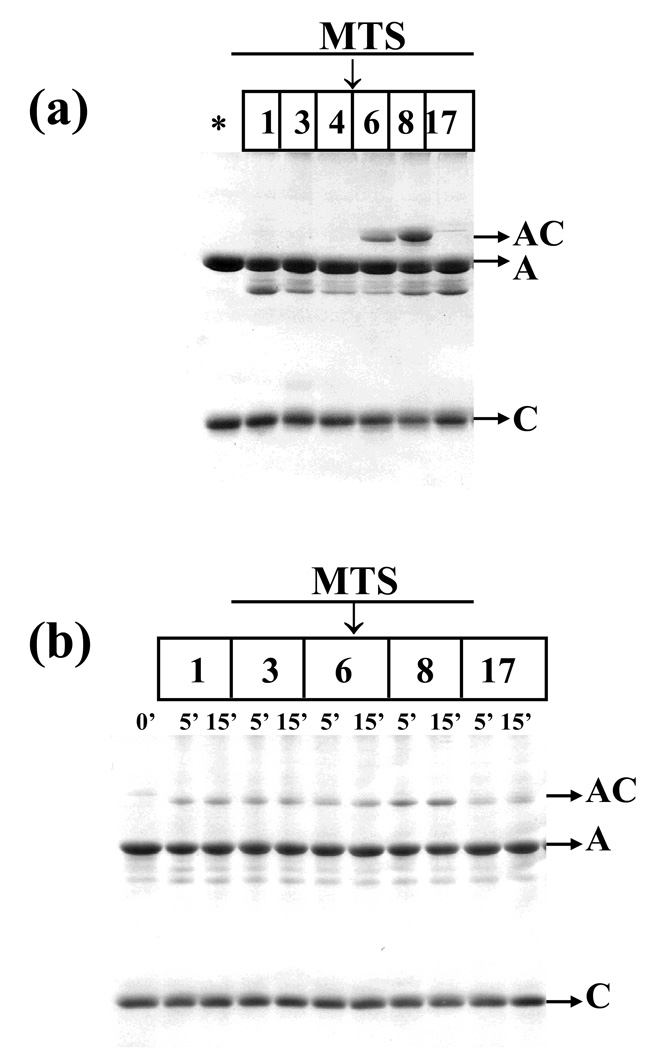
To assess the distance range between cysteine 62 on cofilin and cysteine 345 on actin, MTS reagents of different mean lengths were used as molecular rulers. The shorter reagents, i. e., MTS 1 (5.4 Å), 3 (6.4 Å) and 4 (7.8)(35) did not yield any product with a mass corresponding to the actin-cofilin heterodimer, but the longer reagents, MTS 6 (9.6 Å)(36) and 8 (12.1 Å)(36), efficiently cross-linked cofilin to C345 G-actin. The ‘longest’ reagent used, MTS 17 (19.0 Å), also failed to yield any cross-linking, probably because of its larger size, and/or dynamic states that may be incompatible with bridging Cys 345 on actin to Cys 62 on cofilin (Figure 6(a)). Taken together, the experiments with DTME and MTS reagents allowed an estimation of the distance between residues 62 on cofilin and 345 on actin within the range of 9.6 – 12.1 Å.
Figure 6.
Cross-linking of cofilin to actin mutant C345 with MTS reagents. (a) Cross-linking of actin C345 (5 µM) to WT cofilin (5 µM) with the MTS 1, 3, 4, 6, 8 and 17 reagents (10 µM). The control, in the absence of MTS, is denoted with an asterisk (*). The reaction was carried out for 30 min., on ice. (b) Cross-linking of actin C345 (10 µM) to cofilin mutant cofC1* (10 µM) with MTS 1, 3, 6, 8 and 17 (10 µM). The control, in the absence of MTS, is denoted with an asterisk (*). The reaction was carried out over 5–15 min. in buffer A at room temperature. A, actin; C, cofilin, AC, actin-cofilin heterodimer.
The site of the cross-linking for a minor population of cross-linked species of C345 actin-cofilin, which was present in the DTME reaction (upper AC in Figure 4), was not identified by mass spectrometry analysis because of insufficient amounts of material. In view of the results of C346 actin cross-linking (below) and the flexibility of cofilin’s N-terminal segment we hypothesized that this minor population might correspond to actin C345 cross-linked to the N-terminal segment. To test this possibility, a cofilin mutant containing an extra cysteine residue at the N-terminus, cofC1* was used. This mutant was cross-linked at low efficiency to C345 actin with all MTS reagents tested (MTS 1, 3, 6, 8 and 17) (Figure 6(b)), yielding small amounts of actin-cofilin heterodimer with electrophoretic mobility similar to that of the minor population of DTME cross-linked C345 actin-cofilin species. This result shows that the cross-linking of cysteine 345 to the cofilin N-terminus is indeed possible, but occurs in a small fraction of the actin-cofilin complex population.
Actin residue 346 (subdomain 1, 338–348 helix)
Using the above described protocol also for the cross-linking C346 actin to cofilin with DTME, we observed very efficient production of an actin-cofilin heterodimer, similar to that in the C345 actin reaction (Figure 4). However, in contrast to the C345 mutant, no C346 actin cross-linking to WT cofilin with the MTS reagents was detected, suggesting that cofilin’s native cysteine 62 was not involved in this reaction.
The site of the DTME anchor on cofilin was also mapped using nano-ESI-MS/MS. The TME-containing tryptic peptide was sequenced by acquiring a product ion spectrum on the triply charged precursor ion [M+3H]3+ at m/z 763.4 (Figure 5(b)). Based on the assigned b3 product ion, the cross-linker is localized to the first three residues of the peptide (GSR).
The b3 ion was also observed also in a “top down” MS/MS experiment, in which, after FPLC purification, the full-length TME-modified cofilin was submitted to collision-activated dissociation MS/MS (data not shown). Among the three first residues (GSR), the free N-terminus is most likely the group involved in the cross-linking.
Actin residue 144 (subdomain 3, helix 137–145)
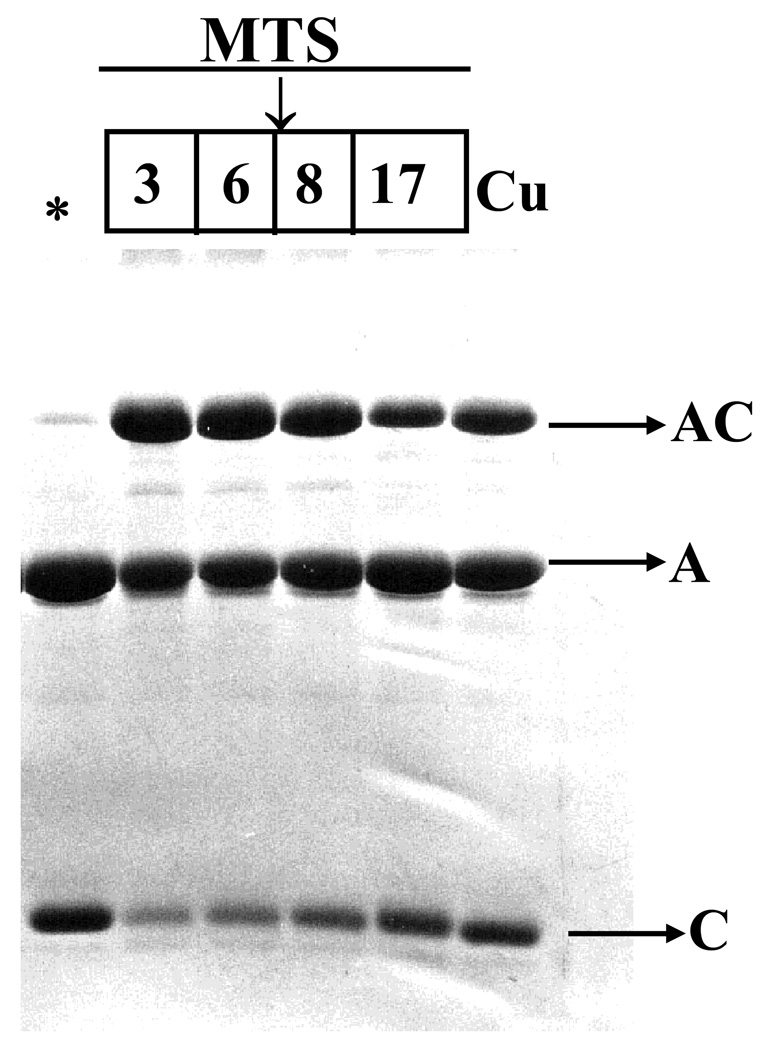
To probe cofilin interaction with actin 137–144 helical region, we used actin C144 and a cofilin mutant, cofC95, with a single reactive cysteine in the α3-helix. Cross-linking with MTS reagents of different length and copper-catalyzed oxidation indicated the proximity of residue 144 on actin to residue 95 on cofilin. As detected by SDS PAGE, a heterodimer of actin-cofilin formed with high efficiency within 5 min of reaction at room temperature. MTS 3 (6.4 Å) and 6 (9.6 Å) gave the highest cross-linking yield. Significant amounts of actin-cofilin heterodimer were also detected in the case of MTS 8 (12.1 Å) and copper-catalyzed air oxidation (zero length cross-link) (Figure 8). These experiments indicated that the distance between residues 144 on actin and 95 on cofilin is shorter than that between cysteine 62 (on cofilin) and cysteine 345 (on actin). These cross-linking results are summarized in Table 2.
Figure 8.
Cross-linking of cofC95 mutant to C144 on G-actin with MTS reagents and upon copper-catalyzed air oxidation. The cross-linking reaction was initiated with the addition of 9 µM of MTS 3, 6, 8, 17 or CuSO4 to the complex (9 µM). The control, without MTS reagents or copper, is marked with an asterisk (*). The reaction was carried out for 5 min. in buffer A, at 22 °C. The cross-linking was stopped with 2 mM NEM. A, actin; C, cofilin, AC, actin-cofilin heterodimer.
Table 2.
Summary of cross-linking reactions
| AA on cofilin | AA on actin | Reagent | Distance range, Å(34 ;36) | Average distance, Å |
|---|---|---|---|---|
| C1 | C374 | CuSO4 | 0 | 0 |
| G1 | o-PDM | 7.67 –10.47 | 9.39 | |
| DTME | 6.68 –16.12 | 12.43 | ||
| G1 | C346 | pPDM | 9.20 – 12.29 | 11.13 |
| DTME | 6.68–16.12 | 12.43 | ||
| C62 | C345 | pPDM | 9.20 – 12.29 | 11.13 |
| DTME | 6.68 – 16.12 | 12.43 | ||
| MTS6 | 3.00 – 12.20 | 9.60 | ||
| MTS8 | 9.00 – 13.80 | 12.10 | ||
| C95 | C144 | CuSO4 | 0 | 0 |
| MTS3 | 5.70 – 7.70 | 6.38* | ||
| MTS6 | 3.00 – 12.20 | 9.60 | ||
| MTS8 | 9.00 – 13.80 | 12.10 | ||
| MTS17 | 2.90 – 24.30 | 19.01* |
Our unpublished data
Actin residue 374 (subdomain 1, C-terminus)
LC-MS/MS analysis was also used to identify the cofilin residue that was cross-linked by DTME to cysteine 374 on WT G-actin. Online HPLC-ESI-MS of TME-cofilin trypsin-cleavage products revealed a unique [M+3H]3+ peptide at m/z 738.1. The molecular weight calculated from this ion is consistent with the N-terminus peptide of cofilin (theoretical monoisotopic mass 2036.01), linked with TME (monoisotopic mass 157.01) and with one maleimide ring hydrolyzed. The fragment ion spectrum yielded an extensive y-ion series across the N-terminal peptide and was consistent with a modification on glycine 1 (Figure 5(c)).
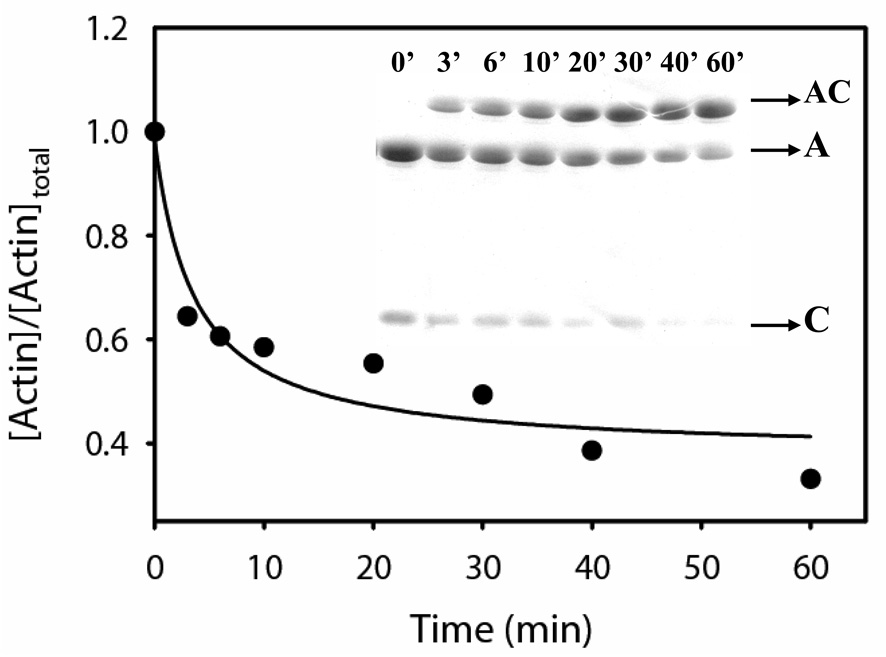
To estimate the proximity between the N-terminus of cofilin and the C-terminus of WT G-actin, we used the cofC1* mutant and monitored disulfide cross-linking between residue 1 on cofilin and 374 on actin upon copper-catalyzed air oxidation (Figure 7). Efficient zero length cross-linking was observed in this reaction, which was 50% completed in approximately 14 min (Figure 7).
Figure 7.
Disulfide cross-linking between cystein 374 on WT-G-actin and cystein 1 on cofC1* mutant. The cross-linking was initiated with the addition of 10 µM CuSO4 to the actin-cofilin complex (10 µM). The reaction was carried out for 60 min. in buffer A, at 22 °C. Aliquots of the sample were withdrawn at selected time points and the reaction was stopped with 2 mM NEM. Reaction products were examined on SDS PAGE (inset) and the amount of uncross-linked actin was estimated using the Scion Image software. A, actin; C, cofilin, AC, actin-cofilin heterodimer.
Docking of cofilin structure to G-actin
To model the G-actin-cofilin complex by docking their atomic structures, the G-actin coordinates were taken from the co-crystal of yeast G-actin-human gelsolin segment I complex (PDB ID 1YAG)(37). The crystal structure of yeast cofilin (PDB ID 1COF)(38) was used for docking to actin. The two residues missing in the original structures on the N-terminus of actin and on the C-terminus of cofilin were added using Insight II (Accelrys Inc., San Diego). Since the N-terminus of cofilin was missing 5 residues, and these amino acids were implicated in cross-linking to actin, multiple models of yeast cofilin N-terminus were created through homology modeling using the S. pombe cofilin crystal structure (PDB ID 2I2Q)(6) and the human cofilin NMR structures (PDB ID 1Q8G and 1Q8X)(18). This resulted in 41 cofilin structures that were used in all subsequent docking simulations. To prepare the actin and cofilin structures for docking, polar hydrogen atoms were added and assigned Kollman united-atom partial charges (39). Atomic solvation parameters were assigned using ‘addsol’ tool of Autodock.
The ‘traditional’ docking protocol relies on multiple docking runs and then clustering of all predicted structures in order to produce one or more viable models. In our case, given the constraints provided by the cross-linking results, we did not rely on consideration of lowest energy or cluster size. Instead of that all structures were analyzed for their agreement with the experimental data. As constraints for docking cofilin to actin we used the distance information obtained for two pairs of cross-linking sites located in helical and/or well structured regions on actin (residues 345, 144) and cofilin (residues 62, 95). A model that represents a group of structures consistent with cofilin Cys 62 - actin Cys 345 and cofilin Cys 95- actin Cys 144 cross-linking is shown on Figure 9 along with the similar model of Wriggers et al (24).
Figure 9.
Models of yeast cofilin bound to G-actin. (a) Representative structure from the group of cofilin-actin docking results that are consistent with WT cofilin-C345 G-actin and cofC95-G-actin C144 cross-links. (b) Wriggers’s et al model of actin-cofilin complex(24). Residues involved in the cross-linking from actin are color coded in red – Cys 374, orange – Leu 346 (partially obscured in this view), yellow – Ile 345 and green – Ser 144 and those on cofilin are in cyan – Ser 2, blue – Cys 62 and purple – Val 95.
Mass spectrometry analysis revealed also that actin residues 346 and 374 could be cross-linked to the N-terminus of cofilin. Considering the flexibility of the N-terminal segment, the distance information obtained for these two sets of cross-linking sites was less helpful for rigid docking purposes, but it shed light on the orientation of cofilin’s N-terminus in the complex.
To further evaluate the fitness of the predicted model, the docked structure was analyzed by the Protein-Protein Interaction server (http://www.biochem.ucl.ac.uk/bsm/PP/server/)(40 ;41). The surface area buried in the predicted complex is within the range of 983 ± 582 Å2, which is consistent with values reported for other protein hetero-complexes (41).
DISCUSSION
The aim of this work was to obtain experimental data that could constrain the computational modeling of the G-actin-cofilin complex structure. Radiolytic oxidation studies of actin and actin-cofilin, have suggested cofilin binding at the subdomain 1 and 2 interface on G-actin, but not at the hydrophobic cleft between subdomain 1 and 3. Our goal was to test this latter model of binding and to obtain distance constraints for independent docking of the actin-cofilin complex. To this end, predictions derived from this model to create several yeast actin and cofilin mutants for fluorescence labeling, and chemical cross-linking combined with MS/MS analysis was used. The distances between cross-linked sites were assessed using reagents of different length as molecular rulers, providing constraints on the structure of the cofilin-G-actin complex. With constraints originating from the cross-linking of residues 144 and 345 on actin, the docking of cofilin structures to G-actin yielded a model of the actin-cofilin interface, with cofilin bound to the hydrophobic cleft between subdomains 1 and 3. This model is similar to the earlier models proposed by W. Wriggers and R. Dominguez based on theoretical studies and molecular dynamics simulation (24 ;25). Fluorescence experiments were consistent with such a docking result.
Extending the cross-linking approach to flexible regions of actin and cofilin presented a difficulty in the attempts to fit all the data into a single model. The rigid docking of cofilin to actin could not take into account the flexibility and possible conformational changes in the C- and N- terminal regions of both proteins and thus, attempts to position the N-terminus of cofilin with respect to actin were unsuccessful. Despite this, the cross-linking of actin residues 346 and 374 to cofilin’s N-terminus indicates that upon complex formation this segment probably changes conformation and faces the “back” of actin.
Cross-linking between actin residue 345 and cofilin residue 62
In the crystal structure of cofilin (38) cysteine 62 should not be accessible to solvent. However, our preliminary experiments detected slow acrylodan labeling of that residue, which was enhanced in the presence of NEM-labeled G-actin. These experiments revealed conformational changes in cofilin upon binding to actin, leading to a partial exposure of cysteine 62. Indeed, the cross-linking of Cys 345 on actin to Cys 62 on cofilin with DTME and MTS reagents was quite efficient (Figure 4 and Figure 6(a)). Our mapping by cross-linking, using a range of MTS reagents and DTME, suggested that cysteine 62 of cofilin is within 9.6 (MTS 6)-12.4 Å (DTME) from cysteine 345 in G-actin, constraining the actin-cofilin interface (see Table 2). The protection of C345 (by cofilin) from acrylodan labeling is consistent with the cross-linking results and indicates proximity of cofilin to helix 337–349 on actin. However, the moderate protection of this cysteine by cofilin and almost complete blocking of its acrylodan modification by GS1 and KabC provide additional evidence that the cofilin binding site on actin is not identical with that for GS1 and KabC (Figure 3(b)).
Cross-linking between actin residue 144 and cofilin residue 95
The α3 helix in cofilin (Val 95 – Ser 104 in yeast cofilin) was suggested to participate in actin binding on the basis of several lines of evidence (16–18 ;27 ;29 ;42), including low efficiency cross-linking of that region to G-actin (29 ;42). A comparison of actin-binding modes for GS1, vitamin D binding protein, and ciboulot allowed Dominguez to hypothesize that cofilin and a number of other actin-binding proteins share a common binding motif, interacting through an hydrophobic α-helix (α3 helix in case of cofilin) with a hydrophobic pocket between subdomains 1 and 3 on actin (25).
The cross-linking between actin mutant C144 and cofC95 with MTS reagents and disulfide bond formation between these residues shows the proximity of cofilin α3-helix to actin helix 137–145 in the complex. The most efficient cross-linking was observed with MTS 3 (6.4 Å) and 6 (9.6 Å). The decreased cross-linking efficiency with longer MTS reagents, 8 (12.1 Å) and 17 (19.0 Å), may indicate that the extended conformation of these cross-linkers does not fit well between the two cysteine residues in the actin-cofilin complex, while the “gauche” conformations of the reagents may not have the appropriate geometry for bridging the 144 and 95 residues (34 ;36). The disulfide cross-linking (catalyzed by copper) reflects the dynamic nature of the complex and confirms that the distance between residues 144 and 95 in the complex is shorter than that between residues 345 and 62.
Cross-linking to the sites located in flexible regions
The activity of cofilins is regulated through a reversible phosphorylation of the serine 3 residue (1). Systematic mutagenesis studies have shown the importance of the N-terminal segment to actin binding (16). The mapping by cross-linking revealed that the C-terminus of actin (cysteine 374) can be covalently attached to the cofilin N-terminus either with DTME, or upon copper catalyzed air oxidation (zero length cross-linking). Cross-linking between residue 346 on actin and the N-terminal region of cofilin with DTME (12.4 Å) was also observed.
Considering these results, it is pertinent to note that the N-terminus of cofilin is missing in the crystal structure of yeast cofilin (38), and appears highly mobile in the NMR structure of human cofilin (18) and in molecular dynamics simulations of yeast cofilin (data not shown). Rigid docking of cofilin to actin could not account for the flexibility of this region, and attempts to dock cofilin structures with different possible conformations of the N-terminal segment did not generate a model consistent with all cross-linking data. Despite this, the structure that is shown in Figure 9 should be consistent with experimental data if the N-terminus was allowed to change its orientation.
The model of G-actin-cofilin complex and its comparison with other existing models
Cofilin docking to actin, as constrained by the cross-linking results yielded a model of the complex shown in Figure 9, that is consistent with the previous model of actin-cofilin generated by molecular dynamics simulation of rabbit actin and yeast cofilin (24). The Cα-Cα distances between cross-linking sites in those models are compared in Table 3. In both models, cofilin binds in the hydrophobic cleft of actin between subdomain 1 and 3, as was previously proposed (24–26). Detailed differences between the two models shown in Figure 9 are beyond the resolution of our cross-linking methods.
Recent consideration of another set of experimental data – the protection of actin residues from radiolytic oxidation by the bound cofilin – favored a different docking model of the actin-cofilin complex, with cofilin bound in the cleft between actin subdomains 1 and 2 (27). This choice was prompted by the lack of significant protection from oxidation of peptides within the cleft between subdomains 1 and 3 in the actin-cofilin complex. In general, such a protection might be expected for specific residues located at a protein-protein interface due to their “burial” and reduced solvent exposure. However, comparison of footprinting results obtained by the same method for actin complexes with cofilin and GS1, which binds in the hydrophobic cleft on actin, allows an alternative interpretation of the data (27 ;43).
A comparison of oxidation rates for the peptides located in the actin hydrophobic cleft in G-actin alone with the rates in actin complexes with GS1 and cofilin shows that both proteins provide a similar degree of protection. Thus, for actin peptide 119–147 (subdomain 3) the ratio of oxidation rates for Ca-ATP-G-actin without and with GS1 was 1.4 versus 1.2 in the case of cofilin (43). Moreover, for the partially overlapping peptides 157–178 and 148–177 in actin’s subdomain 3 the ratios of oxidation rates was 1.6 and 2.1 for GS1 and cofilin, respectively, indicating slightly stronger protection of this area by cofilin. The main difference in the protection effects of GS1 and cofilin on G-actin in radiolytic footprinting experiments was reported for the peptide 337–359 in actin’s subdomain 1: a strong protection by GS1 (oxidation rates ratio 2.75) and no protection by cofilin was observed (43). This observation is in a good agreement with the results of fluorescence experiments on actin mutant C345 (subdomain 1), which confirm that the actin-cofilin interface is different in details from that of actin-GS1 and actin-KabC (Figure 3). In examining these somewhat different effects of GS1 and cofilin on the oxidation of the 337–359 peptide of actin it should be noted that residues L346 and L349, which are primarily involved in actin-GS1 interaction, served as the main probe residues in the footprinting experiments. Based on this and previous work (16 ;27), we can speculate that either those probe residues are not directly involved in cofilin binding and/or the solvent exposure of the probe residues on the peptide 337–359 does not change upon cofilin binding i.e., the actin peptide 337–359 might be a “blind spot” for the radiolytic footprinting in this particular system.
In the synchrotron footprinting experiments performed on G-actin and its complexes, the reported modification rates for the peptides represent the average values of oxidation rates of individual probe residues. It is conceivable that conformational changes induced in the 335–348 helix by cofilin can increase the solvent exposure of some probe residues despite the involvement of this region in cofilin binding. In a hypothetical case when some probe residues become more and other less solvent accessible no protection may be detected for that peptide. That such a possibility exists is apparent from examining the solvent accessibility of surface residues in the complex of actin with vitamin D binding protein (PDB ID 1KXP)(44). While such an analysis (using GETAREA 1.1 software) reveals a significant decrease in solvent accessibility for actin residues I345, L346 and L349, the accessibility of Y337, I341, S348 is increased. The same analysis performed on the docked G-actin-cofilin complex (Figure 9) has not shown a significant decrease in solvent accessibility for probe residues on the actin peptide 337–359. For instance, solvent accessibility is slightly decreased for tyrosine 337 (1.13 fold), but for the other probe residues in that peptide (L346, L349, F352, M355) accessibility is unchanged. Thus, it is difficult to exclude the cleft between subdomains 1 and 3 from being a part of the actin-cofilin interface on the basis of radiolytic protein footprinting data alone.
Probing the actin-cofilin interface by chemical cross-linking provides direct evidence for a cofilin binding site in the hydrophobic cleft between subdomains 1 and 3 on G-actin. According to our model, and consistent with previous results obtained by other approaches(16– 18), the α3-helix and the N-terminal segment of cofilin are involved in actin binding. However, all the cross-linking results obtained for cofilin’s N-terminus could not be accommodated in our model, probably because of the flexibility of that region and the possible conformational changes upon actin binding. Moreover, the possibility that some of the cross-links detected in this region capture different modes of cofilin binding cannot be ruled out. The fact that actin residues 346 and 374, which could be covalently attached to cofilin’s N-terminus with DTME, are facing the opposite side of actin molecule, compared to residues 345 and 144, may suggest that upon actin binding cofilin’s N-terminus changes its conformation and binds to the “back” of the actin molecule. Finally, it should be noted that the experiments carried out in this study did not test an alternative model of cofilin binding to G-actin, between subdomains 1 and 2 (27). The testing of such a binding would require the conformation of model predictions regarding the actin-cofilin interface along the lines of the present study.
MATERIALS AND METHODS
Materials
Bis(methanethiosulfonate) (MTS) cross-linking reagents, MTS1 [1,1-methanediyl bis(methanethiosulfonate)], MTS-3 [1,3-propanediyl bis(methanethiosulfonate)], MTS-4 [1,4-butanediyl bis(methanethiosulfonate)], MTS-6 [1,6-hexanediyl bis(methanethiosulfonate)], MTS-8 [3,6-dioxaoctane-1,8-diyl bis(methanethiosulfonate)] and MTS-17 [3,6,9,12,15-pentaoxaheptadecane-1,17-diyl bis(methanethiosulfonate)], were purchased from Toronto Research Chemicals Inc. (North York, Ontario, Canada). DTME [(Dithio-bis-maleimidoethane)] was obtained from Pierce (a brand of Thermo Fisher Scientific, Rockford, IL) and acrylodan was from Molecular Probes (Eugene, OR). Millipore-filtered water and analytical grade reagents were used in all experiments.
Site-directed mutagenesis
Yeast actin mutants C144 (A144C/C374A), C345 (I345C/C374A) and C346 (L346C/C374A) were created by Dr. V. Korman as previously described (30). Site-directed mutagenesis of yeast cofilin was carried out using the QuikChange kit (Stratagene). The base construct with a substitution of native cysteine 62 in cofilin to serine was created with the following primers (mutated codon underlined): 5’-GAAATTGCCAGAAAACGACAGTCTTTACGCCATTTACG-3’. To create cofilin mutants cofC95 (V95C/C62S), and cofC1* (C1/C62S) that carries an extra cysteine at the N-terminus, the following primers were designed (mutated codons underlined): 5’-CTCCAGACACTGCTCCATGCAGATCTAAGATGGTC-3’ and 5’-CTTTAAGAAGGAGATATATCCATGTGTTCTAGATCTGGTGTTGCTGTTGC-3’.
Proteins
Yeast actin was purified as described (45), with some minor modifications. The protein was eluted from a DNAse affinity column with a G-buffer containing 20–25% sucrose, 50% formamide, 5 mM Tris (pH 7.8 at 22 °C), 0.2 mM CaCl2, 0.3 mM ATP, 1 mM DTT, 0.2 mM PMSF, and was then applied directly to a DEAE column. The inclusion of sucrose improved the quality of actin preparations and the reproducibility of the results. Actin was eluted from the DEAE column in G-buffer (10 mM Tris (pH 7.8 at 22 °C), 0.2 mM CaCl2, 0.2 mM ATP, 1 mM DTT, 0.2 mM PMSF) containing 350 mM KCl.
Yeast cofilin mutants and WT cofilin in pBAT4 vector were expressed in E. coli BL21(DE3) cells at 20 °C (25 °C for WT) with 1 mM IPTG, overnight. Expression level of cofilin mutants was comparable to that of WT cofilin. The protein was recovered form soluble fractions (lysate) using standard procedures (12). For mutant cofilins, the elution profiles from Superdex-75 gel-filtration column were the same as for WT cofilin, suggesting that there was no aggregation. GS1 was expressed and purified essentially as described (46). Actin concentration was measured by the Bradford protein assay, using skeletal rabbit actin as a standard. Cofilin and GS1 concentrations were determined spectrophotometrically, using extinction coefficients E(1%) at 280 nm of 9.2 cm−1 and E(1%) at 280 nm of 1.46 cm−1, respectively.
Actin polymerization assays
Actin polymerization was monitored via light scattering with the PTI fluorometer set at 350 nm for the excitation and emission wavelengths. Actin polymerization was induced by addition 3.0 mM MgCl2 to the protein solution.
Nucleotide exchange assays
The rate of nucleotide exchange in G-actin was monitored through the fluorescence of ε-ATP at 25 °C. One to two µM of ε-ATP-Ca-G-actin was mixed with 0–40 µM of cofilin in G-buffer containing 10 mM HEPES (pH 7.5), 0.2 CaCl2, 5 µM ε-ATP and 1 mM DTT. ε-ATP release was initiated with the addition of ATP (100-fold molar excess over actin). The decrease in ε-ATP fluorescence, upon its release, was followed in the PTI fluorometer with the excitation set at 350 nm and emission at 400–410 nm wavelength (λmax of ε-ATP fluorescence varies for different actin mutants). Dissociation constants, Kd, for actin-cofilin complexes were calculated as described earlier (19 ;31).
Acrylodan labeling and protection experiments
Prior to acrylodan labeling, DTT was removed from G-actin solutions over a Sephadex G-50 spin columns equilibrated with G-buffer containing 10 mM HEPES (pH 7.5), 0.2 CaCl2 and 0.2 mM ATP (Buffer A). G-actin was then incubated with a 3-fold molar excess of acrylodan for 3 hours at 4 °C, after which the reaction was stopped by adding 1 mM DTT. The excess of label was removed from the samples over a Sephadex G-50 gel filtration column. Alternatively, samples were dialyzed overnight against Buffer A, supplemented with 1 mM DTT, and then span for 30 min at 85,000 rpm in a Beckman TLA 110 rotor.
To monitor acrylodan modification of single cysteine residues in actin and its complexes with actin-binding factors, thiol free protein solutions were pre-mixed at the mole ratio of 1:1.5 of actin to ligand. To remove free thiols, G-actin was treated as described above. Cofilin and GS1 were passed through Zeba Desalt Spin Column (Pierce) equilibrated with a thiol free Buffer A. Reactions were initiated by adding aliquots of acrylodan in 100% DMF to the solution. The concentration of organic solvent in the sample did not exceed 1%. After separation of proteins by SDS PAGE, the labeled actin was visualized under UV light. The amount of labeled protein was estimated using Scion Image software.
To estimate the reactivity of native cysteine 62 on WT cofilin, cofilin and its complex with NEM-labeled skeletal G-actin were incubated in the presence of acrylodan at the mole ratio of 1:1 cofilin/complex to reagent. The proteins were then separated on SDS PAGE, and the labeled cofilin was visualized under UV light. Slow acrylodan modification of cofilin’s native cysteine 62 was observed, and it appeared to be increased in the presence of NEM-labeled G-actin (unpublished data).
Actin-cofilin cross-linking
Immediately prior to the reaction, DTT was removed from G-actin samples over a Sephadex G-50 spin column equilibrated with thiol free buffer A or buffer B (10 mM Tris (pH 8.5), 0.2 CaCl2 and 0.2 mM ATP). Cofilin was passed through a Zeba Desalt Spin Column (PIERCE) equilibrated with the same buffer.
Cross-linking with bis(methanethiosulfonate) (MTS) reagents was carried out on ice (30 min), or at room temperature (3–15 min), at molar ratios of 1:1 or 2:1 of MTS to the actin-cofilin complex. Formation of disulfide bonds was catalyzed with the addition of CuSO4 at a mole ratio of 1:1 to actin-cofilin complex. Aliquots were withdrawn from the reaction mixtures at selected time points and free cysteine residues were blocked with 2–10 mM NEM. Cross-linking progress was monitored by SDS-PAGE under non-reducing conditions.
Purification of the DTME-modified species
Thiol free samples of actin and cofilin in buffer B were mixed in 1:1.1 ratio. We observed that alkaline pH (8.3 – 8.5) increased cross-linking efficiency. Reactions were started by adding DTME to the complex at a 1:1 molar ratio. Cross-linking reactions were carried out in buffer B, at 4 °C overnight, to prevent protein denaturation, and were then stopped with 2 mM NEM. Cross-linked and uncross-linked complexes of actin-cofilin were separated from free cofilin by size exclusion chromatography on Superdex 200 equilibrated with a thiol free G-buffer (10 mM Tris (pH 8.3), 0.2 CaCl2 and 0.2 mM ATP). Fractions containing the cross-linked protein were concentrated using an Amicon Ultra-4 centrifugal filter devices with a 10 kDa molecular weight cut-off membrane. To reduce the disulfide bond in DTME, DTT (5 mM) was added to the sample, which was then dialyzed overnight against high salt buffer (10 mM Tris (pH 8.3), 0.2 CaCl2 and 0.4 mM ATP, 5 mM DTT, 1 M NaCl). The resulting TME-modified actin and cofilin were separated by size exclusion chromatography on Superdex 200 equilibrated with 10 mM Tris (pH 8.3), 0.2 CaCl2 and 0.4 mM ATP, 1 mM DTT, 0.5 M NaCl. TME-modified cofilin was concentrated using Vivaspin 500 concentrators (Viva Science, Hannover, Germany) with a 5 kDa molecular weight cut-off membrane PES membrane, and dialyzed overnight against 10 mM (NH4)2CO3 supplemented with 1 mM DTT.
Combined Liquid Chromatography-Electrospray Ionization Mass Spectrometry
FPLC-purified TME-cofilin was subjected to proteolysis with sequencing grade Lys-C or with a combination of Lys-C and trypsin (Roche). Prior to Lys-C digestion, cofilin was denatured with 4 M urea; the urea concentration was reduced to 1 M (by dilution) for trypsin digestion, followed by a Zip Tip (Millipore) clean up.
On line peptide sequencing was accomplished LC-MS/MS on a hybrid quadrupole time-of-flight mass spectrometer QSTAR® XL (QqTOF) (Applied Biosystems, Foster City, CA). The nano-LC was equipped with an LC Packings Jupiter C12 precolumn (150 µm×5 mm) and a LC Packings Jupiter C12 column (75µm×150 mm). The eluents used for the LC were (A) 0.1% formic acid in water and (B) 95% acetonitrile in water containing 0.1% FA. The flow was 230 nL/min, and the following gradient was used: 3% B to 35% B in 5 min, 35% B to 80% B in 45 min and maintained at 80% B for 10 min. The column was re-equilibrated with 3% B for 15 min before the next run. Nanospray ESI-MS/MS experiments were also performed on the QSTAR® equipped with a Protana source. Standard operating conditions were DP1 = 70 V, DP2 = 15 V and focusing potential FP = 400 V, with argon as collision gas (99.999% purity). Calibration was achieved in the tandem MS/MS mode using commercial [Glu]-Fibrinopeptide (Sigma-Aldrich). MS/MS data were analyzed both manually and with Prosight PTM software (https://prosightptm.scs.uiuc.edu).
Docking
AutoDock 3.0 (47) was used to dock cofilin structures to G-actin. Docking of cofilin structures with different possible N-termini was attempted in order to gain a better representation of cross-linking data involving the N-terminus. Grid maps (250 × 250 × 250) were constructed with 0.375 Å spacing centered on the hydrophobic cleft between subdomains 1 and 3 of G-actin. All 41 cofilin structures were docked using the Lamarckian Genetic Algorithm in AutoDock 3.0. The number of individuals in the random population was chosen to be 100 and the maximum number of energy evaluations was 1000000. Elitism, rate of gene mutation, and rate of crossover were set to 1, 0.02 and 0.95 respectively. For each cofilin structure 10 runs were performed resulting in a total of 410 predicted complexes. These structures were clustered by positional root-mean-square deviation (RMSD) cutoff of 6.0 Å and ranked. Overall 69 multi-membered conformational clusters were found. Members of the clusters were visualized using InsightII (Accelrys Inc.) program and examined for their consistency with the cross-linking results.
Table 3.
Comparison of distances in models of the G-actin-cofilin complex.
| Cα-Cα Distances (Å) |
Cofilin aa 62– Actin aa 345 |
Cofilin aa 95– Actin aa 144 |
Cofilin aa 2 – Actin aa 346 |
Cofilin aa 2 – Actin aa 374 |
|---|---|---|---|---|
| Model (Fig. 9) | 17 | 12 | 21 | 31 |
| Wriggers model (24) | 16 | 11 | 14 | 23 |
| Kamal’s model (27) | 37 | 38 | -* | -* |
Missing 5 amino acids on cofilin N-terminus
ACKNOWLEDGMENT
We are grateful to Vicci Korman for providing yeast actin mutants employed in this study and Mai Phan for technical assistance with EM. We thank Mark Chance and Amisha Kamal for valuable comments on the manuscript. This work was supported by grants from USPHS (GM 077190) and NSF (MCB 0316269) to E. R., from NIH (RR 20004) and the US Department of Energy (DE-FC02-02ER63421) to J. A. L., and from the National Institutes of Health GM-067246 to D. S. The UCLA Proteomics Center was established and equipped by a grant to UCLA from the W. M. Keck Foundation.
Abbreviations used
- G-actin
monomeric actin
- F-actin
filamentous actin
- WT
wild type
- DNAse I
deoxyribonuclease I
- GS1
gelsolin segment 1
- KabC
kabiramide C
- LC-MS/MS
liquid chromatography – tandem mass spectrometry
- EM
electron microscopy
- MTS
bis(methanethiosulfonate)
- DTME
dithio-bis-maleimidoethane
- TME
thio-bis-maleimidoethane
- PDB
protein data bank
- p-PDM
N`N`-p-phenylenedimaleimide
- o-PDM
N`N`-o-phenylenedimaleimide
- C345
actin mutant I345C/C374A
- C346
actin mutant L346C/C374A
- C144
actin mutant A144C/C374A
- cofC1*
cofilin mutant C1/C62S
- cofC95
cofilin mutant V95C/C62S
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bamburg JR. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu. Rev. Cell Dev.Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- 2.Bamburg JR, Wiggan OP. ADF/cofilin and actin dynamics in disease. Tr. Cell Biol. 2002;12:598–605. doi: 10.1016/s0962-8924(02)02404-2. [DOI] [PubMed] [Google Scholar]
- 3.Gourlay CW, Ayscough KR. The actin cytoskeleton in ageing and apoptosis. FEMS Yeast Research. 2005;5:1193–1198. doi: 10.1016/j.femsyr.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Du J, Frieden C. Kinetic studies on the effect of yeast cofilin on yeast actin polymerization. Biochemistry. 1998;37:13276–13284. doi: 10.1021/bi981117r. [DOI] [PubMed] [Google Scholar]
- 5.Pavlov D, Muhlrad A, Cooper J, Wear M, Reisler E. Actin filament severing by cofilin. J. Mol. Biol. 2007;365:1350–1358. doi: 10.1016/j.jmb.2006.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 7.Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 1997;136:1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sept D, McCammon JA. Thermodynamics and kinetics of actin filament nucleation. Biophys. J. 2001;81:667–674. doi: 10.1016/S0006-3495(01)75731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galkin VE, Orlova A, Lukoyanova N, Wriggers W, Egelman EH. Actin depolymerizing factor stabilizes an existing state of F-actin and can change the tilt of F-actin subunits. J. Cell Biol. 2001;153:75–86. doi: 10.1083/jcb.153.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galkin VE, Orlova A, VanLoock MS, Shvetsov A, Reisler E, Egelman EH. ADF/cofilin use an intrinsic mode of F-actin instability to disrupt actin filaments. J. Cell Biol. 2003;163:1057–1066. doi: 10.1083/jcb.200308144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bobkov AA, Muhlrad A, Kokabi K, Vorobiev S, Almo SC, Reisler E. Structural effects of cofilin on longitudinal contacts in F-actin. J. Mol. Biol. 2002;323:739–750. doi: 10.1016/s0022-2836(02)01008-2. [DOI] [PubMed] [Google Scholar]
- 13.Bobkov AA, Muhlrad A, Pavlov DA, Kokabi K, Yilmaz A, Reisler E. Cooperative effects of cofilin (ADF) on actin structure suggest allosteric mechanism of cofilin function. J. Mol. Biol. 2006;356:325–334. doi: 10.1016/j.jmb.2005.11.072. [DOI] [PubMed] [Google Scholar]
- 14.De La Cruz EM. Cofilin binding to muscle and non-muscle actin filaments: isoform-dependent cooperative interactions. J. Mol. Biol. 2005;346:557–564. doi: 10.1016/j.jmb.2004.11.065. [DOI] [PubMed] [Google Scholar]
- 15.Kudryashov DS, Galkin VE, Orlova A, Phan M, Egelman EH, Reisler E. Cofilin cross-bridges adjacent actin protomers and replaces part of the longitudinal F-actin interface. J. Mol. Biol. 2006;358:785–797. doi: 10.1016/j.jmb.2006.02.029. [DOI] [PubMed] [Google Scholar]
- 16.Lappalainen P, Fedorov EV, Fedorov AA, Almo SC, Drubin DG. Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO J. 1997;16:5520–5530. doi: 10.1093/emboj/16.18.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guan JQ, Vorobiev S, Almo SC, Chance MR. Mapping the G-actin binding surface of cofilin using synchrotron protein footprinting. Biochemistry. 2002;41:5765–5775. doi: 10.1021/bi0121104. [DOI] [PubMed] [Google Scholar]
- 18.Pope BJ, Zierler G, Kuhne R, Weeds AG, Ball LJ. Solution structure of human cofilin: actin binding, pH sensitivity, and relationship to actin-depolymerizing factor. J. Biol. Chem. 2004;279:4840–4848. doi: 10.1074/jbc.M310148200. [DOI] [PubMed] [Google Scholar]
- 19.Hawkins M, Pope B, Maciver SK, Weeds AG. Human actin depolymerizing factor mediates a pH-sensitive destruction of actin filaments. Biochemistry. 1993;32:9985–9993. doi: 10.1021/bi00089a014. [DOI] [PubMed] [Google Scholar]
- 20.Van Troys M, Dewitte D, Verschelde JL, Goethals M, Vandekerckhove J, Ampe C. Analogous F-actin binding by cofilin and gelsolin segment 2 substantiates their structural relationship. J. Biol. Chem. 1997;272:32750–32758. doi: 10.1074/jbc.272.52.32750. [DOI] [PubMed] [Google Scholar]
- 21.Renoult C, Blondin L, Fattoum A, Ternent D, Maciver SK, Raynaud F, Benyamin Y, Roustan C. Binding of gelsolin domain 2 to actin. An actin interface distinct from that of gelsolin domain 1 and from ADF/cofilin. Eur. J. Biochem. 2001;268:6165–6175. doi: 10.1046/j.0014-2956.2001.02574.x. [DOI] [PubMed] [Google Scholar]
- 22.Blondin L, Sapountzi V, Maciver SK, Renoult C, Benyamin Y, Roustan C. The second ADF/cofilin actin-binding site exists in F-actin, the cofilin-G-actin complex, but not in G-actin. Eur. J. Biochem. 2001;268:6426–6434. doi: 10.1046/j.0014-2956.2001.02592.x. [DOI] [PubMed] [Google Scholar]
- 23.Renoult C, Ternent D, Maciver SK, Fattoum A, Astier C, Benyamin Y, Roustan C. The identification of a second cofilin binding site on actin suggests a novel, intercalated arrangement of F-actin binding. J. Biol. Chem. 1999;274:28893–28899. doi: 10.1074/jbc.274.41.28893. [DOI] [PubMed] [Google Scholar]
- 24.Wriggers W, Tang JX, Azuma T, Marks PW, Janmey PA. Cofilin and gelsolin segment-1: molecular dynamics simulation and biochemical analysis predict a similar actin binding mode. J. Mol. Biol. 1998;282:921–932. doi: 10.1006/jmbi.1998.2048. [DOI] [PubMed] [Google Scholar]
- 25.Dominguez R. Actin-binding proteins - a unifying hypothesis. Tr. Biochem. Sci. 2004;29:572–578. doi: 10.1016/j.tibs.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 26.Hatanaka H, Ogura K, Moriyama K, Ichikawa S, Yahara I, Inagaki F. Tertiary structure of destrin and structural similarity between two actin-regulating protein families. Cell. 1996;85:1047–1055. doi: 10.1016/s0092-8674(00)81305-7. [DOI] [PubMed] [Google Scholar]
- 27.Kamal JKA, Benchaar SA, Takamoto K, Reisler E, Chance MR. Three-dimensional structure of cofilin bound to monomeric actin derived by structural mass spectrometry data. Proc. Natl Acad. Sci. 2007;104:7910–7915. doi: 10.1073/pnas.0611283104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mannherz HG, Ballweber E, Galla M, Villard S, Granier C, Steegborn C, Schmidtmann A, Jaquet K, Pope B, Weeds AG. Mapping the ADF/Cofilin binding site on monomeric actin by competitive cross-linking and peptide array: evidence for a second binding site on monomeric actin. J. Mol. Biol. 2007;366:745–755. doi: 10.1016/j.jmb.2006.11.100. [DOI] [PubMed] [Google Scholar]
- 29.Benchaar SA, Xie Y, Phillips M, Loo RRO, Galkin VE, Orlova A, Thevis M, Muhlrad A, Almo SC, Loo JA, Egelman EH, Reisler E. Mapping the interaction of cofilin with subdomain 2 on actin. Biochemistry. 2007;46:225–233. doi: 10.1021/bi0610754. [DOI] [PubMed] [Google Scholar]
- 30.Korman VL, Anderson SEB, Prochniewicz E, Titus MA, Thomas DD. Structural dynamics of the actin-myosin interface by site-directed spectroscopy. J. Mol. Biol. 2006;356:1107–1117. doi: 10.1016/j.jmb.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 31.Bryan J. Gelsolin has three actin-binding sites. J. Cell Biol. 1988;106:1553–1562. doi: 10.1083/jcb.106.5.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLaughlin PJ, Gooch JT, Mannherz HG, Weeds AG. Structure of gelsolin segment 1-actin complex and the mechanism of filament severing. Nature. 1993;364:685–692. doi: 10.1038/364685a0. [DOI] [PubMed] [Google Scholar]
- 33.Klenchin VA, Allingham JS, King R, Tanaka J, Marriott G, Rayment I. Trisoxazole macrolide toxins mimic the binding of actin-capping proteins to actin. Nat. Struct. Mol. Biol. 2003;10:1058–1063. doi: 10.1038/nsb1006. [DOI] [PubMed] [Google Scholar]
- 34.Green NS, Reisler E, Houk KN. Quantitative evaluation of the lengths of homobifunctional protein cross-linking reagents used as molecular rulers. Protein Sci. 2001;10:1293–1304. doi: 10.1110/ps.51201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loo TW, Clarke DM. Determining the dimensions of the drug-binding domain of human P-glycoprotein using thiol cross-linking compounds as molecular rulers. J. Biol. Chem. 2001;276:36877–36880. doi: 10.1074/jbc.C100467200. [DOI] [PubMed] [Google Scholar]
- 36.Shvetsov A, Stamm JD, Phillips M, Warshaviak D, Altenbach C, Rubenstein PA, Hideg K, Hubbell WL, Reisler E. Conformational dynamics of loop 262–274 in G- and F-actin. Biochemistry. 2006;45:6541–6549. doi: 10.1021/bi052558v. [DOI] [PubMed] [Google Scholar]
- 37.Vorobiev S, Strokopytov B, Drubin DG, Frieden C, Ono S, Condeelis J, Rubenstein PA, Almo SC. The structure of nonvertebrate actin: Implications for the ATP hydrolytic mechanism. PNAS. 2003;100:5760–5765. doi: 10.1073/pnas.0832273100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fedorov AA, Lappalainen P, Fedorov EV, Drubin DG, Almo SC. Structure determination of yeast cofilin. Nat. Struct. Biol. 1997;4:366–369. doi: 10.1038/nsb0597-366. [DOI] [PubMed] [Google Scholar]
- 39.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 40.Jones S, Thornton JM. Protein-protein interactions: A review of protein dimer structures. Prog. Biophys. Mol. Biol. 1995;63:31–59. doi: 10.1016/0079-6107(94)00008-w. [DOI] [PubMed] [Google Scholar]
- 41.Jones S, Thornton JM. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yonezawa N, Nishida E, Iida K, Kumagai H, Yahara I, Sakai H. Inhibition of actin polymerization by a synthetic dodecapeptide patterned on the sequence around the actin-binding site of cofilin. J. Biol. Chem. 1991;266:10485–10489. [PubMed] [Google Scholar]
- 43.Guan JQ, Almo SC, Reisler E, Chance MR. Structural reorganization of proteins revealed by radiolysis and mass spectrometry: G-actin solution structure is divalent cation dependent. Biochemistry. 2003;42:11992–12000. doi: 10.1021/bi034914k. [DOI] [PubMed] [Google Scholar]
- 44.Otterbein LR, Cosio C, Graceffa P, Dominguez R. Crystal structures of the vitamin D-binding protein and its complex with actin: structural basis of the actin-scavenger system. PNAS. 2002;99:8003–8008. doi: 10.1073/pnas.122126299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim E, Wriggers W, Phillips M, Kokabi K, Rubenstein PA, Reisler E. Cross-linking constraints on F-actin structure. J. Mol. Biol. 2000;299:421–429. doi: 10.1006/jmbi.2000.3727. [DOI] [PubMed] [Google Scholar]
- 46.Way M, Pope B, Gooch J, Hawkins M, Weeds AG. Identification of the region in segment 1 of gelsolin critical for actin binding. EMBO J. 1990;9:4103–4109. doi: 10.1002/j.1460-2075.1990.tb07632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]