Abstract
Members of the heat shock protein 70 (Hsp70) family of molecular chaperones are emerging as potential therapeutic targets. Their ATPase activity has classically been measured using colorimetric phosphate-detection reagents, such as quinaldine red (QR). While such assays are suitable for 96-well plate formats, they typically lose sensitivity when attempted in lower volume due to path length and meniscus effects. These limitations and Hsp70’s weak enzymatic activity have combined to create significant challenges in high throughput screening. To overcome these difficulties, we have adopted an energy transfer strategy that was originally reported by Zuck et al. (Anal. Biochem. 2005, 342:254–259). Briefly, white 384-well plates emit fluorescence when irradiated at 430 nm. In turn, this intrinsic fluorescence can be quenched by energy transfer with the QR-based chromophore. Using this more sensitive approach, we tested 55,400 compounds against DnaK, a prokaryotic member of the Hsp70 family. The assay performance was good (Z′ ~ 0.6, CV ~8%) and at least one promising new inhibitor was identified. In secondary assays, this compound specifically blocked stimulation of DnaK by its co-chaperone, DnaJ. Thus, this simple and inexpensive adaptation of a colorimetric method might be suitable for screening against Hsp70-family members.
Keywords: phosphate, malachite green, ATPase, molecular chaperone, fluorescence assay
INTRODUCTION
Members of the heat shock protein 70 (Hsp70) family of molecular chaperones are ubiquitously expressed and remarkably well conserved. In both eukaryotes and prokaryotes, Hsp70s play important roles in protein quality control, including during folding, transport and turnover.1 In addition, they have been implicated in cancer2 and a number of protein-misfolding disorders, such as Parkinson’s disease.3 In prokaryotes, the Hsp70 ortholog, DnaK, has been found to be essential for virulence in Staphylococcus aureus.4 Despite these promising observations, a scarcity of drug-like inhibitors has limited in-depth exploration of this chaperone as a therapeutic target.5
Members of the Hsp70 family are composed of two domains: a substrate-binding domain (SBD) and a nucleotide-binding domain (NBD).6 The SBD accommodates short stretches of peptide and it seems to have a preference for the hydrophobic residues that might be exposed in misfolded or incompletely folded proteins.7 Binding of the SBD to its substrates is regulated, in part, by the nucleotide state of the adjacent NBD: the ATP-bound form has relatively weak affinity for substrates, while the ADP-bound form binds tightly.8 However, the intrinsic ATPase activity of these proteins is slow; for example, Escherichia coli DnaK, has a Vmax of ~1 pmol ATP/μg enzyme/minute.9 This modest turnover allows regulation by co-chaperones, such as DnaJ.10 DnaJ belongs to a family of co-chaperones that bind Hsp70s through a conserved J-domain. This protein-protein contact accelerates ATP turnover through an allosteric mechanism.11
In envisioning strategies for inhibiting members of the Hsp70 family, two of their activities become apparent as potential targets.5 One approach is to inhibit binding of the SBD to its substrates. This strategy is exploited by certain insect-derived, anti-bacterial peptides12 and a fluorescence polarization-based assay to identify similar compounds has been developed.13 A separate strategy is to block nucleotide turnover.14; 15 This goal might be accomplished by either directly competing with nucleotide16 or by inhibiting the protein-protein interactions with the stimulatory co-chaperones. For example, we recently employed the well-known malachite green (MG) reagent to detect ATP hydrolysis by the DnaK-DnaJ combination.17.18 Because the stimulatory activity of the J-domain dominates the phosphate signal, we anticipated that inhibitors found by this method would preferentially target the protein-protein contact. Indeed, we found new inhibitors in pilot screens that specifically block DnaK-DnaJ interactions19 and the resulting, first-generation compounds have been used in a variety of disease models to reveal potential roles of Hsp70s.20–22
To identify additional chemical scaffolds, we sought to screen larger compound collections in high throughput ATPase assays. Unfortunately, our attempts to further miniaturize the absorbance platforms to low volume 384-well microtiter plates were frustrated by significantly decreased sensitivity. Guided by the work of Zuck and coworkers,23 we explored whether sensitivity could be enhanced using energy transfer methodology. We were particularly interested in studying DnaK, as this chaperone has been implicated as an emerging anti-bacterial target.4 Further, DnaK’s weak ATPase activity makes it a particularly challenging test case. Briefly, we found that the fluorescence method improved sensitivity for phosphate and permitted screening of over 55,000 compounds with good assay performance (Z′ ~ 0.6, CV ~ 8%). These experiments yielded at least one new inhibitor of DnaK, which appears to block stimulation by DnaJ.
MATERIALS AND METHODS
Reagents
Unless otherwise specified, all reagents were purchased from Sigma (St. Louis, MO). E. coli DnaK was purified according to published procedures.18 E. coli DnaJ was purified as previously described.24
Small Molecule Libraries
The MicroSource MS2000 library contains ~2,000 bioactive compounds with a minimum of 95% purity. Briefly, the collection includes 958 known therapeutic drugs, 629 natural products and derivatives, 343 compounds with reported biological activities and 70 compounds approved for agricultural use. The University of Michigan Center for Chemical Genomics (CCG) small molecule library consists of 16,000 Maybridge HitFinder, 13,000 ChemBridge, 20,000 ChemDiv, 3,000 NCI, 450 NIH Clinical Collection (NCC) compounds and ~20,000 natural product extracts. The activity of promising compounds was confirmed using repurchased samples from original vendors. Compound 3c was purchased from Cayman Chemical Inc. (Ann Arbor, MI). Compounds were used without further purification.
High Throughput ATPase Assay – Absorbance Method
The assay procedure was adopted from previous reports with modifications where indicated.18 All components other than compounds were added by a Multidrop dispensor (Thermo Fisher Scientific, Inc., Waltham, MA). Stock solutions of 0.05% w/v quinaldine red (QR), 2% w/v polyvinyl alcohol, 6% w/v ammonium heptamolybdate tetrahydrate in 6 M HCl and water were mixed in a 2:1:1:2 ratio to prepare the QR reagent. This reagent was prepared fresh prior to each experiment. For compound screening, a stock solution of DnaK and DnaJ was prepared in assay buffer (100 mM Tris-HCl, 20 mM KCl, and 6 mM MgCl2, 0.01% Triton X-100, pH 7.4) so that the final concentration of DnaK was 0.4 μM and DnaJ was 0.7 μM (unless noted). This solution (10 μL) was then added to each well of a 384-well clear plate (Thermo Fisher Scientific, Inc., Waltham, MA). To this solution, 0.4 μL of either compound (1.5 mM) or DMSO was added to each well by Biomek HDR (Beckman, Fullerton, CA). Finally, 4 μL of a 7 mM ATP solution was added to begin the reaction. The plates were then incubated for 3 hrs at 37 °C. After incubation, each well received 40 μL of the QR reagent, allowing 2 min of reaction time, and then quenched by addition of 32% w/v solution of sodium citrate (4 μL). The plates were then incubated for an additional 15 min at 37 °C before measuring absorbance at 530 nm on a PHERAstar plate reader (BMG Labtech, Cary, NC).
High Throughput ATPase Assay – Fluorescence in White Plates
The QR reagent was prepared exactly as indicated above. All components other than compounds were added by a Multidrop dispensor (Thermo Fisher Scientific, Inc., Waltham, MA). The DnaK-DnaJ stock solution was prepared so that the final concentration of DnaK was 0.4 μM and DnaJ was 0.7 μM (unless noted). This solution (5 μL) was then added to each well of a 384-well opaque, white, low-volume, non-sterile, polystyrene 384-well plates (Greiner Bio-One North America Inc., Monroe, NC). To this solution, 0.2 μL of either compound (1.5 mM) or DMSO was added to each well by Biomek HDR (Beckman, Fullerton, CA). Finally, 2 μL of a 3.5 mM ATP solution was added to begin the reaction. The plates were then incubated for 3 hrs at 37 °C. After incubation, each well received 15 μL of the QR reagent, allowing 2 min of reaction time, and then quenched by addition of 32% w/v solution of sodium citrate (2 μL). These plates were incubated for 15 min at 37 °C and the fluorescence intensity measured (excitation 430 nm, emission 530 nm) on a PHERAstar plate reader. Standard curves were obtained using stock solutions of dibasic potassium phosphate.
Tryptophan Fluorescence Assay
The method for measuring binding to DnaK was carried out as previously described.25 Briefly, DnaK (5 μM) in 1 mM ATP was incubated with the indicated concentration of compound 3c in a total volume of 25 μL for 20 min at 37 °C. The emission spectrum between 300 - 450 nm was recorded (excitation at 290 nm) using a SpectraMax M5 microplate reader (Molecular Devices, Sunnyvale, CA).
Enzyme-linked Immunosorbant Assay
The procedure for DnaK binding to luciferase was adapted from a previous report.26 Briefly, firefly luciferase (0.2 mg/mL) was first incubated with 0.1 μM trypsin at 37 °C for 1 hr in HEPES buffer (40 mM HEPES, 8 mM MgCl2, 20 mM NaCl, 20 mM KCl, 0.3 mM EDTA, pH 7.2), the reaction was quenched with 1 mM PMSF and diluted to 5 μg/mL with PBS buffer (pH 7.4). An aliquot (50 μL) was then added to 96-well plates (ThermoFisher brand, clear, non-sterile, flat bottom). After 1 hr of incubation at 37 °C, the wells were washed three times with 100 μL PBS-T (0.05% Tween-20). A solution of DnaK (at the indicated concentrations), compound 3c, and 1 mM ATP was pre-incubated in HEPES with 0.01% Tween for 30 min. From this solution, 50 μL was added to wells and allowed to bind for 30 min at room temperature. After washing three times with PBS-T, 5% non-fat milk (w/v) in 100 μL TBS-T was used to block the non-specific binding sites. Primary antibody was then added (1:3000 dilution of rabbit anti-DnaK serum in TBS-T, 50 μL/well) and the plates were incubated for 1 hr at room temperature. Wells were then washed three times with TBS-T, followed by addition of HRP-conjugated secondary (1:3000 dilution of goat anti-rabbit serum in TBS-T, 50 μL/well). After a 1 hr incubation, the wells were washed three times and signal developed for 20 min using the TMB kit from Cell Signaling Technology (Danvers, MA). The absorbance was measured using a SpectraMax M5 microplate reader (Molecular Devices, Sunnyvale, CA) at 450 nm.
RESULTS
Fluorescent, Energy Transfer Method Has Enhanced Sensitivity for Inorganic Phosphate
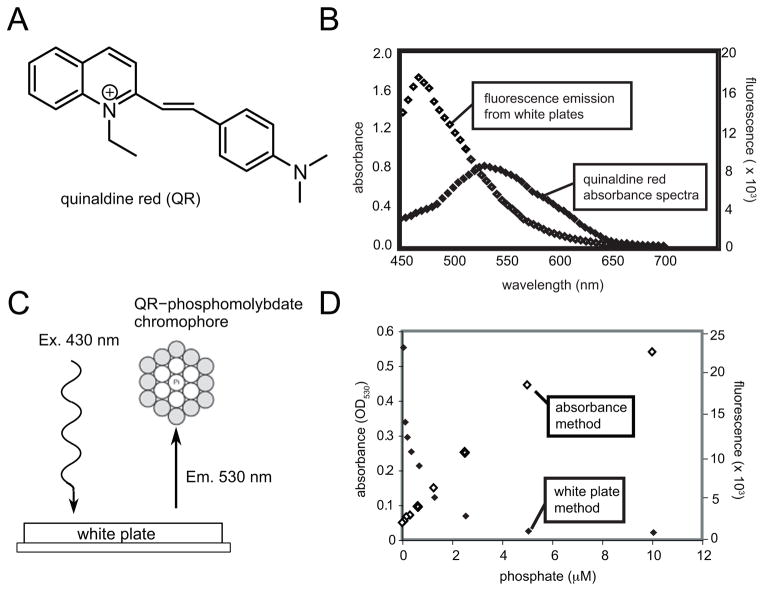
The primary technical challenge in performing absorbance assays in high-density formats, such as 384- and 1536-well plates, is that the sensitivity is typically decreased ~2.5 fold by the restrictive well geometry.27; 28 In addition, absorbance assays demand flat, clear-bottomed microtiter plates that consume more reagents than other well geometries (e.g. round, concave). However, absorbance assays are often inexpensive and robust, so there is interest in finding ways to adopt them to higher density. One promising approach was reported by Zuck et al., who successfully converted a QR-based phosphate assay for use in 384-well format (Fig. 1A).23 They observed that when white, opaque microtiter plates are irradiated at 430 nm, they emit fluorescence in a broad range between approximately 450 and 600 nm (Fig. 1B). Because the QR-complex absorbs light at 530 nm, the emission from wells containing phosphate is quenched; thus, this simple modification converts the absorbance assay into a fluorescence-quenching format (Fig. 1C). Zuck and colleagues found that this strategy improves sensitivity because both the fluorescence emission and excitation are absorbed by the chromophore.23 Further, the Walt group has explored the mechanistic underpinnings of a similar process, and they found that both radiative and non-radiative energy transfer events contribute to improved sensing.29
FIG. 1.
Model for converting an absorbance assay into a fluorescence quenching method. (A) Chemical structure of quinaldine red (QR). (B) Overlap between QR-phosphomolybdate chromophore and intrinsic fluorescence of white opaque 384-well plates. When irradiated at 430 nm, white plates emit broadly between 450 and 550 nm. (C) Schematic representation of white plate method. Fluorescence emission from white plates is quenched by QR in the presence of molybdate and inorganic phosphate (spheres). (D) Comparison of sensitivity for phosphate between absorbance and fluorescence methods. Results are the average of at least triplicates and error bars are standard error of the mean (some bars are smaller than the symbols).
Before adapting this platform for use against DnaK, we were interested in discerning the lowest concentration of phosphate that could be robustly detected. Accordingly, we prepared phosphate standard solutions and directly compared the sensitivity of the absorbance- and fluorescence-based methods. For these experiments, we found that the linear detection range of the absorbance method was approximately 0.5 to 5 μM (Fig. 1D), in agreement with previous reports.17 We found that the white plates gave a linear range of approximately 0.06 to 0.6 μM, an approximately 10-fold improvement in sensitivity.
Fluorescence Method Has Superior Performance in Pilot Screens
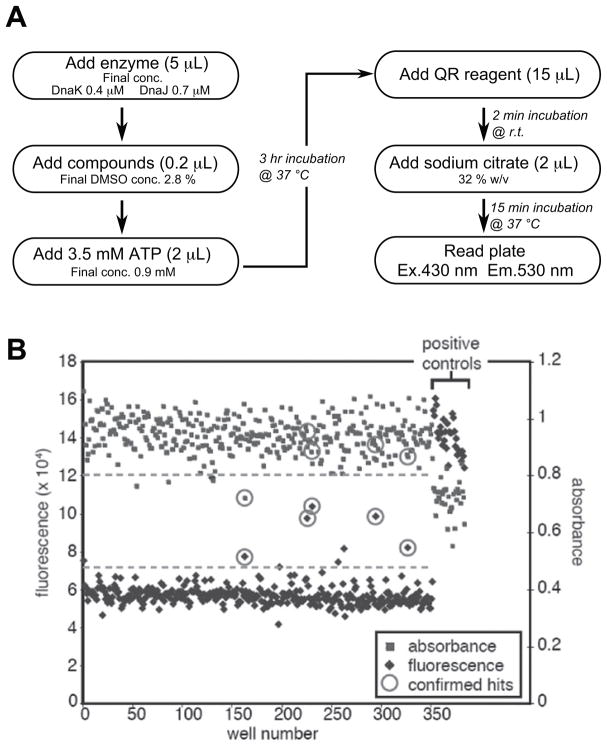
To our knowledge, a high throughput screening application of this fluorescence method has not yet been reported. Therefore, we first performed a pilot screen in 384-well plates to gauge its performance. Briefly, we found that the combination of 0.4 μM DnaK and 0.7 μM DnaJ gave a signal that was linear for at least 2–3 hours when incubated with 1 mM ATP (Supplemental Fig. 1). From these experiments, we developed a protocol that was adopted from the previous 96-well version (Fig. 2A).18 For calculation of Z′ scores, we defined the negative control as the signal from enzyme plus DMSO and the positive control as buffer plus DMSO.
FIG. 2.
Comparison of screening performances between absorbance and fluorescence methods. (A) Detailed protocol for fluorescence quenching method in 384-well plates. (B) Representative data from the side-by-side pilot screen for absorbance method and fluorescence method. Each dotted line represents three times standard deviation from the negative control. Data points for known inhibitors (as determined from previous 96-well assays) are circled. Fluorescence based method successfully identified all of the five known inhibitors whereas absorbance based method only detected one. The fluorescence method yielded ~4-fold higher signal-to-noise ratio than the absorbance method.
Using these conditions, we screened the MS2000 collection of bioactive compounds. This library is known to contain five DnaK inhibitors; thus, it could be used to probe the assay performance.21 Also, we directly compared the absorbance and energy transfer methods by splitting the reactions into either clear plates or low volume, white plates. Using this side-by-side approach, we confirmed our previous observations that the absorbance-based assay is a relatively poor HTS method, with a Z′ score of approximately 0.2. Moreover, this method only identified one of five known inhibitors and it gave two false positives (Fig. 2B). In contrast, the fluorescence method yielded a Z′ score of 0.6 and it correctly identified all five known inhibitors. We also confirmed that the fluorescence method could accurately replicate the IC50 of one inhibitor (Supplemental Fig. 1C). Together, these results suggest that the fluorescence method is suitable for screening in low volume 384-well plates.
High Throughput Screen for Inhibitors of DnaK’s ATPase Activity
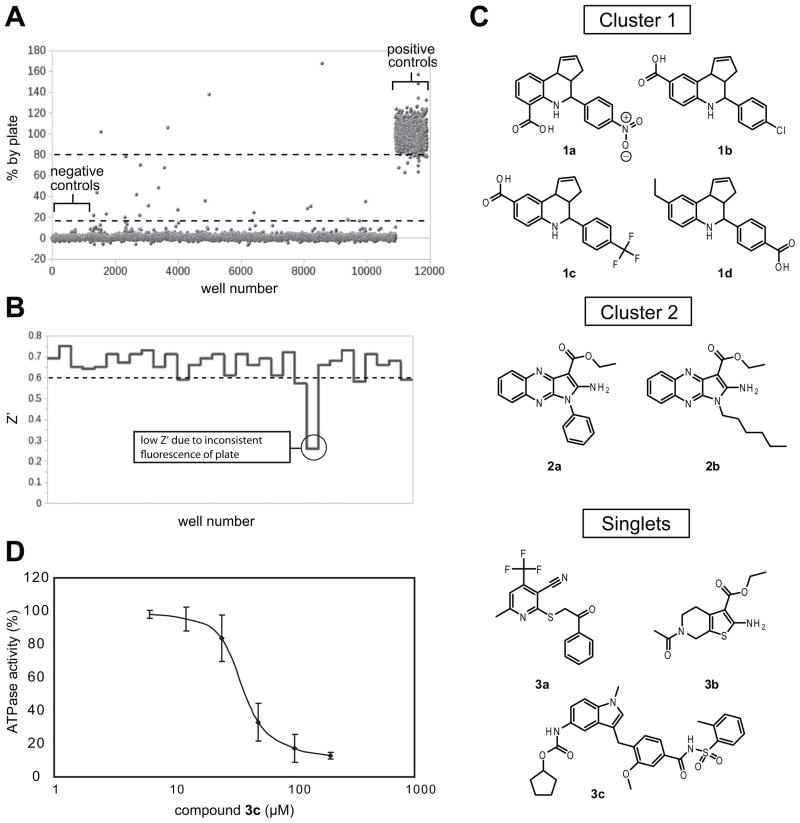
Based on the promising results from the pilot experiments, we screened an additional 55,400 small molecules and natural product crude extracts against the ATPase activity of DnaK-DnaJ (Fig. 3A). In these experiments, we found that the Z′ factors ranged from 0.10–0.75 with an average of 0.58 (Fig. 3B). Interestingly, we found that the occasional poor Z′ values were partially attributable to plates with intrinsic fluorescence that were either significantly above or below average (Supplemental Fig. 2). Because these microtiter plates were not originally manufactured for use in fluorescence applications, we expect that the amount of fluorescent material may be unevenly distributed, resulting in occasional plates with higher or lower signal. These unsatisfactory plates (Z′ value < 0.4) were excluded from further analyses (3 % of total plates), increasing the average Z′ score to 0.60. More recently, we have started “pre-screening” fresh, empty plates to remove these outliers.
FIG. 3.
Screening of 55,400 samples. (A) Representative data from the screening campaign. Positive controls (buffer plus DMSO) and negative controls (enzyme plus DMSO) are shown. Some examples of false positives are clearly shown above the 100% line. (B) Representative Z′ values by plate from the screen. The average Z′ for the entire screening was 0.58. We observed occasional low Z′, which is likely due to plates’ irregular fluorescence properties (circled). (C) Chemical structures of representative active compounds. A total of 36 inhibitors were identified in this screen; 26 cluster I compounds, 6 cluster II and 4 singlets. The structures of the re-tested compounds are shown. (D) Inhibition curve for compound 3c. IC50 value was calculated to be 37 ± 1 μM. The assay was performed in triplicates and error bars represent standard error of mean.
Samples were screened at a single concentration (between 10 to 40 μM) against DnaK-DnaJ, with the addition of 0.01% Triton X-100 to minimize false discovery of aggregators. From these screens, samples that exhibited effects of 35% or higher by plate were considered inhibitors. Samples with signals at least 3-times the standard deviation from the negative controls and samples on their given plate were also included. Of 55,400 samples tested, 598 fell into this category (hit rate: 1.1 %). In our previous studies, activators of Hsp70s have also proven to be powerful chemical tools.20; 21 With the aim of potentially expanding the number of activators, the HTS results were reviewed for samples with signals of −2.5 standard deviations or lower. Using this modest criterion, 122 unique samples were identified as activators (hit rate: 0.22 %).
To confirm their activities, the samples were subjected to confirmatory retesting in duplicate. Prior to this experiment, 10 compounds containing a heavy metal and 11 compounds with a molecular weight above 650 Da were excluded, because they were expected to have unfavorable properties. In addition, the natural product extracts were excluded and their analysis will be reported elsewhere. Thus, a total of 508 unique compounds (400 inhibitors and 108 activators) were assayed and 73 inhibitors confirmed. None of the activators confirmed upon retesting, although it should be noted that they only exhibited weak activity in the primary screen and the assay conditions were optimized to identify inhibitors. The Z′ factors for the confirmation assay ranged from 0.65 and 0.70.
The confirming structures were subsequently clustered to determine if series were present. Clustering at 65% + similarity with the fingerprints (Unity) and clustering algorithms (Optisim) by Benchware DataMiner produced 55 clusters. It has been shown that a compound with 85% or greater structural similarity to an active compound will have a ~30% probability of also being active.30 Therefore, we analyzed our internal database of ~150,000 compounds and retrieved structures that have 85% or greater similarity to any of the confirmed inhibitors. Based on this analysis, we selected 76 compounds that belong to three largest clusters for testing in dose dependence assays. These assays were performed using a 2-fold dilution series of eight compound concentrations (1 to 125 μM). Of the 127 compounds tested, 70 showed dose dependent inhibition curves. In order to minimize false positives, these 70 compounds were then evaluated for autofluorescence. Briefly, compounds were excluded if their intrinsic fluorescence (Ex: 430 nm, Em: 530 nm) at 5 μM was at least 10% of the positive control or if their dose dependence plateau was above the positive control (see examples in Fig. 3A). These filters excluded 34 samples, leaving 36 unique compounds as potential DnaK-DnaJ inhibitors. These compounds were clustered to reveal six unique chemical scaffolds. Finally, each of these compounds was manually reviewed for the possession of reactive groups such as Michael acceptors, epoxides, and free thiols. However, none of them had such functional groups.
To further evaluate the identified actives, we repurchased four examples that belong to the largest cluster (cluster I, containing 26 of the 36 remaining compounds); 1a (4-(4-nitrophenyl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-6-carboxylic acid) (ChemBridge, cat. 5585430), 1b (4-(4-chlorophenyl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-8-carboxylic acid) (ChemDiv, cat. 2374-0013), 1c (4-(3-(trifluoromethyl)phenyl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-8-carboxylic acid) (ChemDiv, cat. 5408-1849), 1d (4-(4-ethylphenyl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-8-carboxylic acid) (ChemDiv, cat. 6415-0967) (Fig. 3C). Unexpectedly, none of these compounds were inhibitors upon re-testing. These samples were subsequently subjected to a side-by-side assay against the original stock solutions. Interestingly, the two series had significantly different inhibitory profiles; the “old” samples were highly active whereas the repurchased compounds were inactive. Despite these differences, analysis by mass spectrometry and 1H NMR suggested that both sets of compounds were relatively pure and that they had the correct molecular formulae. One potential clue to this discrepancy came from observations made during the assay setup, in which we noticed differences in the color of these samples. Specifically, over the span of 5 days at room temperature in DMSO, the re-purchased compounds would eventually become slightly colored. Moreover, these aged samples now had anti-DnaK ATPase activity upon re-testing. Based on the reported synthesis of this scaffold31 and the presence of a free acid in the active structures, we suspected that the color difference (and perhaps the activity differences) might arise due to metal chelation. Indeed, treatment with EDTA inhibited the color change and extraction of the “active” form with an EDTA solution led to a loss of its activity (data not shown). Thus, we concluded that the active sample appears to involve a metal and no further investigation was pursued.
Compounds belonging to the other clusters were studied in a similar manner. Briefly, we repurchased two compounds from cluster II [2a (ethyl 2-amino-1-phenyl-1H-pyrrolo[2,3-b]quinoxaline-3-carboxylate) (ChemDiv, cat. 5122-1769), 2b (ethyl 2-amino-1-hexyl-1H-pyrrolo[2,3-b]quinoxaline-3-carboxylate) (ChemDiv, cat. 6256-4951)] and the three singlets that were available for resupply [3a (6-methyl-2-((2-oxo-2-phenylethyl)thio)-4-(trifluoromethyl)nicotinonitrile) (ChemDiv, cat. 3076-0200), 3b (ethyl 6-acetyl-2-amino-4,5,6,7-tetrahydrothieno[2,3-c]pyridine-3-carboxylate) (ChemDiv, cat. K813-0112), 3c (cyclopentyl (3-(2-methoxy-4-((o-tolylsulfonyl)carbamoyl)benzyl)-1-methyl-1H-indol-5-yl) carbamate) (Cayman Chemical Inc., cat. 10008282)]. We confirmed the structure of these compounds by mass spectrometry and then determined their IC50 values. Compounds 2a, 2b, 3a and 3b had poor activity (> 500 μM) and were not further pursued. However, compound 3c inhibited ATPase activity with an IC50 value of 37 ± 1 μM (Fig. 3D). Previously identified, allosteric Hsp70 inhibitors also have relatively modest, micromolar IC50 values.5 Thus, the activity of 3c is comparable to some of the known compounds.
Compound 3c Binds DnaK, Favors High Affinity Binding to Luciferase and Blocks DnaK’s Stimulation by DnaJ
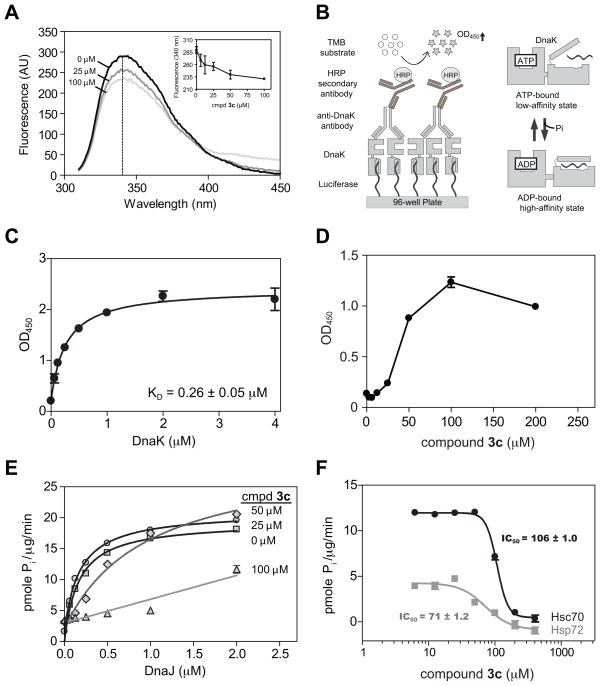
To characterize compound 3c in secondary assays, we first investigated its ability to directly bind DnaK. Using tryptophan fluorescence, we found that the compound bound the chaperone with an estimated KD of 13 ± 1.7 μM, in relatively good agreement with the IC50 value from the ATPase assay (Fig. 4A). Next, we wanted to explore how compound 3c might influence the structure of DnaK and, specifically, its interactions with substrates. As mentioned above, the ATP-bound form of DnaK has relatively poor affinity for substrate, while the ADP-bound form has enhanced affinity (Fig. 4B).8 To test whether compound 3c might change DnaK’s apparent affinity for substrate, we measured binding of the chaperone to the model substrate, firefly luciferase (Fig. 4B). By this method, the affinity of DnaK for luciferase was 260 ± 50 nM (Fig. 4C) and addition of compound 3c significantly improved the apparent affinity (Fig. 4D).
FIG. 4.
Compound 3c binds DnaK and blocks DnaJ-stimulated ATPase activity. (A) Tryptophan fluorescence was used to measure binding of compound to DnaK. Protein (5 μM + 1 mM ATP). and compound were pre-incubated for 20 minutes at 37 °C prior to measurement of intrinsic fluorescence (ex 290 nm; em 300–450 nm). Inset: the results of the dose dependence experiments are the average of triplicates and the error bars represent standard error of mean. The dose dependence experiments were carried out at 340 nm (dotted line). (B) On the left is a schematic of the ELISA for DnaK binding to a model substrate, firefly luciferase. On the right is a schematic of the ADP-bound form of DnaK, which binds tighter than the ATP-bound form to substrates. (C) Measurement of DnaK binding to luciferase, using the ELISA approach. The results are the average of duplicates and the error bars represent standard error of mean. (D) Compound 3c favored the high-affinity form of DnaK. The chaperone (0.05 μM DnaK + 1 mM ATP) was treated with 3c at the indicated concentrations for 20 minutes prior to the ELISA. (E) Compound 3c specifically blocked DnaJ-mediated stimulation of ATP turnover by DnaK. The DnaK concentration was 0.6 μM and ATPase activity was measured using the absorbance version of the assay. The results are the average of triplicates and the error bars represent standard error of mean. (F) Compound 3c also inhibited the DnaJ-stimulated ATPase activity of bovine Hsc70 and human Hsp72, as measured using the absorbance version of the assay. The protein concentrations were: Hsc70 or Hsp72 (0.6 μM) and DnaJ (1.0 μM). The results are the average of triplicates and the error bars represent standard error of mean.
Because much of the signal from the ATPase assay is due to the stimulatory activity of the co-chaperone DnaJ, we hypothesized that compound 3c might alter the interaction between DnaK and DnaJ. To explore that model, we determined the K0.5 of DnaJ to estimate the strength of the protein-protein interaction. In the absence of inhibitor, we found that the K0.5 of DnaJ was 0.17 ± 0.01 μM (Fig. 4E). Addition of low levels (25 μM) of compound 3c did not significantly alter this value (K0.5 = 0.18 ± 0.01 μM). However, higher concentrations (50 or 100 μM) increased the K0.5 to 0.96 ± 0.16 μM and greater than 2 μM, respectively (Fig. 4E). These results suggested that binding of compound 3c to DnaK might interrupt its interactions with DnaJ.
Finally, members of the Hsp70 family are highly conserved, with DnaK sharing nearly 50% sequence identity with human Hsp72 (HSPA1A) and bovine Hsc70 (HSPA8). To test whether compound 3c might inhibit the ATPase activity of these related family members, we carried out dose dependence experiments in the presence of DnaJ. We found that this compound had an IC50 of 71 ± 1.2 μM for human Hsp72, and an IC50 of 106 ± 1.0 μM against bovine Hsc70 (Fig. 4F), showing that compound 3c also inhibits the mammalian isoforms.
DISCUSSION
Absorbance assays are widely used to monitor the activity of ATPases, GTPases, phosphatases and kinases. They are widely used, in part, because they are robust, easy to use and relatively inexpensive. However, these assays are typically performed in cuvettes or medium-density (i.e. 96-well) microtiter plates. While these formats are suitable for some studies, they are not preferred for high throughput screens, especially in low volume. Adopting a method developed by Zuck et al., 23 we have successfully converted a QR-based absorbance assay to 384-well format and screened a collection of >55,000 compounds. Although this general method was published in 2005, it remains largely under-evaluated in the screening literature. Rather, other phosphate-detection technologies, such as Phosphate Sensor (Invitrogen, Carlsbad, CA) and Transcreener™ ADP2 kit (Bellbrook Laboratories, Madison, WI) have been more widely used.32 In our hands, this simple and inexpensive (approximately 1–2 cents/well) fluorescence method was robust and had good assay performance against a weak ATPase. However, it must be emphasized that there is significant potential for interference from autofluorescent or colored compounds, yielding false positives. Also, it should be clearly noted that occasional plates, from multiple manufacturers, produced very poor Z′ values due to their inconsistent fluorescence (see Supplemental Fig. 2). Despite these important concerns, we suspect that this procedure may be useful in certain settings, especially when cost is a primary concern.
Hsp70 family proteins have emerging potential as therapeutic targets; however, only a handful of relatively modest inhibitors are known.5 Clearly, one challenge is the weak enzymatic activity of these chaperones, which hindered our own efforts to screen for new inhibitors in low volume, 384-well format. Using the energy transfer methodology, we embarked on a screening strategy against E. coli DnaK in combination with its co-chaperone DnaJ. Screening of 55,400 molecules identified compound 3c, with an IC50 value of 37 ± 1 μM (see Fig. 3C) and an affinity of 13 ± 1.7 μM (see Fig. 4A). Compound 3c is a known leukotriene receptor antagonist and an FDA-approved asthma drug, marketed by Astra-Zeneca as zafirlukast (Accolate®), thus it is likely not a selective DnaK inhibitor. However, this scaffold might still be a revealing chemical probe. Specifically, it might help illuminate the currently cryptic mechanisms of allostery in the chaperone complex. For example, we found that compound 3c selectively suppressed stimulation by DnaJ, while having little effect on the intrinsic ATPase activity (see Fig 4). Further, this compound did not block binding of DnaK to luciferase and, rather, it significantly enhanced this interaction (see Fig 4D). The high affinity form of DnaK is a poor partner for DnaJ,10 which might partially explain the observed behaviors. Future structural studies are expected to provide more insight into the mechanism of this compound and, more broadly, the pathways of allosteric regulation in the DnaK-DnaJ complex.
Supplementary Material
Acknowledgments
The authors thank H. Larch and D. Walt for critical and insightful comments. This work was supported by grants from ThermoFisher Scientific Inc., the Alzheimer’s Association (IIRG-07-60067), the NIH (NS059690) and NSF (MCB-0844512) to J.E.G.
References
- 1.Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 2.Ciocca DR, Calderwood SK. Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones. 2005;10:86–103. doi: 10.1379/CSC-99r.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Witt SN. Hsp70 molecular chaperones and Parkinson’s disease. Biopolymers. 2010;93:218–228. doi: 10.1002/bip.21302. [DOI] [PubMed] [Google Scholar]
- 4.Singh VK, Utaida S, Jackson LS, Jayaswal RK, Wilkinson BJ, Chamberlain NR. Role for dnaK locus in tolerance of multiple stresses in Staphylococcus aureus. Microbiology. 2007;153:3162–3173. doi: 10.1099/mic.0.2007/009506-0. [DOI] [PubMed] [Google Scholar]
- 5.Patury S, Miyata Y, Gestwicki JE. Pharmacological targeting of the Hsp70 chaperone. Curr Top Med Chem. 2009;9:1337–1351. doi: 10.2174/156802609789895674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertelsen EB, Chang L, Gestwicki JE, Zuiderweg ER. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc Natl Acad Sci U S A. 2009;106:8471–8476. doi: 10.1073/pnas.0903503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flynn GC, Pohl J, Flocco MT, Rothman JE. Peptide-binding specificity of the molecular chaperone BiP. Nature. 1991;353:726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- 8.Mayer MP, Schroder H, Rudiger S, Paal K, Laufen T, Bukau B. Multistep mechanism of substrate binding determines chaperone activity of Hsp70. Nat Struct Biol. 2000;7:586–593. doi: 10.1038/76819. [DOI] [PubMed] [Google Scholar]
- 9.Russell R, Wali Karzai A, Mehl AF, McMacken R. DnaJ dramatically stimulates ATP hydrolysis by DnaK: insight into targeting of Hsp70 proteins to polypeptide substrates. Biochemistry. 1999;38:4165–4176. doi: 10.1021/bi9824036. [DOI] [PubMed] [Google Scholar]
- 10.Szabo A, Langer T, Schroder H, Flanagan J, Bukau B, Hartl FU. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc Natl Acad Sci U S A. 1994;91:10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang J, Maes EG, Taylor AB, Wang L, Hinck AP, Lafer EM, et al. Structural basis of J cochaperone binding and regulation of Hsp70. Mol Cell. 2007;28:422–433. doi: 10.1016/j.molcel.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kragol G, Lovas S, Varadi G, Condie BA, Hoffmann R, Otvos L., Jr The antibacterial peptide pyrrhocoricin inhibits the ATPase actions of DnaK and prevents chaperone-assisted protein folding. Biochemistry. 2001;40:3016–3026. doi: 10.1021/bi002656a. [DOI] [PubMed] [Google Scholar]
- 13.Kang Y, Taldone T, Clement CC, Fewell SW, Aguirre J, Brodsky JL, et al. Design of a fluorescence polarization assay platform for the study of human Hsp70. Bioorg Med Chem Lett. 2008;18:3749–3751. doi: 10.1016/j.bmcl.2008.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogel M, Bukau B, Mayer MP. Allosteric regulation of Hsp70 chaperones by a proline switch. Mol Cell. 2006;21:359–367. doi: 10.1016/j.molcel.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 15.Swain JF, Dinler G, Sivendran R, Montgomery DL, Stotz M, Gierasch LM. Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol Cell. 2007;26:27–39. doi: 10.1016/j.molcel.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Massey AJ, Williamson DS, Browne H, Murray JB, Dokurno P, Shaw T, et al. A novel, small molecule inhibitor of Hsc70/Hsp70 potentiates Hsp90 inhibitor induced apoptosis in HCT116 colon carcinoma cells. Cancer Chemother Pharmacol. 2009 doi: 10.1007/s00280-009-1194-3. [DOI] [PubMed] [Google Scholar]
- 17.Carter SG, Karl DW. Inorganic phosphate assay with malachite green: an improvement and evaluation. J Biochem Biophys Methods. 1982;7:7–13. doi: 10.1016/0165-022x(82)90031-8. [DOI] [PubMed] [Google Scholar]
- 18.Chang L, Bertelsen EB, Wisén S, Larsen EM, Zuiderweg ER, Gestwicki JE. High-throughput screen for small molecules that modulate the ATPase activity of the molecular chaperone DnaK. Anal Biochem. 2008;372:167–176. doi: 10.1016/j.ab.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 19.Wisen S, Bertelsen EB, Thompson AD, Patury S, Ung PM, Chang L, et al. Binding of a Small Molecule at a Protein-Protein Interface Regulates the Chaperone Activity of Hsp70-Hsp40. ACS Chem Biol. doi: 10.1021/cb1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koren J, 3rd, Jinwal UK, Jin Y, O’Leary J, Jones JR, Johnson AG, et al. Facilitating Akt clearance via manipulation of Hsp70 activity and levels. J Biol Chem. 2010;285:2498–2505. doi: 10.1074/jbc.M109.057208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jinwal UK, Miyata Y, Koren J, 3rd, Jones JR, Trotter JH, Chang L, et al. Chemical manipulation of hsp70 ATPase activity regulates tau stability. J Neurosci. 2009;29:12079–12088. doi: 10.1523/JNEUROSCI.3345-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang AM, Morishima Y, Clapp KM, Peng HM, Pratt WB, Gestwicki JE, et al. Inhibition of Hsp70 by methylene blue affects signaling protein function and ubiquitination and modulates polyglutamine protein degradation. J Biol Chem. 2010 doi: 10.1074/jbc.M109.098806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuck P, O’Donnell GT, Cassaday J, Chase P, Hodder P, Strulovici B, et al. Miniaturization of absorbance assays using the fluorescent properties of white microplates. Anal Biochem. 2005;342:254–259. doi: 10.1016/j.ab.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 24.Chang L, Thompson AD, Ung P, Carlson HA, Gestwicki JE. Mutagenesis reveals the complex relationships between ATPase rate and the chaperone activities of Escherichia coli heat shock protein 70 (HSP70/DNAK) J Biol Chem. 2010 doi: 10.1074/jbc.M110.124149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ha JH, McKay DB. Kinetics of nucleotide-induced changes in the tryptophan fluorescence of the molecular chaperone Hsc70 and its subfragments suggest the ATP-induced conformational change follows initial ATP binding. Biochemistry. 1995;34:11635–11644. doi: 10.1021/bi00036a040. [DOI] [PubMed] [Google Scholar]
- 26.Wawrzynow A, Zylicz M. Divergent effects of ATP on the binding of the DnaK and DnaJ chaperones to each other, or to their various native and denatured protein substrates. J Biol Chem. 1995;270:19300–19306. doi: 10.1074/jbc.270.33.19300. [DOI] [PubMed] [Google Scholar]
- 27.Kreusch S, Schwedler S, Tautkus B, Cumme GA, Horn A. UV measurements in microplates suitable for high-throughput protein determination. Anal Biochem. 2003;313:208–215. doi: 10.1016/s0003-2697(02)00460-8. [DOI] [PubMed] [Google Scholar]
- 28.Lavery P, Brown MJ, Pope AJ. Simple absorbance-based assays for ultra-high throughput screening. J Biomol Screen. 2001;6:3–9. doi: 10.1177/108705710100600102. [DOI] [PubMed] [Google Scholar]
- 29.Yuan P, Walt DR. Calculation for Fluorescence Modulation by Absorbing Species and Its Application to Measurements Using Optical Fibers. Analytical Chemistry. 1987;59:2391–2394. [Google Scholar]
- 30.Martin YC, Kofron JL, Traphagen LM. Do structurally similar molecules have similar biological activity? J Med Chem. 2002;45:4350–4358. doi: 10.1021/jm020155c. [DOI] [PubMed] [Google Scholar]
- 31.Kiselyov A, Smith L, Armstrong R. Solid support synthesis of polysubstituted tetrahydroquinolines via three-component condensation catalyzed by Yb(OTf)(3) Tetrahedron. 1998;54:5089–5096. [Google Scholar]
- 32.Rowlands M, McAndrew C, Prodromou C, Pearl L, Kalusa A, Jones K, et al. Detection of the ATPase activity of the molecular chaperones Hsp90 and Hsp72 using the TranscreenerTM ADP assay kit. J Biomol Screen. 15:279–286. doi: 10.1177/1087057109360253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.