Abstract
microRNAs (miRNAs) are the subject of enormous interest. They are small non-coding RNAs that play a regulatory role in numerous and diverse cellular processes such as immune function, apoptosis and tumorigenesis. Several virus families have been shown to encode miRNAs, and an appreciation for their roles in the viral infectious cycle continues to grow. Despite the identification of numerous (>225) viral miRNAs, an in depth functional understanding of most virus-encoded miRNAs is lacking. Here we focus on a few viral miRNAs with well-defined functions. We use these examples to extrapolate general themes of viral miRNA activities including autoregulation of gene expression, avoidance of host defenses, and a likely important role in maintaining latent and persistent infections. We hypothesize that although the molecular mechanisms and machinery are similar, the majority of viral miRNAs may utilize a target strategy that differs from host miRNAs. That is, many viral miRNAs may have evolved to regulate viral-encoded transcripts or networks of host genes that are unique to viral miRNAs. Included in this latter category are a likely abundant class of viral miRNAs that may regulate only one or a few principal host genes. Key steps forward for the field are discussed, including the need for additional functional studies that utilize surgical viral miRNA mutants combined with relevant models of infection.
Introduction
Discovery of miRNAs
Originally called small temporal RNAs (stRNAs) for the roles the founding members played in regulating the proper timing of developmental events, miRNAs were first discovered in 1993 by Ambros and colleagues in the laboratory model organism nematode Ceanorhabditis elegans (C. elegans) (Lee et al., 1993). Using genetic studies, it was demonstrated that a small RNA derived from a hairpin structure had potent negative trans regulatory ability by binding to the 3’ UTR of other transcripts (Reinhart et al., 2000). Originally thought to be an oddity of nematodes, it was later demonstrated that at least some C. elegans miRNAs are conserved with other metazoans including humans (Pasquinelli et al., 2000). In 2001, three landmark papers were published in a single issue of Science that described over a hundred miRNAs (some evolutionarily conserved) that are found in diverse metazoan cells (Lagos-Quintana et al., 2001; Lau et al., 2001; Lee and Ambros, 2001). These studies launched a revolution in our understanding of gene regulation that continues to expand with currently over 1000 papers on miRNAs being published each year (source ISI web of Science, over 1000 papers published each year since 2008 with “microRNA” used as the topic search term).
Biogenesis
Because viral miRNAs mostly utilize the same processing and effector machinery as host miRNAs, and excellent reviews have already be written on this subject (Bartel, 2004; Carthew and Sontheimer, 2009; Kim, 2005), we provide only a brief overview of biogenesis here. miRNAs are derived from primary transcripts (pri-miRNAs), typically hundreds to thousands of nucleotides long, that undergo a series of endonucleolytic processing and shuttling events to give rise to a final ~22 nucleotide effector molecule. Most pri-miRNAs are transcribed by RNA polymerase II (pol II); hence, similar to protein coding mRNAs, their transcription is regulated by proteinaceous transcription factors, they are processed to the display typical 7-methyl guanosine cap structures as well as polyA tails, and they can undergo splicing reactions. The vast majority of pri-miRNAs contain an ~80 nucleotide hairpin secondary structure that can be intronic or exonic. The hairpin portion of the pri-miRNA is recognized by Microprocessor, a multi-protein nuclear complex containing the RNaseIII-like enzyme Drosha. Drosha-mediated double-stranded cleavage of a bottom portion of the stem (away from the terminal loop) “liberates” a ~60 nucleotide hairpin called a precursor miRNA (pre-miRNA), which is recognized by the RAN-GTPase Exportin5 complex and exported to the cytoplasm (Lee et al., 2003; Zeng et al., 2005).
Once in the cytoplasm, the terminal loop portion of the pre-miRNA is removed by the RNaseIII-like enzyme Dicer (Bernstein et al., 2001; Hutvagner et al., 2001), leaving a transient, partially-complementary, double-stranded siRNA-like molecule. One strand of this molecule becomes stably incorporated into the multi-protein RNA induced silencing complex (RISC), where it serves to guide RISC to the mRNA transcript targets. RISC-bound miRNAs bind to mRNA targets with partial base pair complementarity, typically in the 3’ untranslated region (UTR) (Grimson et al., 2007). RISC-bound transcripts produce less protein due to molecular mechanisms which are still ill defined (Bartel, 2009). Frequently, but not always, this inhibition is associated with transcript destabilization of sufficient magnitude to be picked up in high-throughput analysis methods such as microarrays (Lim et al., 2005). Hence, analysis of mRNA steady state levels can sometimes be utilized to fish out miRNA-targeted transcripts. Although uncommon in animals, some viral and most plant miRNAs bind to their mRNA targets with perfect complementarity, and similar to siRNAs, cause a specific, irreversible endonucleolytic cleavage event in the target transcript. One unresolved mystery is why this mode of miRNA-mediated regulation is so rarely used by animal host miRNAs.
Some key functions of host miRNAs and their mode of action
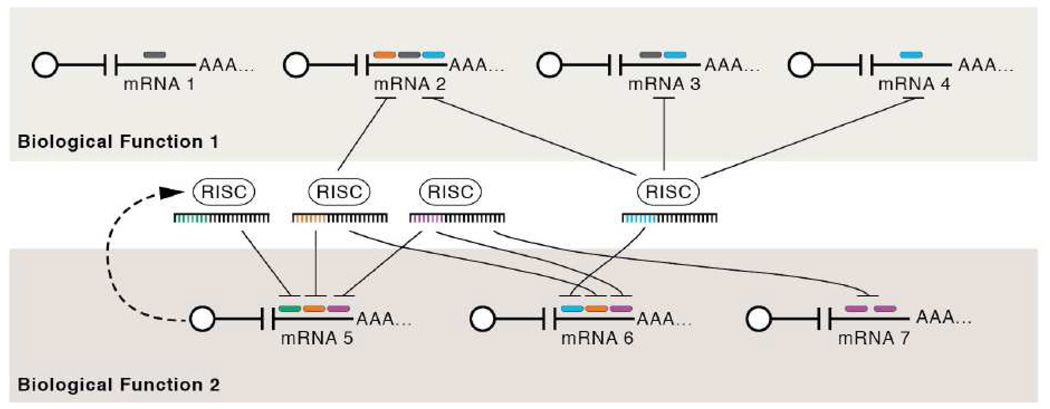
Originally identified as modulators of developmental timing, host miRNAs have now been shown to affect numerous other diverse processes including (just to name a few) cell fate determination (Johnston and Hobert, 2003), neuronal plasticity (Schratt et al., 2006), cholesterol metabolism (Krutzfeldt et al., 2005), and processes of obvious “virological relevance” such as the innate and adaptive immune responses ((Lodish et al., 2008) and references therein), apoptosis ((Subramanian and Steer, 2010) and references therein), cell cycle (Kedde et al., 2010), and tumorigenesis ((Medina and Slack, 2008) and references therein). Bioinformatics and experimental approaches have led to a model whereby ~30–75% of different mRNA transcripts are regulated by miRNAs, with each miRNA potentially regulating hundreds of transcripts (Bartel, 2009). This sets up a complex web of potential regulatory interactions. For example, the human genome encodes over 900 pre-miRNAs (Griffiths-Jones, 2006), allowing for some mRNAs to be regulated by multiple miRNAs. In this model, a network of specific transcripts and the multiple miRNAs they interact with, combine to modulate a particular biological outcome (Figure 1).
Figure 1. Model for microRNA-mediated regulation.
mRNAs are depicted as white circles (the 5’ cap) extending to the 3’ poly A tails (AAA…), miRNAs are depicted as being associated with the RNA induced silencing complex (RISC) with their important target binding determinant region (the “seed” region, nucleotides 2–8) depicted in color. The respective miRNA/RISC docking site is shown as a color-matched bar in 3’ UTR of the mRNA transcripts. This hypothetical example demonstrates several points thought to be common in many miRNA-regulated networks. mRNAs 1–4 play a role in biological process 1, mRNA 5–7 play a role in biological process 2. Note that several different miRNAs can regulate a single mRNA and conversely several different mRNAs are regulated by one miRNA. In addition, the same mRNA target can contain multiple miRNA docking sites for the same miRNA, as shown for mRNA 7. Combined, these features allow for fine-tuning regulation of gene expression. Typically, the more miRNAs regulating a transcript, the greater the degree of repression. A single miRNA can regulate hundreds of mRNA targets and can play a role in regulating different biological functions. This role can serve to regulate the amount of or the timing of gene expression. mRNA 5 can trigger a negative feedback loop of its own expression as it induces increased autoregulatory miRNA levels. Positive feed-forward loops (not depicted) also occur. Note, the potential for extensive cross talk between miRNAs and targets (and this is likely a gross oversimplification as there are greater than 900 human miRNAs known with ~hundreds of different ones being expressed in any single cell type at a given time). Furthermore, conservative estimates suggest 30%, but perhaps as many as 70% of transcripts (or more) are regulated by miRNAs. The overall picture emerging is that of complex, interlaced, regulatory networks that can serve as rheostats to finely regulate gene expression. Virus-encoded miRNA regulation offers several advantages to the virus including: the ability to regulate hundreds of transcripts, non-immunogenicity (since no proteins are required to be made by the virus), and a relatively small amount of genomic space.
Although our understanding of what dictates specific interactions between miRNA and target is still imperfect, it is clear that for many miRNA-target pairs, the 5’ end of a miRNA plays an especially important role in dictating target specificity. Nucleotides 2–8, or minimally 2–7 (numbering starting from the 5’ end of the miRNA) comprise the so-called “seed” region (reviewed in (Bartel, 2009)). The seed region often binds to target mRNAs in their 3’ UTR with perfect complementarity. Thus, miRNA overexpression or inhibition, combined with cDNA microarray analysis and computational analysis, can identify some bona fide miRNA-mRNA target interactions. Of course, there are caveats to this approach including the observation that not all miRNA-target interactions display perfect seed complementarity (Didiano and Hobert, 2006) and not all miRNAs bind to their targets in the 3’UTR (Grey et al., 2010). Furthermore, not all miRNA-target interactions result in robust, measurable decreases in target transcript steady state levels. Despite the numerous exceptions, mounting evidence suggests that many miRNA-target interactions are mediated by perfect seed complementarity between miRNA and the 3’UTR of target transcripts (Grimson et al., 2007).
Identification of viral miRNAs
Most viral miRNAs had initially been identified by a protocol previously developed for the identification of host-encoded miRNAs, a procedure that involves RNA size fractionation, ligation of linkers, reverse transcription, concatamerization, and Sanger sequencing. We and others have also developed computational approaches that rely on commonalities in the predicted secondary structures of pre-miRNAs to identify miRNA-encoding loci specifically in viral genomes (Grundhoff et al., 2006; Pfeffer et al., 2005; Sullivan et al., 2005). While such ab initio prediction approaches often produce significant numbers of false positives that have to be eliminated experimentally, they have the advantage of being able to identify the less abundantly expressed miRNAs which frequently had been overlooked in the original cloning protocol. However, with the advent of massively parallel sequencing technologies it is now possible to explore libraries of cloned small RNAs with unprecedented depth; as a consequence, the bioinformatic prediction of potential pre-miRNA hairpin structures is nowadays rarely used for other than confirmatory purposes.
The viruses that encode miRNAs
(For this section please refer to Figure 2 and Table 1). In 2004, Pfeffer and colleagues published the first report describing the cloning of viral-encoded miRNAs from a cell line infected with Epstein-Barr virus (EBV) (Pfeffer et al., 2004). EBV is a member of the Herpesviridae in the gamma sub-family and is associated with several tumors in its human host, including Burkitt’s lymphoma, Hodgkin’s disease and nasopharyngeal carcinoma. Notably, EBV also efficiently transforms primary B cells in vitro, giving rise to the so-called lymhoblastoid cell lines (LCLs). The proliferation of LCLs is driven by the expression of at least 9 viral proteins, some of which (e.g. latent membrane protein 1, LMP1) also exhibit transforming potential in mice. However, most EBV-associated tumors express a more restricted repertoire of viral proteins (in Burkitt’s lymphoma, usually only a single viral protein without known oncogenic potential is expressed), and the pathogenesis of these tumors hence is likely to be much more complex (reviewed in (Bornkamm, 2009)).
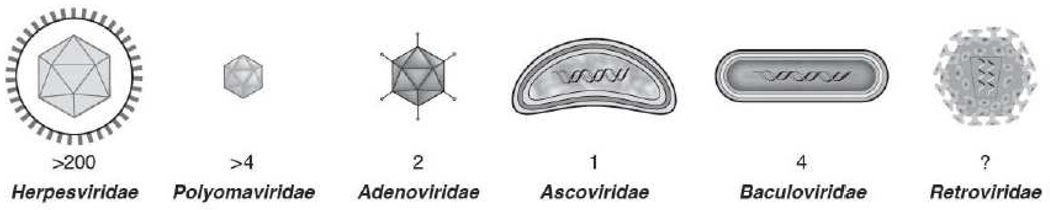
Figure 2. Virus families that encode microRNAs or microrna-like molecules.
Depicted are cartoon diagrams of virions (not exactly to scale) from virus families that have been reported to encode miRNAs. Indicated is the number of distinct pre-miRNAs reported for each family. The Herpesviridae encode the most distinct pre-miRNAs, with greater than two hundred from at least 15 different viruses. All herpesviruses that are known to encode miRNAs, express multiple pre-miRNAs (from a ~7 to greater than 25). Several members of the Polyomaviridae are known to encode a single pre-miRNA. Some members of the adenoviridae encode two non-canonical pre-miRNA-like molecules, called Virus Associated (VA) RNAs. miRNA biogenesis from VA RNAs does not utilize Drosha. These atypical pre-miRNAs are pol III transcripts that are inefficiently processed into miRNAs (less than 1% processing efficiency), but nonetheless make up the most abundant miRNAs present in the cell due to the sheer abundance of the VA RNA precursor (10 ^7 – 10 ^8 copies per cell). Currently, two insect viruses, an ascovirus and a baculovirus, are known to encode a pre-miRNA. There have been several controversial reports purporting that HIV, a member of the retroviridae, encodes pre-miRNAs. These reports have not been independently verified and their existence is the subject of some debate (Lin and Cullen, 2007). See also Table 1.
Table 1.
Virus-encoded miRNAs or miRNA-like molecules.
| Virus Family or Subfamily |
Virus Species | Pre-miR hairpins |
Mature miRs |
|---|---|---|---|
| Alpha- herpesvirinae | Herpes Simplex Virus 1 | 16 | 25 |
| Herpes Simplex Virus 2 | 18 | 24 | |
| Herpes B virus | 3 | 3 | |
| Herpesvirus of turkeys | 17 | 28 | |
| Infectious laryngotracheitis virus | 7 | 10 | |
| Bovine herpesvirus 1 | 10 | 12 | |
| Marek’s disease virus type 1 | 14 | 26 | |
| Marek’s disease virus type 2 | 18 | 36 | |
| Beta- herpesvirinae | Human cytomegalovirus | 11 | 17 |
| Mouse cytomegalovirus | 18 | 28 | |
| Gamma- herpesvirinae | Epstein Barr virus | 25 | 44 |
| Rhesus lymphocryptovirus | 36 | 50 | |
| Kaposi’s sarcoma-associated herpesvirus | 12 | 25 | |
| Rhesus monkey rhadinovirus | 15 | 25 | |
| Mouse gamma herpesvirus 68 | 15 | 28 | |
| Polyomaviridae | Simian virus 40 | 1 | 2 |
| JC polyomavirus | 1 | 2 | |
| BK polyomavirus | 1 | 2 | |
| Mouse polyomavirus | 1 | 2 | |
| Merkel cell polyomavirus | 1 | 2 | |
| SA12 | 1 | 2 | |
| Ascoviridae | Heliothis virescens ascovirus | 1 | 1 |
| Baculoviridae | Bombyx mori nucleopolyhedrosis virus | 4 | 4 |
| Adenoviridae | Human adenoviruses types 2 and 5, others? | 2* | 3 |
pre-miRNA-like molecules
Initially, Pfeffer and colleagues identified 5 EBV-encoded pre-miRNAs in B95-8 cells, an LCL generated by the in vitro infection of marmoset B cells (Pfeffer et al., 2004). Subsequent studies revealed numerous additional pre-miRNAs encoded by EBV such that, today, EBV is known to encode a total of 25 pre-miRNAs (Cai et al., 2006b; Grundhoff et al., 2006; Zhu et al., 2009), which produce at least 44 mature miRNA species (see Table 1). Indeed, most of these miRNAs had been missed in the initial study due to the fact that they are located in a region that is deleted from the genome of the B95-8 EBV strain. Since the B95-8 laboratory strain of EBV efficiently transforms primary B cells, the deleted miRNAs are clearly not required for growth transformation in vitro. However, one cannot conclude from this observation that these miRNAs do not contribute to tumorigenesis in vivo; the fact that the known field strains as well as tumor-derived EBV strains retain the deleted miRNA-coding segment points towards an important function of this genomic region.
With the benefit of hindsight it may not be surprising that EBV was found to encode miRNAs, since nearly all herpesviruses that have been reported investigated appear to do so. Indeed, more than 95% of the viral miRNAs known today are of herpesvirus origin (see Table 1). This superiority in numbers is also due to the fact that, even considering their considerable genome size, individual herpesviruses tend to express rather large numbers of miRNAs (some family members encode clusters with more than 30 pre-miRNAs). Taken together, this suggests that miRNAs play an important role in the herpesviral lifecycle. Since herpesviruses exhibit a biphasic lifecycle and establish long-lasting latent infections, one important benefit they could gain from employing miRNAs is the ability to regulate host and/or viral gene expression without expressing proteinaceous factors that may elicit an antigenic immune response. This hypothesis is further corroborated by the fact that most herpesviruses express their miRNAs during latency, even though this is a stage of the viral lifecycle, which is otherwise highly restrictive for viral gene expression.
The discovery of EBV-encoded miRNAs was followed by reports from several groups describing the identification of a total of 12 pre-miRNAs encoded by Kaposi sarcoma-associated herpesvirus (KSHV) (Cai et al., 2005; Grundhoff et al., 2006; Pfeffer et al., 2005; Samols et al., 2005). KSHV encodes 12 different pre-miRNAs that give rise to 25 different readily-detectable miRNAs. Like EBV, KSHV belongs to the gamma-herpesvirus subfamily, whose members are frequently associated with tumors in their natural host. KSHV is considered the etiologic agent of Kaposi’s sarcoma, a highly vascularized lesion comprised mostly of endothelial cells that afflicts mainly immuncompromised individuals. KSHV infection is also associated with rare B cell hyperproliferative disorders, including Primary Effusion Lymphoma (PEL) and some forms of Multicentric Castleman’s Disease (MCD) (see (Ganem, 2010; Wen and Damania, 2010) for recent reviews). Since gamma herpesvirus-associated tumorigenesis is always driven by latently infected cells, an attractive hypothesis is that modulation of host gene expression by viral miRNAs may causally contribute to tumorigenic processes. As discussed later, although so far not proven in animal models, at least for some viral miRNAs, the circumstantial evidence for this theory is very strong.
Additionally, like some other Herpesviruses (Buck et al., 2007; Dolken et al., 2007; Umbach et al., 2008), KSHV encodes several miRNA derivatives that are encoded exactly antisense relative to the abundant pre-miRNAs (Lin et al., 2010; Umbach and Cullen, 2009). Interestingly, the KSHV antisense miRNAs are only detectable in cells undergoing lytic replication, and are likely derivative of recently described longer non-translated RNAs that span much of the KSHV genome (Chandriani et al., 2010; Lin et al., 2010). However, in contrast to other viral antisense miRNAs that are fairly abundant (Buck et al., 2007; Dolken et al., 2007; Umbach et al., 2008), the KSHV antisense miRNAs are of extremely low abundance, thus, their functional relevance remains uncertain. Because of the low abundance of these miRNAs, we do not include them in Table 1 listing the known viral miRNAs.
As is evident from the list in Table 1, herpesvirus miRNAs are not only found in the gamma-, but also in the alpha- and beta herpesvirus subfamilies. With one notable exception (see miR-155 mimics – viral analogues of a human oncomir?), alpha- and beta herpesviruses do not cause tumors in their hosts. They are nevertheless important pathogens of animals and humans. For example, the neurotropic alpha herpesviruses Herpes Simplex Virus 1 and 2 (HSV-1 and -2) cause oral or genital herpes lesions, but may also lead to encephalitis or meningitis. The beta herpesvirus human cytomegalovirus (HCMV) represents the most significant viral cause of birth defects in industrialized countries and frequently causes life-threatening disease in immunocompromised individuals. While not tumorigenic, it is conceivable that viral miRNAs enhance the virulence of these pathogens and thus contribute to disease development.
In addition to the herpesviruses, numerous members of the polyomavirus family, as well as adenoviruses, an ascovirus, and a baculovirus have been shown to encode miRNAs or miRNA-like molecules (Aparicio et al., 2006; Cantalupo et al., 2005; Hussain et al., 2008; Seo et al., 2009; Seo et al., 2008; Singh et al., 2010; Sullivan et al., 2005; Sullivan et al., 2009). These findings predict that the viruses most likely to encode miRNAs will have DNA genomes, nuclear life cycles, and an ability to undergo latent and/or persistent infections. Additionally, there have been several reports suggesting HIV, an RNA virus whose genome is reverse transcribed and incorporated into the host DNA, may also encode miRNAs. However, these reports are controversial, with other labs unable to detect expression of these putative miRNAs (Lin and Cullen, 2007; Pfeffer et al., 2005). Finally, low or high-throughput sequencing efforts of RNA from cultured cells infected with any of several different RNA viruses, including HCV, YFV, WNV, and Influenza A failed to identify viral-encoded miRNAs (Pfeffer et al., 2005; Umbach et al., 2010).
Given the potential benefits afforded to the viral life cycle by viral-encoded miRNAs, it remains an enigmatic question as to why more virus families do not utilize this regulatory strategy. miRNAs take up relatively little genomic space, presumably are non-immunogenic, and each has the potential to regulate numerous host and/or viral transcripts. In addition, one might predict that evolving inverted hairpins should be a fairly common occurrence in evolutionary time. If true, why have viral-encoded miRNAs not been discovered in more virus families? Aside from the obvious reason that many viruses have yet to be surveyed, there are at least two reasons that could account for this: lack of access to nuclear miRNA processing machinery and the destabilizing effects of miRNA processing on RNA genomes. However, at least in theory, neither of these barriers are insurmountable (discussed in detail below).
If viruses that replicate in the cytoplasm (e.g. Pox, Picorna, Flavi,) are to encode miRNAs, they will necessarily have to bypass the need for nuclear Microprocessor cleavage. Two ways this could happen include: 1) These viruses could encode a protein with a Drosha-like function (of note, although no virus-encoded miRNA-processing proteins have yet been described, the principal hypothesis is supported by the recent identification of a Closterovirus-encoded RNAse III homolog even though this protein has a different function in inhibiting host RNAi activity (Kreuze et al., 2005)), and 2) miRNA-like molecules could be produced by transcripts whose ends come together to form a stem loop structure that is cleaved directly by Dicer. In support of this later scenario is the existence of endogenous mirtrons, short introns generated by canonical splicing of pol II-derived transcripts, that form a hairpin structure which mimics a Drosha product (Okamura et al., 2007; Ruby et al., 2007). Additionally, adenovirus expresses miRNA-like molecules that are derived from the pol III transcribed VA RNAs, which fold into structures that are processed by Dicer but not Drosha (Andersson et al., 2005; Lu and Cullen, 2004). While in both of these examples, the RNAs are initially transcribed in the nucleus, they nonetheless support the possibility that short RNAs transcribed in the cytoplasm could directly interact with Dicer to be processed into functioning miRNAs. Of note, since they bypass the microprocessor, such structures need not display the fundamental structural features of canonical pre-miRNA hairpins. This concept is exemplified by the fact that the VA RNAs represent a multi-stem structure that only mimics the basal part of a bona-fide pre-miRNA stem, including the typical 3’-OH overhang required for Exportin 5 transport and Dicer cleavage. Consequently, if such miRNA mimics exist in RNA viruses, they may well be missed by any prediction algorithm trained to pick up typical pre-miRNA stem-loops.
Viruses with RNA genomes may be at a fitness disadvantage if they were to encode regions that are prone to endonucleolytic cleavage by either Dicer or Drosha. For example, this could result in reductions in genome/antigenome copy number, or mRNA production. In spite of this theoretical barrier, retroviruses, a flavivirus, and influenza can be engineered to express biologically active miRNAs or miRNA-like molecules (Rouha et al., 2010; Shapiro et al., 2010; Varble et al., 2010). Notably, because flaviviruses have a lifecycle that is restricted to the cytoplasm, Drosha-independent mechanisms must exist to produce these miRNA-like small RNAs. Thus, at least in a laboratory setting, it is clear that viruses with RNA genomes can express miRNAs (or miRNA-like molecules) and still be grown to reasonably high titers.
Finally, we note that some viruses that would seem perfect candidates for encoding miRNAs apparently do not. Deep sequencing of small RNAs from human trigeminal ganglia infected with both Varicella Zoster Virus (VZV) and Herpes Simplex 1 (HSV-1) readily identified HSV-1 miRNAs but failed to uncover any VSV miRNAs (similar to the HSVs, VZV is also a neurotropic alpha herpesvirus most well known for causing chickenpox and shingles) (Umbach et al., 2009). A previous computational analysis (Pfeffer et al., 2005) also suggested VZV may not encode pre-miRNAs (it should, however, be noted that this analysis was performed with a stringent prediction algorithm that also missed several authentic pre-miRNAs in other herpesvirus genomes). Taken together, the above may indicate that possibly some herpesviruses do not encode miRNAs. Similarly, low throughput sequencing of RNA from human papillomavirus 31 (HPV 31)-infected cultured cells failed to identify any HPV-encoded miRNAs (Cai et al., 2006a). While neither the absence of computationally predicted pre-miRNAs nor the inability to detect mature miRNAs in infected cells rule out with certainty that these viruses might encode miRNAs, (for example, miRNAs may be only expressed in tissues where different promoters are active or novel RNA processing events occur), these results suggest that some viruses with nuclear replication, DNA genomes, and latent/persistent lifecycles, do not encode miRNAs. If true, then just as some members of the herpesvirus family do and some do not encode miRNAs, it remains a formal, albeit purely speculative, possibility that some strains of papillomaviruses encode miRNAs.
Functions of viral miRNAs
Overview
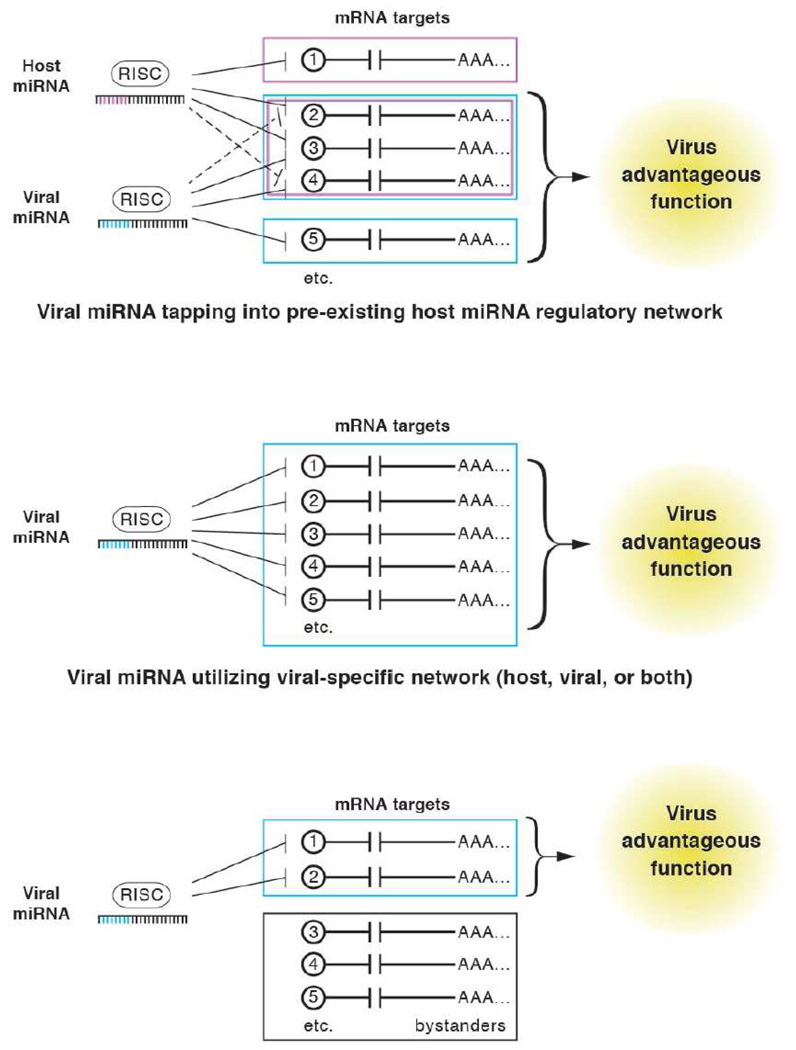
A primary goal of the field is, once a new viral-encoded miRNA is discovered, to identify its functions and relevant mRNA targets. To accomplish this, one must weed-out the “bystander” mRNAs, which can be targeted by a viral miRNA, but provide no fitness advantage to the virus (Figure 3). Remember, although expression of both a typical miRNA as well as a non-natural small RNAs can affect hundreds of mRNA targets, these target pools are of a fundamentally different quality: Endogenous miRNAs have co-evolved with their target sites over millions of years, and it therefore can be reasonably assumed that cellular miRNAs are part of complex networks in which many target interactions are of physiological relevance (Figure 1). However, such assumptions cannot be made for artificial small RNAs or for miRNAs of viral origin, as the host is unlikely to evolve and maintain a network of targets for a parasite (as discussed later, an exception from this rule may be viral mimics of cellular miRNAs, or miRNAs which target special mRNA elements). It is thus likely that, although viral miRNAs may regulate many transcripts in an experimentally reproducible and robust manner, a significant number of these interactions will represent the equivalent of “off target” effects of siRNAs. While readily tolerated during infection, these “neutral selection” negatively-regulated transcripts may provide little insight into the relevant functions of a miRNA. In addition, as multiple studies have now been published that implicate viral miRNAs as playing an important role in maintaining latent/persistent infection (see below), it becomes imperative that viral miRNAs are studied in relevant model systems that recapitulate all modes of infection. For some viral-encoded miRNAs, this necessitates in vivo models. In particular, those viral miRNAs that function to evade the adaptive immune response underscore the importance of animal models of infection. Engineered mutant viruses, defective for miRNA expression, represent a powerful approach to understanding viral miRNA function. So far, relatively few viral miRNAs have any identified mRNA targets with a confirmed functional role. Even fewer (n = 2) have been studied in the context of a deleted virus in vivo (Dolken et al., 2010; Sullivan et al., 2009). The first in vivo phenotype reported for an engineered miRNA mutant virus (Dolken et al., 2010) underscores the power of authentic infection systems, although an understanding of the targets of these miRNAs remains incomplete (discussed in detail in the “Functions-first” section below). Ultimately, the marriage of additional surgical mutant strains with relevant models of infection should be most informative.
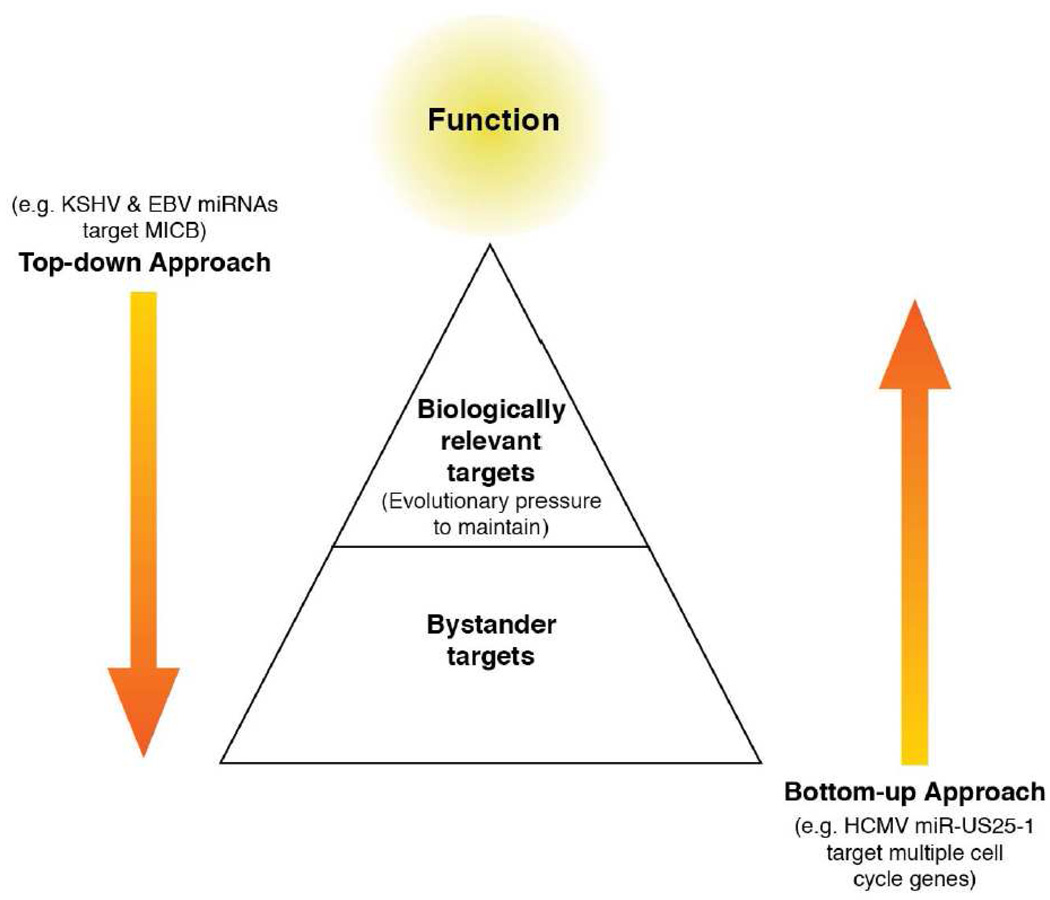
Figure 3. Strategies for determining virus-encodes microRNA function and their relevant targets.
Most strategies for determining the function and relevant targets of virus-encoded miRNAs can be grouped into two different approaches. First, in the “Bottom-up” approach, lists of putative targets can be generated by computational methods or by changes cDNA expression or RISC association studies, performed in the presence of increased or decreased activity of the miRNA. The challenge from these approaches is to glean the function of these miRNAs in the context of infection. To accomplish this, one must weed out the “bystander” targets from the functionally relevant targets. The second approach, the “Top-down” approach”, involves first determining whether a virus-encoded miRNA affects a virologically relevant function, and then to use this information to help identify the relevant target(s). The challenge with such an approach is developing relevant functional screens that can accommodate a reasonable amount of throughput.
Strategies for determining viral-encoded miRNA function
There are four predominant strategies that have been used to uncover viral miRNA functions. The first three (antisense targets, computational prediction, & comparative cDNA expression profiling/RISC pull down), describe “Bottom-up” approaches, whereby candidate targets are first identified, confirmed, and then dissected to predict function (see Figure 3). The fourth, describes a “Top-down” approach, whereby phenotypes or putative functions are first identified, confirmed in the context of infection, and then utilized to identify relevant targets. Bottom-up approaches can be particularly advantageous as a starting point in the absence of any preconceived hypotheses of function. However, each bottom-up strategy can have drawbacks. Antisense transcripts sometimes are not regulated in a siRNA-like manner (see below), and computational and comparative cDNA expression studies suffer from false positives and neutral “bystander” targets that are indeed targeted by the miRNA of interest, but provide no selective advantage to the virus (“hitching a ride,” so to speak, with the functional targets that are drivers of selection). Finally, “functions-first” strategies are a powerful approach when a particular viral–advantageous function is suspected and readily assayed with reasonable throughput. However, this approach also has drawbacks- such as the possibility of investing a large commitment of resources and time and coming up with a screen that produces no new insight to the viral miRNA of interest. Obviously, all four strategies represent only starting points to advance hypotheses. Ideally, models emerging from these strategies must be tested in the context of infection in a relevant cell type. Listed below are examples of viral miRNA functions gleaned from each strategy. An overview of known targets of cellular or viral targets is given in tables 2 and 3, respectively (please note that, with a few exceptions, we have listed only targets confirmed (at least) minimally by demonstrating that target sites confer repression in reporter assays, that mutation of the target site alleviates repression and, for cellular targets, that endogenous protein levels are down-regulated upon ectopic miRNA expression). The columns labeled ‘Lead’ indicate whether computational prediction or cDNA profiling via microarrays or RISC pull down was used to identify candidates, or whether an informed approach was used to analyze a specific subset of suspected targets, e.g. antisense transcripts or cellular or viral genes known to be implicated in certain cellular processes such as apoptosis.
Table 2.
Cellular Targets of viral miRNAs.
| Target | Virus | miRNA(s) | Proposed function |
Confir mation * |
Lead | Reference(s) |
|---|---|---|---|---|---|---|
| PUMA | EBV | miR-BART5 | anti- apoptotic |
1,2,3,5 | prediction | Choy et al., 2008 |
| SMAD2 | MDV1 | miR-M3 | 1,2,3 | informed | Xu et al., 2010 | |
| THBS1 | KSHV | miR-K1 miR-K3-3p miR-K6-3p miR-K11 |
1,2,3 | microarray | Samols et al., 2007 | |
| TWEAKR | KSHV | miR-K12-10a | 1,2,3,4, 5 |
microarray | Abend, Uldrick, and Ziegelbauer, 2010 | |
| BCLAF1 | KSHV | miR-K5 miR-K9 miR-K10a/b |
Inhibit caspase, may facilitate lytic cycle |
1,2,3,4, 5 |
microarray | Ziegelbauer, Sullivan, and Ganem, 2009 |
| MICB | hCMV KSHV EBV |
miR-UL112 miR-K12-7 miR-BART2- 5p |
immune escape |
1,2,3,4 | prediction / informed |
Stern-Ginossar et al., 2007 Nachmani et al., 2009 |
| CXCL16 | mCMV | miR-M23-2 | 1,2,3,6 | informed | Dolken et al., 2010a | |
| CXCL-11 | EBV | mir-BHRF1-3 | immune modulation |
5 | informed | Xia et al., 2008 |
| p21 | KSHV | miR-K1 | prevent cell cycle arrest |
1,2,3,5 | microarray | Gottwein and Cullen, 2010 |
| CCNE2 H3F3B TRIM28 |
hCMV | miR-US25-1 | block cell cycle to prevent apoptosis? |
1,2,3 1,2 3,4 |
RISC/miR NA pull down |
Grey et al., 2010 |
| C/EBPbeta p20 (LIP) |
KSHV | miR-K12-3 miR-K12-7 |
paracrine growth promotion? |
3 | informed / prediction |
Qin et al., 2010 |
| Gemin8 | KSHV | miR-K12-4-3p | ? | 1,2,3 | RISC pull down |
Dolken et al., 2010b |
| BACH1 Fos |
KSHV | miR-K12-11 | mimic cellular miR-155 |
1,2,3 1,2,3,5 |
microarray |
Skalsky et al., 2007 Gottwein et al., 2007 |
| PU.1 | MDV1 | miR-M4 | 1,2,3 | informed | Zhao et al., 2009 | |
| GPM6B RREB1 c-Myb MAP3K7I P2 PU.1 C/EBP |
MDV1 | miR-M4 | 1,2 | informed / prediction | Muylkens et al., 2010 | |
| Rbl2 | KSHV | miR-K12-4-5p | increased DNA methylation? |
1,2,3 | informed | Lu et al., 2010b |
| MAF | KSHV | miR-K12-1 miR-K12-6-5p miR-K12-11 |
trans- / de- differentiatio n |
1,2,3,4, 5 |
microarray / prediction |
Hansen et al., 2010 |
| IkappaB- alpha |
KSHV | miR-K12-1 | promote latency |
1,2,3,4, 6 |
informed | Lei et al., 2010 |
| NFIB | KSHV | miR-K12-3 | 1,2 | prediction | Lu et al., 2010a | |
| Dicer | EBV | miR-BART6 | 1,3 | prediction | Iizasa et al., 2010 |
numbers indicate that the following observations were made: Upon ectopic miRNA expression (1) luciferase reporters and/or target protein expression constructs are repressed and (2) Target site mutation in luciferase reporters and/or expression constructs leads to de-repression. (3) Endogenous protein levels are down regulated upon ectopic expression of the miRNA or upon (4) de novo infection with live virus. In infected cells, treatment with specific (5) miRNA inhibitors results in up-regulation of endogenous protein levels. (6) knockout virus mutants were investigated in vivo or in vitro.
Table 3.
Viral Targets of viral miRNAs.
| Target | Virus | miRNA(s) | Proposed function |
Confir mation * |
Lead | Reference(s) |
|---|---|---|---|---|---|---|
| LT-Ag | SV40 JCV BKV PyV MCPy V |
miR-S1 miR-J1 miR-B1 miR-P1 miR-M1 |
contribute to immune evasion by limiting viral antigen expression? |
3,5 1,3,4 1,3 3,4,5 1 |
informed |
Sullivan et al., 2005 Seo et al., 2008 Seo, Chen, and Sullivan, 2009 Sullivan et al., 2009 |
| LMP-2a | EBV | miR-BART22 | 1,2 | prediction | Lung et al., 2009 | |
| LMP1 | EBV | miR-BART1- 5p miR-BART16 miR-BART17- 5p |
prevent apoptosis |
1,2 | informed | Lo et al., 2007 |
| BALF5 | EBV | miR-BART2 | prevent lytic replication / promote latency |
1,2,3 | informed | Barth et al., 2008 |
| RTA | KSHV | miR-K9* | 1,2,4 | informed | Bellare and Ganem, 2009 | |
| IE72 (UL123, IE1) |
hCMV | miR-UL112-1 | 1,2 | prediction | Grey et al., 2007 | |
| UL114 | hCMV | miR-UL112-1 | 1,2,5 | informed | Stern-Ginossar et al., 2009 | |
| ICP0 ICP4 |
HSV-1 | miR-H2-3p miR-H6 |
1,2 | informed | Umbach et al., 2008 | |
| ICP34.5 ICP0 |
HSV-2 | miR-2 miR-3 |
1,2 | informed | Tang, Patel, and Krause, 2009 | |
| UL28 UL32 |
MDV1 | miR-M4 | 1,2 | prediction | Muylkens et al., 2010 | |
| IE1 | hCMV | miR-UL112-1 | modulate lytic replication |
1,2,5 | prediction | Murphy et al., 2008 |
| ICP34.5 | HSV-2 | miR-1 | control neurovirule nce |
1 | informed | Tang et al., 2008 |
| DNA polymerase I |
HvAV | miR-1 | regulate viral replication |
1 | informed | Hussain, Taft, and Asgari, 2008 |
numbers indicate that the following observations were made: Upon ectopic miRNA expression (1) luciferase reporters and/or target protein expression constructs are repressed and (2) Target site mutation in luciferase reporters and/or expression constructs leads to de-repression. In infected cells, (3) specific cleavage products are observed for miRNAs that are antisense to their proposed target. Inhibition of authentically expressed miRNAs in infected cells via specific (4) miRNA inhibitors results in up-regulation of the target. (5) Knockout virus mutants were investigated in vivo or in vitro.
1. Antisense regulation of mRNA targets
Perhaps the most straightforward examples of determining viral miRNA function occur when a viral miRNA is encoded on the opposite strand of a coding mRNA transcript. In this situation, the miRNA would be predicted to result in a siRNA-like cleavage of the target mRNA if both are temporally co-expressed. miRNAs with perfect complementarity targets are commonly found in plants, but for incompletely understood reasons, this strategy is rarely utilized in animals (Bartel, 2004). Interestingly, numerous animal virus miRNAs have been identified that are encoded with perfect complementary, antisense to protein-coding transcripts. Notably, the first report describing a viral miRNA showed that EBV encodes a miRNA (miR-BART2) complementary to the BALF5 polymerase-coding transcript (Pfeffer et al., 2004). Two other studies, one that pre-dated this discovery (Furnari et al., 1993), and one subsequent study (Barth et al., 2008), lend support to the model that EBV miR- BART2 cleaves the polymerase-encoding transcript. Since the polymerase encoded by BALF5 is part of the lytic lifecycle (gamma herpesviruses recruit cellular polymerases to replicate their genomes during latency) it has been proposed that the negative regulation by miR-BART2 may promote latent infection. However, as BALF5 is efficiently silenced at the transcriptional level in latently infected cells, the functional relevance of the cleavage event remains enigmatic; perhaps it serves as a second line defense against untimely BALF5 expression. Interestingly, the closely related rhesus lymphocryptovirus (rLCV) encodes an orthologous miRNA that has diverged in sequence (Walz et al., 2010). Due to its conserved location, however, it is expected to have retained the ability to cleave the antisense-transcribed BALF5 transcripts.
Several members of the polyoma virus family (small viruses with DNA genomes, associated with several human diseases in immunocompromised patients with clinical outcomes including tumorigenesis, organ rejection, and dementia), encode miRNAs late during lytic infection that are complementary to the early transcripts (Cantalupo et al., 2005; Seo et al., 2009; Seo et al., 2008; Sullivan et al., 2005; Sullivan et al., 2009). miRNAs from four different polyomaviruses, including Simian Virus 40 (SV40), JCV, BKV, and murine Polyomavirus (muPyV) have been shown to direct the cleavage of the early transcripts that encode the large and small tumor antigen (T antigen) proteins (Seo et al., 2009; Seo et al., 2008; Sullivan et al., 2005; Sullivan et al., 2009). Additionally, Merkel Cell Polyomavirus (MCV) encodes a miRNA that is complementary to the early mRNAs, and negatively regulates reporters that contain portions of the early mRNAs (Seo et al., 2009). The strongest evidence that this autoregulation of viral transcripts is functionally relevant comes from our studies of a mutant SV40 virus that fails to express the miRNA. Compared to wildtype, this virus encodes increased early protein (both large and small T antigens) levels (Sullivan et al., 2005). Additionally, cells infected with the mutant virus are more susceptible to in vitro cytotoxic T cell (CTL)-meditated lysis (when co-cultured with CTL clones reactive for a specific T antigen epitope). A mutant strain of muPyV, dl1013, does not produce the viral miRNA due to a small deletion. Like the SV40 miRNA mutant, dl1013 expresses increased levels of the T antigen protein (compared to wild type virus) during infection of cultured cells (Sullivan et al., 2009). Furthermore, like SV40, early transcript cleavage fragments are detectable that bear the molecular hallmarks of miRNA-mediated RISC cleavage. However, both the SV40 mutant and the muPyV mutant viruses replicate to wild type levels in cultured cells. These data argue that despite being preserved in laboratory strains throughout decades of passage, the functional role of the miRNA is not essential for infection of cultured cells. Furthermore, infection of mice with miRNA mutant muPyV shows an indistinguishable phenotype from wild type virus in assays of acute infection and CTL reactivity in vivo. Thus, while the conserved nature of the miRNA-mediated autoregulation of the T antigens in diverse members of the Polyomaviridae suggests an important role in the virus life cycle, it seems this role is not measurable by standard assays of laboratory infection. The muPyV miRNA example, which is the first viral miRNA to be studied in a mutant context in vivo, lends support to the notion that the functional roles of viral miRNAs are likely to serve as a subtle regulators– similar to their host counterparts. Ongoing experiments revolve around assaying for differences in persistence, shedding, and transmission of the muPyV miRNA mutant.
Although at least four well-characterized viral miRNAs that direct cleavage of complementary mRNA targets exist, (Barth et al., 2008; Pfeffer et al., 2004; Seo et al., 2008; Sullivan et al., 2005; Sullivan et al., 2009), there are interesting exceptions to this mode of antisense regulation. In HSV-1 infection, miR-H2-3p is encoded antisense to ICP0 (an immediate early transcription factor involved in initiating full progression of lytic replication). However, miR-H2-3 does not direct cleavage of this mRNA transcript (Umbach et al., 2008). Indeed, miR-H2-3 negatively regulates protein expression from the ICP0 transcripts, but does so in a manner that does not result in turnover of the mRNA transcript. The mechanism for how this occurs is currently unknown. In addition, HCMV miR-UL112 is encoded antisense to and with perfect complementarity to the viral UL114 transcript encoding DNA glycosylase (Grey et al., 2005; Pfeffer et al., 2005). Stern-Ginossar et al. report that ectopic expression of miR-UL112 prior to infection (with a virus expressing an epitope-tagged version of UL114), results in reduced protein levels of tagged UL114 during infection (Stern-Ginossar et al., 2009). It should be noted that these experiments did not examine whether a specific cleavage event was detectable in the tagged UL114 transcript, and the decrease observed could be partially explained by pleiotropic effects of other viral transcript targets of miR-UL112 (Grey et al., 2007). However, despite perfect complementarity of miRNA and target as well as robust expression of both mRNA and miRNA transcripts during acute infection, Grey and Nelson report that mRNA cleavage is not observed, even though reporters engineered with a perfect complementary site to miR-UL112 are susceptible to negative regulation (Grey and Nelson, 2008). For these reasons, Grey and Nelson speculate that the secondary structure of the regions flanking the miR-UL112 complementary site in UL114 might act to prevent cleavage. This strategy could allow a virus to encode miRNAs antisense to mRNA transcripts without altering viral gene expression from transcripts lying on the other strand. While key questions remain, these combined results suggest that in some contexts, miR-112 may be able to direct negative regulation of UL114, perhaps in a cleavage-independent manner, similar to HSV miR-H2-3 and ICP0. Future studies regarding the activities of miR-H2-3p and miR-UL112 should be informative not only to herpes virus gene regulation, but also more broadly to the optimal design of small interfering RNAs (siRNAs).
Finally, we note that there are several additional examples where virus-encoded miRNAs negatively regulate perfectly complementary mRNA targets, but it is not yet clear if this regulation is due to siRNA-like cleavage (Tang et al., 2008; Xia et al., 2008). Of particular interest is a host target, CXCL-11. As shown from miRNA inhibition studies in infected cells, EBV-miR-BHRF1-3 negatively regulates CXCL-11 (Pfeffer et al., 2004; Xia et al., 2008). Because CXCL-11 is an interferon-inducible, pro-immunomodulatory cytokine, this example might represent an immune evasion strategy of the virus that is also relevant to EBV-associated tumorigenesis. Although one may have expected that more viral miRNAs might have evolved perfect complementarity towards their target, so far, CXCL-11 is the unique example of a viral miRNA that has adopted this strategy.
2. Computational identification of miRNA targets
A rapid method for formulating hypotheses as to the targets/functions of a virus-encoded miRNA is to use one of several popular target prediction programs (Enright et al., 2003; Kertesz et al., 2007; Kiriakidou et al., 2004; Krek et al., 2005; Lewis et al., 2003; Miranda et al., 2006; Rehmsmeier et al., 2004). Almost all of these algorithms operate on the common principle that target recognition should occur primarily through binding of the seed region; hence they search for transcripts with regions that are complementarity to this 6–7 nt signature. Although most algorithms also consider additional parameters (e.g. auxiliary base pairing of distal miRNA sequences, sequence context or structural accessibility of the target site) to rank the putative targets, because of the rudimentary understanding of the molecular events which govern target recognition, all suffer from rather large numbers of false positive predictions. For cellular miRNAs, the most effective means to distinguish functional target sites from false positives (by far) is consideration of evolutionary conservation: as many of the miRNAs are conserved between different species, so are their target sites. However, as discussed above, because the host is unlikely to conserve target sites for the benefit of a virus, such a filter would only have utility for the minority of virus miRNAs that may mimic evolutionary conserved host miRNAs. This likely accounts for the fact that, although computational approaches have been used with great success for identifying host miRNA-target pairs, there are far fewer examples of un-informed bioinformatic predictions yielding functional insights into virus-encoded miRNAs. Nevertheless, some notable exceptions exist in which this approach was utilized successfully (see below).
Computational prediction identifies host-encoded targets
To our knowledge, there are only three examples in which cellular targets were identified by computational analysis without any further functional or phenotypic leads. The first is the p53 up-regulated modulator of apoptosis (PUMA), a pro-apoptotic factor which is repressed by the EBV-encoded miR-BART5 (Choy et al., 2008). PUMA was identified as a top-ranking target of miR-BART5 by the RNAhybrid (Rehmsmeier et al., 2004) and miRANDA (Enright et al., 2003) algorithms. Lending support to the idea that the observed repression is of physiological relevance is the observation that PUMA expression levels are low in nasopharyngeal carcinoma (NPC), an EBV-positive tumor that expresses high levels of the BART-miRNAs. Specific inhibition of miR-BART5 in tumor-derived cell lines furthermore leads to apoptosis, and this phenotype can be partially reversed by introducing an siRNA directed against PUMA. Hence, at least in NPC, miR-BART5 may contribute to tumorigenesis by antagonizing apoptotic stimuli resulting from constitutive transcription of the PUMA locus. The authors also note that the target site in the PUMA transcript is conserved in other vertebrate species; however, as this is not a cellular target site, for the reasons discussed above, this observation alone does not allow any conclusions as to its functional importance. It is, however, interesting to note that the related rLCV encodes a miR-BART5 orthologue (miR-rL1-8) has a conserved seed sequence (Cai et al., 2006b), and therefore would be expected to repress PUMA expression in its natural host, the rhesus macaque. A recent study, however, observed that miR-rL1-8 was unable to repress the 3’-UTR of the rhesus transcript, indicating that this function in fact may not be a conserved feature of miR-BART5 (Riley et al., 2010).
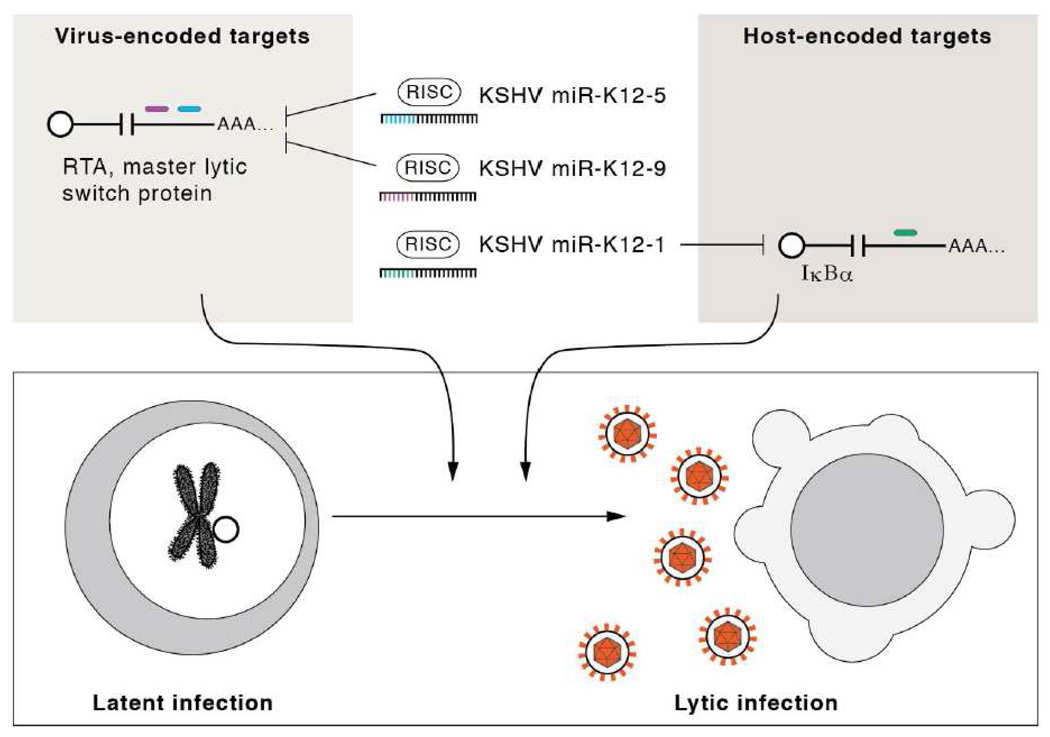
The second example is the identification of the major histocompatibility complex class I– related chain B (MICB) transcript as a target for the Human Cytomegalovirus (HMCV)-encoded miR-UL112 (Stern-Ginossar et al., 2007). MICB was predicted as a top target by the RepTar algorithm (Stern-Ginossar et al., 2007) and was confirmed as a direct target of miR-UL112 using luciferase assays. Importantly, endogenous protein levels of MICB are down-regulated by ectopic miR-UL112 overexpression as well as by infection with live virus, whereas a mutant virus defective for miR-UL112 expression also fails to repress MICB. MICB is a stress-induced ligand of the natural killer (NK) cell activating receptor NKG2D; its repression would be predicted to be of great benefit for HCMV as the protein is critical for the elimination of virus-infected cells by NK cells. Indeed, fibroblasts infected with miR-UL112 defective virions not only fail to downregulate MICB but are also much more susceptible to NK-cell mediated cytotoxicity, further underlining the functional relevance of these findings. In a surprising twist, by systematic functional testing of EBV and KSHV miRNAs, Nachmani et al. later showed that EBV as well as KSHV also express miRNAs with the ability to suppress MICB expression (Nachmani et al., 2009). Since the active miRNAs do not show any sequence homology and also engage different target sites within the MICB transcript, this would mean that several viruses have independently evolved mechanisms to counteract this important mediator of innate immunity.
The last example of bioinformatically predicted targets is a study that used the DIANA-microT program (Kiriakidou et al., 2004) to identify the miRNA-processing enzyme Dicer as a target of the EBV-encoded miR-BART6-5p (Iizasa et al., 2010). Although 3’-UTR luciferase reporters as well as endogenous Dicer levels are repressed by ectopic miR-BART6 expression, the functional consequences of this regulation remain unclear. Downregulation of Dicer would be expected to result in a widespread repression of miRNAs, which indeed transient overexpression of miR-BART6-5p in HeLa cells seems to induce. Whether miR-BART6-5p also leads to similar changes in authentically infected cells is unknown. The authors argue that a recent study which has reported global down-regulation of host-miRNAs in de novo-EBV infected cells (Godshalk et al., 2008) argues for this possibility; however, this study used the B95-8 strain of EBV, which lacks the entire miR-BART6 precursor due to a large deletion. Nevertheless, Iizasa et al. show that introduction of an antagomir (chemically-modified specific antisense oligonucleotide miRNA inhibitor) against miR-BART6-5p into latently infected cells leads to upregulation of several latent and lytic genes, which may point towards an important role of this miRNA in the regulation of viral latency.
Computational prediction identifies virus-encoded targets
In spite of the disadvantage of using computational prediction with mostly non-evolutionarily conserved viral miRNAs, there is at least one advantage to studying viral-encoded targets of miRNA– there are substantially fewer potential targets that are encoded by viral genomes. Grey et al. was one of the first reports to utilize this strategy with success on a miRNA encoded by HCMV (Grey et al., 2007). Using a computational strategy that relied in part on comparing putative HCMV miRNA targets to similar targets in the chimpanzee, Grey et al. show that miR-UL112-1 target three viral transcripts. Lending credence to the functional relevance of this observation, exogenous pre-expression of miR-UL112-1 results in reduced virus replication. Thus, some viral miRNAs function to autoregulate viral gene expression. Interestingly, this same miRNA also negatively regulates a component of the host innate anti-viral immune response (discussed in the previous section).
Currently, there are two examples of insect viruses that encode miRNAs, Bombyx mori nucleopolyhedrosis virus (BmNPV) and Heliothis virescens ascovirus (HvAV). BmNPV is a member of the baculovirus family, whose members are DNA viruses and include those that have been developed for possible use as biopesticides, and more recently, have been utilized to grow abundant amounts of recombinant, post-translationally-modified eukaryotic proteins. Four BmNPV miRNAs have been identified using computational and deep sequencing approaches (Singh et al., 2010). All BmNPV miRNAs are of unknown function. HvAV is a member of the ascovirus family. Ascoviruses have DNA genomes, infect invertebrate insects, and are evolutionarily distantly related to the Iridoviridae. Bioinformatic analysis was used to identify the viral DNA polymerase as a target of HVaV-miR-1(Hussain et al., 2008). Similar to HCMV miR-UL112-1, exogenous pre-expression of this miRNA results in reduced virus DNA replication. Thus, divergent virus families utilize virus-encoded miRNAs to autoregulate gene expression. The likely functional importance of autoregulation of viral gene expression is further discussed below (see “Lessons learned” section, below).
3. cDNA profiling to identify mRNA targets
There have been numerous studies that have utilized cDNA expression profiling to identify viral miRNA targets. In this approach, a viral miRNA of interest is either exogenously-expressed or has its function blocked or removed (by antisense inhibitors or mutation within the virus genome). Cells are then either directly profiled for changes in mRNA steady state levels, or alternatively, some component of the RISC complex is precipitated, and changes in the levels of mRNAs recruited to the RISC are assayed. The subset of differentially regulated/precipitated transcripts is then interrogated for the presence of potential target sites using computational methods. This approach represents an excellent beginning strategy, particularly when no functional insights regarding a particular miRNA’s function are suspected. However, divining functions from a long list of candidate targets often requires extensive additional experiments. Therefore, confirmatory experiments are usually performed on a small subset of the originally identified candidates only. Most often such candidates are selected because they are suspected to be of importance for the viral lifecycle, such as for example known pro-apoptotic factors or genes that are involved in immune responses. While it is reasonable to assume that such transcripts represent the most relevant targets among the candidates, one should keep in mind that this also introduces a strong bias in an otherwise unbiased cDNA expression or RISC screening approach. In the following section we highlight a few reports that utilized profiling strategies, with a particular emphasis on those that provide functional insights in the context of infection.
A recent study from Grey et al. used two different positive enrichment strategies to identify mRNA targets of miR-US25-1, one of the most highly expressed miRNAs made during HCMV lytic infection (Grey et al., 2010). Here, a stable cell line expressing an epitope-tagged component of RISC (Myc-Argonaute 2) was used in conjunction with a construct that expresses miR-US25-1. Additionally, a biotinylated synthetic mimic of miR-US25-1 was used to pull out RISC-bound transcripts. This combined strategy identified numerous (approximately 15) transcripts that scored in both assays. Many of the targets play a role in cell cycle control, such as cyclin E2. Perhaps the most striking finding from this study was that most of the target transcripts are bound by RISC in their 5’ UTRs. This was determined initially from a bioinformatic approach that identified enrichment of seed matches predominantly in the 5’ UTR of the transcripts enriched for RISC binding in the presence of miR-US25-1. Chimeric luciferase reporters confirmed direct regulation of the 5’ UTRs of two transcripts including cyclin E2. This is the first report of a virus miRNA targeting the 5’UTR of an mRNA, and opens the possibility that other miRNAs (host or viral) may utilize a similar mechanism. Importantly, infection with wild type virus results in increased recruitment of the cyclin E2 transcript to RISC, and infection with a miR-US25-1 deletion mutant virus results in higher levels of cyclin E2. Although most of the remaining targets await experimental confirmation, the identification of multiple targets involved in a single process such as cell cycle regulation makes miR-US25-1 unique amongst the other viral miRNAs with known functions. Most virus-encoded miRNA are expected to regulate one or a few targets to effect a particular function, or more rarely, tap into pre-existing host miRNA seed- target networks (discussed in depth in the Viral miRNAs - tapping into host miRNA regulatory networks? section, below). As the latter is apparently not the case for miR-US25-1 (there is no recognizable sequence homology to any of the currently known human miRNAs), it is tempting to speculate that miR-US25-1 could exploit the presence of a hypothetical sequence element that may be common to the 5’-UTR of transcripts encoding cell-cycle related factors.
The virus with most published reports utilizing cDNA profiling to identify miRNA targets and functions is Kaposi’s Sarcoma-associated herpesvirus. As described above, similar to EBV, KSHV infection is associated with several tumor entities. Unlike EBV, however, KSHV does not encode a single dominant oncogene, and the virus also does not exhibit strong transforming activity in vitro. Perhaps reflecting this more modest behavior in experimental systems, Kaposi’s sarcoma represents a hyperproliferative disease much more than a true neoplastic tumor (see (Ganem, 2010) for more in-depth information on this disease). The proliferating component in KS lesions are the so-called spindle cells, cells of endothelial origin which are, for the most part, latently infected with KSHV. However, these cells require exogenous growth factors for continued proliferation and survival, and hence are dependent on a KS microenvironment that is rich in immunoinfiltrates and cytokines. Interestingly, cells that undergo lytic replication release significant amounts of cyto- or chemokines as a result of endogenous host gene expression as well as due to the expression of host genes that have been captured by the virus. Since a significant proportion of KSHV-positive cells (~1–5%) in KS lesions undergo lytic replication, it is thought that these lytically reactivated cells greatly contributes to KS pathogenesis via paracrine mechanisms. Thus, besides the identification of host genes that are regulated by the virus, determining how viral gene products help to maintain latent infection, and which virus gene products control the switch to lytic infection is of paramount importance to understanding KSHV-associated disease.
Indeed, among the cellular KSHV miRNA targets that have so far been identified, there are several genes that play a role in the regulation of the switch from latent-to-lytic infection (see Tables 2 and 3, also discussed below in “Lessons learned” section). In addition, several targets have been identified that may play an important role in the etiology of KSHV-associated disease. For example, Thrombospondin is an anti-angiogenic factor frequently mutated in various cancers, and has previously been found to be downregulated in KS lesions. Indeed, exogenous expression of 10 of the 12 KSHV miRNAs downregulates Thrombospondin levels (Samols et al., 2007), indicating that multiple KSHV miRNAs cooperate to repress this cellular factor. Given the highly vascularized nature of KS lesions, it is possible that miRNA-targeting during latent infection plays a dominant role in mediating this phenotype. A more recent study identified the TNF-like weak inducer of apoptosis (TWEAK) receptor (TWEAKR) as a direct target of miR-K12-10a (Abend et al., 2010). In addition to demonstrating repression in luciferase assays, Abend and colleagues also showed that KSHV infection leads to downregulation of the protein in endothelial cells. Furthermore, miR-K10a-mediated downregulation of TWEAKR prevents TWEAK-induced caspase activation, and alleviating TWEAKR repression in infected cells via a specific miR-K10a inhibitor reverses this effect. Interestingly, this repression may not be only beneficial for the infected cell in which the actual down-regulation occurs, but also for the surrounding cells: as downregulation of TWEAKR inhibits pro-inflammatory cytokine release by primary endothelial cells in response to TWEAK treatment, this mechanisms may serve to dampen immune responses in KS lesions.
Another example is Bcl2-associated factor (BCLAF1), a protein that interacts with several different members of the BCL family. Like the BCL family, it appears that BCLAF1 can have either pro or anti apoptotic functions. Several KSHV-encoded miRNAs can inhibit BCLAF1 expression during infection, and under some conditions of growth for cultured cells, KSHV miRNA inhibition of BCLAF1 can result in decreased cleavage of PARP-1 (Ziegelbauer et al., 2009). However, repression of BCLAF1 by the KSHV miRNA does not inhibit apoptosis, and might function to modulate apoptosis-independent functions of PARP-1. Interestingly, BCLAF1 expression can foster decreased lytic activation of cells latently infected with KSHV. Thus, KSHV miRNAs that target BCLAF1 can alter the balance between latent/or lytic infection in a pro or inhibitory fashion, at least under some experimental conditions (discussed in greater detail in the “A predominant role of viral miRNAs may be during latent or persistent infection” section). Another example includes the cell cycle regulatory proteins, p21. P21 is activated by several different signaling pathways, most notably p53. P21 negatively regulates cell cycle progression. Gottwein and Cullen have shown that KSHV miR-K12-1 binds to the 3’UTR of p21 mRNA and direct repression of protein expression (Gottwein and Cullen, 2010). Ectopic expression of this miRNA can block p21-mediated cell cycle arrest. Importantly, this repression is functionally relevant in cells latently infected with KSHV, as inhibition results a slightly higher percentage of cell that undergo p21-mediated arrest. Thus, numerous cellular targets of KSHV miRNAs have been identified that play a role in the virus life cycle and likely in the diseases associated with infection of this virus.
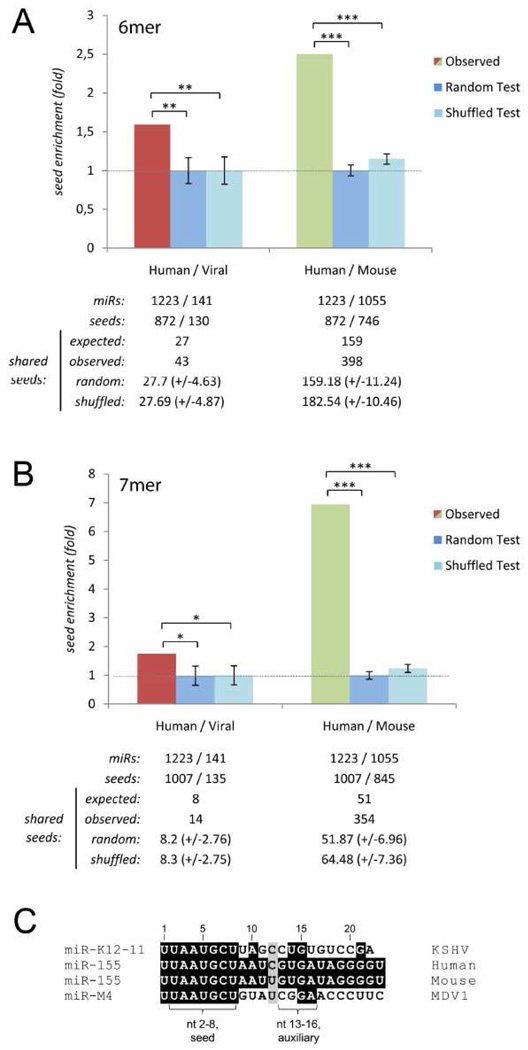
Two different labs have utilized an array-profiling strategy to demonstrate that KSHV miR-K12-11 has numerous cellular targets that likely play a role during infection. Compared to the vast majority of virus-encoded miRNAs, miR-K12-11 is atypical in that it shares extensive identity (11/22) including a perfect seed match with a host miRNA (miR-155). Both the Renne and Cullen groups have independently shown that ectopic expression of either miR-K12-11 or miR-155 results in downregulation of a common set of host mRNA transcripts (Gottwein et al., 2007; Skalsky et al., 2007). Furthermore, chimeric luciferase reporters containing the 3’ UTRs of several of these targets were used to demonstrate direct regulation. Hence, miR-K12-11 may have evolved to mimic a cellular miRNA. Because miR-155 not only plays an important role in B cell development but is also considered to have oncogenic potential, the implications of these findings will be discussed separately in the section Viral miRNAs - tapping into host miRNA regulatory networks?.
4. “Functions-first” screening
“Functions-first” approaches have the obvious advantage of starting with an already known phenotype of relevance, preferably one that has been observed in authentic infectious model systems. These approaches can be extremely powerful, especially if the phenotype can be observed in vitro. In these cases, individual miRNAs or combinations thereof can be tested for their ability to reproduce the phenotype. In the reverse approach, specific miRNA inhibitors or mutant viruses that are defective for miRNA expression can be employed to demonstrate the absence of the phenotype in infected cells. Cellular or viral factors known to be involved in particular pathways tied to the phenotype in question can then be systematically tested for their downregulation by specific miRNAs. Of course, depending on the number of suspected factors and the complexity of the phenotype, the identification of the responsible target transcripts may still represent a challenging task.
A recent example of a “functions-first” approach is a study that investigated KSHV-induced phenotypic changes in endothelial cells (Hansen et al., 2010). Previous studies had shown that KSHV infection of primary lymphatic or blood endothelial cells (BEC or LEC, respectively) induces a transcriptional reprogramming such that the resulting cells are of an intermediate phenotype and express markers of both lineages (Carroll et al., 2004; Hong et al., 2004; Wang et al., 2004). This observation is of interest because the spindle cells in KS lesions likewise express LEC as well as BEC markers, suggesting that KSHV induces a partial de- or trans-differentiation during tumorigenesis. In order to elucidate the potential role of viral miRNAs during this process, Hansen and colleagues subjected LECs which had been transduced with a recombinant lentivirus expressing 10 of the 12 KSHV-encoded miRNAs to cDNA profiling. Analysis of the negatively regulated transcripts with the PITA algorithm (Kertesz et al., 2007) led to the identification of the cellular transcription factor MAF (musculoaponeurotic fibrosarcoma oncogene homolog), which was experimentally confirmed to be a direct target of several KSHV-encoded miRNAs. Indeed, MAF is expressed in a LEC-specific manner and suppresses several BEC markers, hence contributing to LEC identity. Importantly, not only does authentic KSHV infection of LECs result in the repression of MAF expression, but the phenotype can also be prevented by treatment of the infected cells with specific inhibitors of the viral miRNAs, or by ectopic expression of a MAF cDNA lacking the miRNA target sites. Hence, the study by Hansen et al. represents an excellent example of a “functions first” approach in which a phenotype observed in infected cells could be successfully tied to a distinct target of virally encoded miRNAs.
The precise miRNA targets that mediate another KSHV-induced phenotype are less well defined. Qin et al. observed that KSHV infection or introduction of an expression construct encoding 10 KSHV-encoded pre-miRNAs led to increased release of interleukins 6 and 10 in macrophages (Qin et al., 2010). The authors also identified the transcriptional repressor C/EBPbeta p20 (LIP) as a potential target transcript and demonstrated down regulation of endogenous LIP levels after ectopic miRNA expression. Although siRNA knockdown of LIP also led to increased IL-6 and IL-10 release, whether this transcript is a direct target of KSHV miRNAs, and whether it is indeed repressed and contributes to cytokine production in KSHV-infected cells remains unknown. Nevertheless, the findings by Qin et al. suggest that viral miRNA-expression in infected monocoytes may contribute to KS pathogenesis via paracrine mechanisms.
Xu et al. systematically tested miRNAs encoded by Marek’s disease virus 1 (MDV1), a member of the alpha herpesvirus family, for their ability to inhibit apoptosis (Xu et al., 2010). As further discussed in the section “Viral miRNAs - tapping into host miRNA regulatory networks?”, MDV1 is an atypical alpha herpesvirus that can induce tumors in its natural host. Out of 14 miRNAs known to be encoded by MDV1, miR-M3 in particular was able to promote cell survival after cisplatin treatment. The authors used TargetScan (Lewis et al., 2003) to identify promising miR-M3 target candidates, and chose the transcription factor SMAD2 (mothers against decapentaplegic homologue 2) because of its well-known role in transforming growth factor (TGF)-beta signaling and apoptosis. Indeed, SMAD2 was confirmed as a target that is directly repressed miR-M3, and rescue of SMAD2 expression using a construct lacking the target sites also antagonized the anti-apoptotic effects of the miRNA. The observation that MDV1-induced splenic tumors displayed markedly lower SMAD2 levels than non-tumor splenic tissues furthermore corroborates the assumption that anti-apoptotic functions of MDV1-encoded miRNAs contribute to tumorigenesis.
Two recent studies have used viral mutants to investigate miRNA functions in primary in vivo or in vitro infection systems. Dölken and colleagues (Dolken et al., 2010) investigated a mutant of mouse cytomegalovirus (MCMV). Like HCMV, MCMV encodes a number of miRNAs (see Table 1) that are, however, not related in sequence (Buck et al., 2007; Dolken et al., 2007). The authors introduced point mutations in the precursor sequences of the two most abundantly expressed viral miRNAs, miR-M23-2 and miR-M21-1. These precursors are transcribed from opposite strands of the same genomic locus; therefore 5 nucleotide exchanges were sufficient to simultaneously destroy both hairpin structures and abrogate mature miRNA expression. Infection of mice demonstrated that the resulting mutants were specifically impaired in their ability to mount a persistent infection in salivary glands. Furthermore, attenuation was only observed in mice able to mount an effective natural killer (NK) cell response, and could be reversed by the combined depletion of natural killer (NK) cells and CD4- positive (but not cytotoxic / CD8-positive) T-cells. This is especially interesting considering that MCMV infection in salivary glands is known to be controlled by the concerted action of the above immune cell types rather than by cytotoxic T cells (Polic et al., 1998; Polic et al., 1996). Hence, some MCMV- encoded miRNAs may have evolved to mediate immune escape in specific organs. While the molecular basis of the escape remains to be unraveled (the chemokine CXCL16 was identified as a putative target of miR-M23-2, but is unlikely to be solely responsible for the phenotype), the study by Dölken and colleagues demonstrates the power of investigating miRNAs functions in the context of an authentic in vivo infection. It is unlikely that the phenotype would have been noticed by any other approach.
Another study by Seto et al. (Seto et al., 2010) systematically eliminated the miRNAs of the B95-8 strain of EBV. As discussed above, the fully transforming B95-8 strain harbors a deletion that removes all but 8 of the EBV-encoded miRNAs. These miRNAs reside at two different loci: three (miRs-BHRF1-1 to 3) are found flanking the BHRF1 open reading frame, whereas five (miRs-BART-1, 2, 3, 4 and 15) remain at the BART locus, the region which is affected by the genomic deletion. By selectively scrambling the genomic sequences encoding only the mature miRNAs, Seto and colleagues generated surgical mutants which either lack the BHRF-miRNAs, or in which additionally the BART miRNAs had been eliminated as well; the latter represents the first herpesviral mutant in which all miRNAs have been summarily obliterated. While both mutants were principally able to transform primary B cells in vitro, closer examination revealed that cultures infected with the mutated viruses consistently contained fewer cells in S-phase, especially during the early phase of infection. Additionally, these cells exhibited higher apoptotic rates than those infected with the parental strain. Since the phenotype was equally observed in both mutants, this observation argues for an important role of the BHRF1 miRNAs in providing an advantage during the early stages of primary B cell infection, a phase of the viral lifecycle which is critical for the establishment of long term latency. So far, however, the cellular targets that mediate this phenotype are unknown. A surprising finding of the study was that neither cells infected with the mutant lacking all miRNAs, nor with another mutant in which the genetic deletion of B95-8 cells had been repaired, exhibited altered levels of lytic replication. This was contrary to expectations based on the fact that, as discussed in other sections of this review, several studies have proposed a role of herpesvirus miRNAs in promoting viral latency. The study by Seto et al. thus may indicate that herpesviruses differ in the extent to which they employ miRNAs in order to control latency, and underscores the necessity to investigate viral mutants in relevant infection systems.
Finally, one of the more striking “functions-first” approaches involves the ability of a single KSHV or EBV-encoded miRNA to evade the Natural Killer (NK) cell innate antiviral response (Figure 3). Mandelboim and colleagues had previously used a computational prediction approach to a specific NK ligand, MICB as a target of an HCMV miRNA (as discussed above in the “computational identification of miRNA targets section) (Stern-Ginossar et al., 2007). In a subsequent manuscript, a functional screen was conducted of EBV and KSHV miRNAs to identify those that could downregulate MICB surface expression (Nachmani et al., 2009). Interestingly, the binding sites in the MICB 3’ UTR were different for all three different (HCMV, KSHV, or EBV) viral miRNAs, suggesting this is an example of convergent evolution.
Lessons learned
1. A predominant role of viral miRNAs may be during persistent/latent infection
Viruses that can form long-term infections without clearance by the host immune response are said to be able to form establish “persistent” infections. The ability to form persistent infections is crucial to fostering spread of the virus and various virus families accomplish this via different, and sometimes ill-defined mechanisms. Viral latency is a form of persistence whereby the infected cell does not produce any viral progeny; rather, the viral genome persists in a state of restricted viral gene expression (in its extreme form, this can occur in the absence of any viral proteins) that allows (and often also fosters) host cell survival and serves as a successful immune evasion strategy. During latent infection, the host cell maintains the viral genome but does not actively produce infectious virions. A hallmark of latency is its reversibility, occasionally resulting in full viral gene expression and culminating with the production of infectious virions. The herpesviruses have a well-defined latent infectious cycle and it is notable that the majority of viral miRNAs were discovered from cells latently infected with herpesviruses. This observation argues for a likely important role of the herpesviral miRNAs in maintaining latent infection.
Multiple studies conducted with KSHV miRNA mutant viruses have uncovered a role for the KSHV miRNAs in controlling lytic replication. Both Lei et al. and Lu et al. utilized a similar BAC mutant strategy in which 10 of the 12 KSHV pre-miRNAs were deleted (Lei et al., 2010; Lu et al., 2010b). Both studies reported that infection with these mutant viruses led to an increase in the transcript levels of the master lytic switch protein, RTA. RTA is both necessary and sufficient to initiate the full cascade of viral lytic gene expression. Consequently, cells infected with these mutant viruses had increased expression of lytic transcripts. Lei et al. further showed a modest, approximately two-fold increase in virion production in the miRNA mutant, as measured by qRT-PCR of encapsidated genomes. Both studies implicate host targets of the miRNAs as playing a role in the increased lytic phenotypes that are observed. Lei et al. argue for miRNA-mediated increased NFKappa B signaling, which is known to decrease the propensity of lytic activation (de Oliveira et al., 2010), while Lu et al. implicate host DNA methyltransferases in possibly playing a role in the phenotype observed. Additionally, Lu and Rana et al. (Lu et al., 2010a) investigated the ability of KSHV-encoded miRNAs to suppress expression of RTA. miR-K12-3 was found to be able to suppress RTA but apparently does not target the 3’-UTR of the viral transcript: The authors noted that a computationally predicted target of miR-K12-3, the cellular transcription NFIB (nuclear factor I/B), had been previously shown to be able to reactivate KSHV (Yu et al., 2007). NFIB was shown to bind to the ORF50 promoter, and the 3’-UTR of the transcript furthermore can be directly targeted by miR-K12-3. However, it is presently unclear to what extent miR-K12-3 contributes to the maintenance of latency in infected cells. In contrast, Lu et al. also suggest that miR-K12-5 can directly regulate the RTA transcript via its 3’ UTR. Interestingly, Bellare and Ganem have reported that a different miRNA, KSHV miR-K12-9*, also can directly regulate the 3’ UTR of the RTA transcript (Bellare and Ganem, 2009). The authors systematically tested 17 KSHV-specific antagomir inhibitors and observed that only inhibition of miR-K12-9* led to an appreciable up-regulation of lytic cycle induction in infected cultures; reporter assays confirmed that miR-K12-9* mediates repression via a single target site in the 3’-UTR of the RTA transcript. It is unclear why miR-K12-5 scored in one study and KSHV miR-K12-9* scored in the other, but robust miRNA-mediated regulation of RTA would clearly be expected to affect lytic induction. The miRNA-meditated direct effects on RTA reported in both Lu et al. and Bellare and Ganem are nevertheless subtle. The mechanisms thus may serve to repress those few RTA transcripts that are produced at basal level in latently infected cells. Indeed, we and others, have previously observed that the RTA promoter carries bivalent histone marks and thus exists in a pre-activated state throughout latency (Gunther and Grundhoff, 2010; Toth et al., 2010). Such promoters are expected to be especially sensitive to occasional/stochastic firing, and our findings hence support the hypothesis that a post-transcriptional mechanism is required to counteract from low-level RTA transcription during latency. It should be noted, that two other studies have demonstrated that selective antisense inhibition of a subset of the KSHV miRNAs has the opposite effect, whereby under these experimental conditions, some miRNAs actually function to promote lytic replication (Lu et al., 2010a; Ziegelbauer et al., 2009). Thus, there is much to sort out, such as the relative contribution of the individual miRNAs, as well as the various reported host and viral miRNA targets that are most important. However, it seems likely (as determined from the BAC mutant studies), that a function the KSHV miRNAs as a whole, is to negatively regulate the initiation of lytic replication– at least under some circumstances (Figure 4).
Figure 4. The role of virus-encoded miRNAs in promoting KSHV latency.
Mutants of KSHV that delete 10 of the 12 pre-miRNA have an increased potential to undergo spontaneous lytic replication, thus implicating the KSHV miRNAs as playing a role in regulating lytic replication (Lei et al., 2010; Lu et al., 2010b). Three different miRNAs encoded by KSHV likely play a particularly important role in this process. Two miRNAs target the viral-encoded RTA, the master lytic switch transactivating protein that is both necessary and sufficient to trigger lytic replication in KSHV-infected cells. A third miRNA targets the host-encoded IkappaBalpha, an inhibitor of NF Kappa B signaling. Lower NF Kappa B signaling is associated with increased lytic activation for several gamma herpesviruses (de Oliveira et al., 2010). Thus, KSHV miRNA-mediated inhibition of an inhibitor of NF Kappa B promotes latency. We note a few KSHV miRNAs also play a “anti-lytic induction” role in some contexts (Lu et al., 2010a; Ziegelbauer et al., 2009), (not depicted). miRNA-RISC complexes and mRNAs are depicted as described in the legend for Figure 1. In the cartoon for latently-infected cells, the viral episome is depicted as circle attached to the chromosome.
As mentioned in above sections, a particular viral miRNA may target host transcripts, viral transcripts, and in some cases, both. Regulation of both host and viral transcripts can play a profound role in staving off the host immune response and promoting cell survival in persistently-infected cells. Viral miRNAs from three families, the herpesviruses, the polyomaviruses and ascoviruses can negatively regulate viral gene expression (autoregulation). This mode of autoregulation, in theory, can help maintain a persistent infection by 1.) lowering specific antigen presentation of the targeted gene product to the adaptive immune response, or 2.) by preventing completion of the lytic cycle. For some herpesviruses, a bioinformatic study suggests that viral autoregulation is a predominant function of the viral miRNAs (Murphy et al., 2008). The increased propensity of the KSHV miRNA mutants to enter into the lytic replication cycle (as described above) supports the notion that other herpesviruses likely utilize viral miRNAs to optimize the decision to switch to lytic replication. In particular, latent infection with the human herpes simplex viruses (HSV1 and HSV2) results in expression of several viral-encoded miRNAs but no viral proteins. Some of these HSV miRNAs have the evolutionarily conserved ability to downregulate expression of some viral transcripts required for lytic infection (Tang et al., 2008; Umbach et al., 2008; Umbach et al., 2009). Similarly, pre-expression of an HCMV miRNA leads to decreased viral DNA replication (Grey et al., 2007), thus, it is possible that HCMV also utilizes viral miRNAs to curtail lytic replication. This model might also apply to the Polyomaviridae. Polyomaviruses form lifelong, persistent infections, however the exact cell types and the mechanisms involved are poorly defined. Several members of the polyomavirus family encode miRNAs that can autoregulate early gene products (the T antigens) (as described above in the “Antisense regulation of mRNA targets” section). As the T antigens are required to initiate lytic replication, inhibiting their function could dampen virus production in some cell types and thus promote persistent infection in vivo. If this model were correct, then there would have to be a mechanism in some cellular contexts of expressing the Polyomaviridae miRNAs during persistence that is independent of the late protein products (which would be immunogenic and toxic if expressed in persistently-infected cells). Currently, this topic is the subject of ongoing studies (AG and CSS, unpublished observations). On the other hand and in contrast to KSHV, an EBV mutant virus in which all miRNAs were deleted did not display an increased propensity to undergo lytic replication in cultured cells (Seto et al., 2010). This suggests that despite the similarities in life cycles between the human gamma herpesviruses, the viral-encoded miRNAs have evolved different functionalities regarding control of the exit from latency into lytic replication. Thus, in practice, the exact contribution of viral miRNAs to persistent infection awaits experiments in the appropriate model system for each individual virus.
2. A common role of viral miRNAs in evading host defenses?
Almost all viruses encode accessory proteins that are not directly involved in the replication process and serve no structural role in the assembly of virions. Typically, these regulatory proteins are involved in combating host defenses in the ongoing “cat and mouse” game of the evolution virus-host interactions. Therefore, it is not surprising that some viral miRNAs have been associated with similar activities in preventing host defenses including the cytotoxic T cell response, the natural killer cell response, and apoptosis (reviewed in (Sullivan, 2008)). Autoregulatory viral miRNAs could indirectly evade the immune response by lowering the antigenicity of viral proteins or the overall amount of virus replication. A possible example of this is the polyomavirus SV40. Cells infected with an SV40 miRNA mutant virus were more susceptible (compared to wildtype-infected cells) to cytotoxic T cell-mediated lysis in vitro (Sullivan et al., 2005). However, in vivo experiments with a similar miRNA mutant murine Polyomavirus (muPyV) failed to detect a similar function (Sullivan et al., 2009). Thus, it remains uncertain as to the role of polyomaviral miRNA-mediated autoregulation in vivo. As discussed above, miRNAs encoded by HCMV, KSHV, EBV, and MCMV have been associated with evasion of the NK cell response. Similarly, divergent herpesviruses have been shown to encode miRNAs that have anti-apoptotic functions (see Tables 2 & 3). It seems probable that some of the remaining virus-encoded miRNAs of unknown function may be involved in similar immune evasion activities. However, as the muPyV example demonstrates, it is imperative that ultimately the relevance of these activities is confirmed in vivo.
3. Conservation of viral miRNAs?
The experimental evidence gathered so far suggests that virally-encoded miRNAs appear to mediate evolutionarily conserved functions (e.g. immune evasion, cell cycle control, defense against apoptosis, promotion of latency) in a considerable number of viral species. Considering this, it is surprising that the miRNAs themselves show little primary sequence conservation. A notable exception from this rule are some miRNAs that are conserved between viral species that also show considerable global sequence identity. Chief among these are EBV and the related rhesus lymphocryptovirus (rLCV). Both viruses share similar biological properties (including their association with lymphomas in their natural host) and, despite of being separated by more than 13 million years of evolution, have retained ~ 60–65% of sequence identity. Out of 25 and 34 pre-miRNAs known to be encoded by EBV and rLCV, respectively, 22 show significant sequence similarity such that they can be traced back to a common ancestor (Cai et al., 2006b; Walz et al., 2010). However, only 14 of these also produce mature miRNAs with a conserved seed sequence, whereas the remaining nucleotides have diverged. Likewise, the human alpha-herpesviruses HSV1 and 2, which share approximately 60% sequence identity, encode at least 16 and 18 pre-miRNAs, respectively. Of the 9 precursors that are recognizably of common origin, 6 produce at least one mature miRNA species with a conserved seed. In addition, members of the Polyomaviridae including SV40, SA12, BKV, and JCV all express miRNAs that are similar enough (~65% identity or higher) to suggest a common ancestor.
We have recently performed a bioinformatic study to interrogate all fully sequenced gamma herpesvirus genomes for the presence of further putative candidates, including not only known but also predicted pre-miRNA hairpins in our analysis (Walz et al., 2010). This analysis shows that evolutionary conservation of viral pre-miRNA sequences is indeed rare. At present, it is difficult to understand what the low degree of conservation among viral miRNAs means. Clearly, this feature distinguishes viral miRNAs from their cellular counterparts, which have maintained extensive sequence identity (in most cases not only limited to the seed) over millions of years. It is, however, possible that viral miRNAs may have conserved important functions despite of the absence of sequence conservation. For example, miRNAs which autoregulate viral gene expression may have co-evolved with the viral genome and thus, although (or rather because) having diverged in sequence, still recognize identical targets. By the same token, miRNAs in viral species that infect different host species (such as EBV and rLCV) may have acquired changes in order to maintain complementarity to host mRNA targets. This hypothesis seems especially attractive since most viral miRNAs do not mimic cellular miRNAs (see below). While targets sites for cellular miRNAs are (for the most part) evolutionarily conserved due to their co-evolution, there should be less or no evolutionary pressure for the host to maintain a site that is exclusively used by a viral miRNAs (unless the site has some other, host-related function which necessitates its conservation). It follows that it is the viral miRNA itself that would have to undergo adaptive changes if the target sites changes throughout evolution (not too unlikely an event considering that most sites are located in the 3’-UTR of the target transcript). The ultimate proof for above scenario would be the identification of a transcript with homologous but diverged target sites in related host species (for example humans and rhesus monkey) that are matched by an equally diverged miRNA pair of common origin in viruses that infect those host species (for example ebv-miR-BART2-5p and rlcv-miR-L1-33 (Walz et al., 2010)). The observation of such ‘adaptive’ evolution would be a strong argument for the functional importance of a target; note that this is different from the recent observation of three unrelated viral miRNAs encoded by HCMV, EBV and KSHV (all human viruses) that target MICB via different sites, which would represent a case of convergent evolution rather than preservation of a function adopted by a common ancestral miRNA. In fact, the rLCV homologue of the EBV miRNA (miR-BART2-5p) that targets two sites in the human MICB transcript (Nachmani et al., 2009) has acquired a nucleotide change in its seed sequence (Walz et al., 2010). However, both target sites are perfectly conserved in the rhesus genome and the rLCV miRNA thus may have lost the ability to target this transcript.
4. Viral miRNAs - tapping into host miRNA regulatory networks?
The observation that several miRNAs of viral origin share sequences with host miRNAs has received much attention. Clearly, by emulating host miRNAs viral miRNAs could gain access to the large and intricate networks of target transcripts maintained by the host (Figure 5). If so, it is surprising that there is only one known example of a an entire pre-miRNAs that has (possibly) been captured from the host (Waidner et al., 2009), despite of the fact that herpesviruses have frequently pirated host cell genes and subverted them to their own purposes. However, a short seed sequence may also be evolved independently; one would expect that a considerable number of such viral “mimics” of host miRNAs exist. In fact, a survey of all viral and host miRNA sequences currently deposited in miRBase (release 16) seems to somewhat support this notion. For example, among the 141 known miRNAs of human herpes- and polyomaviruses, there are 43 instances of unique 6mer seed sequences that are shared with human miRNAs (Figure 6A, further explained below). However, given the limited complexity of a 6 nucleotide motif, one needs to consider the possibility that some seed sharing may occur by chance. A recent study which has analyzed EBV-encoded miRNAs in nasopharyngeal carcinoma has taken this consideration into account and has found, nevertheless, that EBV miRNAs with a shared seed are approximately 2 fold more abundant than expected based on a purely probabilistic model (Chen et al., 2010). We have performed a similar analysis for all human herpes- and polyomavirus miRNAs deposited in miRBase and come to a similar conclusion. For this purpose, we extracted all human miRNA sequences as well as human viral miRNA sequences and, in each contingent, identified unique 6mer seed sequences (i.e. nt 2 – 7 of the mature miRNA). As noted below the bar graph in Figure 6A (left panel), there are 1223 human miRNAs, which all together code for 872 unique seeds (the number of unique seeds is lower since many cellular miRNAs belong to families that share a common seed). Hence, of the 46=4096 possible combinations for 6mer seeds, 872/4096=21% are being used by human miRNAs. In other words, for any randomly selected 6mer, there is a substantial probability (p=0.21) that it can also be found in a human miRNA. Accordingly, when considering the 130 seeds which were used by the 141 viral miRNAs analyzed, one would expect that a considerable 0.21×130=27 seeds are shared by pure coincidence. However, we observed that a total of 43 seeds were shared between viral and human miRNAs, corresponding to a significant ~1.6 fold enrichment over the expected random number (Figure 6A). We further considered the possibility that there might be a general or positional bias in the nucleotide composition of the seed sequences, which could potentially falsify the results as the expect values are calculated assuming an unbiased composition. Therefore, we generated test sets based on viral miRNA sequences in which either the seed sequences were completely randomized (dark blue bars), or in which nucleotides were shuffled according to their position (light blue bars) which should preserve any potential bias, and again compared the seeds to those of human miRNAs. Each test was performed 10,000 times to calculate enrichment mean and standard deviation values. As shown in Figure 6A, no enrichment was observed in either control test, and the actually observed seed enrichment was significantly higher in both cases. For comparison, we performed the same analysis for human and mouse miRNAs (Figure 6A, right panel); in this case, the seed enrichment was 2.5 fold and highly significant. Interestingly, a slight enrichment (~1.2 fold) was observed in the shuffled test, indicative of a mild sequence bias that did not, however, affect the statistical significance of the virus-host shared seed enrichment.
Figure 5. A hypothesis: most virus-encoded miRNAs will regulate a target mRNA network substantially different than their host counterparts.
With a few rare, important exceptions, most virus miRNAs are not predicted to tap into existing host miRNA-target regulatory networks (Top panel). Some virus-encoded miRNAs have been shown to regulate a fairly large number of mRNA transcripts involved in a particular process, despite the fact that these miRNAs share no seed sequence identity with host miRNAs. This implies that sometimes, virus-encoded miRNAs are able to tap into viral-specific, miRNA-mRNA transcript networks (Middle panel). Finally, we propose the hypothesis that the major function of many virus-encoded miRNAs will be to regulate a small number of key (host or viral) transcripts (Bottom panel). Despite the fact that exogenous expression of such miRNAs will result in changes in the steady state levels of numerous (~hundreds) of transcripts, only a small minority of these need be of functional relevance and the rest may simply be bystanders. Parsing out the relevant targets is the major challenge of “Bottom-up” approaches (described in Figure 3).
Figure 6. Seed sharing amongst viral and host miRNAs.
A, B: Statistical analysis of seed sharing. We analyzed the sharing of 6mer (nt. 2–7 of the mature miRNA sequence, top panel) or 7mer (nt. 2–7, bottom panel) seeds between human and human herpes- or polyomavirus miRNAs (left columns in each panel), or for comparison between human and mouse miRNAs (right columns in each panel). All sequences were curated from the September 2010 release 16 of miRBase. We first identified the total number of unique seeds in each entity, then calculated the number of shared seeds that would be expected to occur by mere chance. The red and light green bars show the relative enrichment of the number of actually observed number of shared seeds over the calculated expect value. As a control, we performed two tests using random and shuffled seed sequences (dark and light blue columns, respectively; see text for further details). P-values for the hypothesis that differences between the observed enrichment and either test set are statistical significant (one-tailed hypergeometric test) are indicated by asterisks (*: p < 0.5, ** p < 0.1, ***: p < 0.01). C: Alignment of the KSHV encoded miR-K11 (top), human and murine miR-155 (center) and miR-M4 encoded by MDV1 (bottom). Nucleotides that are conserved between both cellular mir-155 orthologues as well as matching nucleotides in viral miRNAs are shown in white on black background. Nucleotides in viral miRNAs that match residues that are not conserved between the cellular mir-155 orthologues are shown black on gray background. The seed region (nts. 2–8) and the region which is considered to be of special importance for auxiliary base pairing (nts. 13–16) (Bartel, 2009) are marked underneath the alignment.
While it thus appears that 6mer seeds shared with host miRNAs are enriched, most canonical target sites pair with an extended seed comprising nucleotide 2–8 of the miRNA, i.e. a 7mer (Bartel, 2009). We hence repeated our analysis with such 7mer motifs (Figure 6B), and observed that, although fewer (14) viral 7mer seeds are shared with human miRNAs, the number is still significantly higher than the 8 expected by pure chance. As expected, a similar analysis of seeds shared between mouse and human miRNAs shows a substantially higher (~7fold) enrichment over the expected value. Taken together, the above indicates that, while a minority of viral miRNAs may indeed have evolved to mimic cellular miRNAs, a significant proportion (>50%) of shared seeds will occur by chance, and the percentage of viral miRNAs that are conserved is dwarfed compared to the host miRNAs. Hence, in each case, additional evidence is required before common functionality can be inferred. So far, compelling evidence only exists in a single case: that of miR-155, which shares extended seed identity with two viral miRNAs.
miR-155 mimics – viral analogues of a human oncomir?
Seed sharing between the KSHV-encoded miR-K11 and human miR-155 was first noted in a review article by Nair and Zavolan (Nair and Zavolan, 2006). Subsequently, two groups independently performed cDNA profiling to demonstrate that ectopic expression of either miR-K12-11 or miR-155 results in downregulation of a common set of host mRNA transcripts (Gottwein et al., 2007; Skalsky et al., 2007), reviewed in (McClure and Sullivan, 2008). Several of the candidates were further confirmed as direct targets using chimeric luciferase reporters. miR-155 was originally identified as the only known functional product of the bic (B cell integration cluster) gene, a common integration site of avian leukosis virus (ALV) in virus-induced B cell lymphomas (Clurman and Hayward, 1989). Since then, miR-155 has been found to be over-expressed in many lymphomas as well as acute myeloid leukemia (AML) and solid tumors of the breast, colon and lung (see (Teng and Papavasiliou, 2009) for a recent review on this topic). The observation that forced expression of miR-155 in B cells or hematopoietic stem cells leads to pre-B-cell lymphoproliferation which can progress to frank lymphoma and AML-like disease, respectively, has further bolstered the notion that mir-155 represents a so-called “oncomiR”, i.e. the functional equivalent of a proto-oncogene. A major physiological role of miR-155 appears to be in the hematopoietic compartment, where it shapes the inflammatory and immune response by regulating the activity and differentiation of B cells, T cells and macrophages; for example, mice deficient in miR-155 are immunocompromised and display severe defects in B- and T-lymphocyte maturation as well as antigen presentation by dendritic cells (Rodriguez et al., 2007).
Given the above, it is conceivable that KSHV, a virus that naturally infects B cells, may have evolved a mimic of miR-155, perhaps to modulate host cell differentiation in order to more efficiently gain access to the memory B-cell compartment, which is believed to be a long-term reservoir of latent infection. Under certain (yet to be defined) conditions, however, the constitutive expression in latently infected cells may also promote cellular transformation and hence result in virally induced tumorigenesis. A striking observation is that the alpha herpesvirus MDV1, a virus which is associated with lymphomas in chickens, also encodes a miRNA that shares seed identity with miR-155 (Morgan et al., 2008). Remarkably, the closely related MDV2 not only lacks tumorigenic potential, but also a putative miR-155 mimic (indeed, MDV1 is the only known alpha herpesvirus with a profound tumor association in its natural host). As observed for its KSHV-encoded counterpart, several cellular targets were shown to be commonly regulated by miR-M4 and cellular miR-155 (Muylkens et al., 2010; Zhao et al., 2009). Strikingly, although the lymphotropic EBV does not encode a miR-155 mimic of its own, the viral LMP-1 protein strongly induces expression of the cellular miR-155 in latently infected B-cells (Gatto et al., 2008; Yin et al., 2008).
Taken together, although formal proof in animal models is still missing, the available experimental as well as circumstantial evidence very strongly argues for an important role of KSHV-miR-K11 and MDV1-miR-M4 during virus-induced tumorigenesis. The extent to which these miRNAs phenocopy individual miR-155 functions remains to be determined and, likewise, although a number of shared targets of miR-155 and its viral mimics have been identified, additional experiments will be required to demonstrate their functional importance during the normal viral lifecycle as well as virus-induced disease.
Conclusions
While still in the early days of understanding the biology of virus-encoded miRNAs, much progress has been made in the past few years. A picture is emerging whereby viral miRNAs will generally (but not always) undergo transcription and biogenesis similar to host miRNAs. Those viruses most likely to encode miRNAs will have DNA genomes, and have a nuclear replication cycle. Unlike host miRNAs, it seems many virus-encoded miRNAs have evolved to regulate viral transcripts or small networks of host genes via viral-specific miRNA target binding sites. There are, however, notable exceptions whereby some viral miRNAs will mimic host miRNAs and likely regulate numerous host transcripts that comprise the network of genes regulated by a particular host miRNA. While caution is warranted to ensure that the functions uncovered for virus-encoded miRNAs are not “pigeon-holed” by pre-conceived notions of viral gene function, a picture is emerging where common functions of viral miRNAs across divergent families will include: autoregulation of virus gene expression, regulation of viral persistence/latency, alteration of the cell cycle, and avoidance of host defenses. Because a majority of virus-encoded miRNAs have been indentified in cells latently-infected with herpesviruses, it is possible that inhibiting these miRNAs might one day provide a strategy for clearing latent infections– a “holy grail” of the herpesvirus field. A combination of “bottom-up” and “top-down” approaches will continue to unveil new putative targets and functions and for viral miRNAs. It is imperative that these putative functions be validated in a relevant infectious context. It is striking that, to date, only one example of an in vivo function for a virus-encoded miRNA has emerged. Thus, a firm understanding of some virus-encoded miRNAs awaits the development of the appropriate model systems for those viruses. In a relatively short time frame, virus-encoded miRNAs have added significantly to our understanding of some viruses. In the near future, this understanding will continue to grow, incorporating new viruses, new functions, and possibly new insights of therapeutic relevance.
Acknowledgements
This work was supported by grant RO1AI077746 from the National Institutes of Health and a UT Austin Institute for Cellular and Molecular Biology fellowship to CSS. AG’s work is supported by a grant from the Deutsche Forschungsgemeinschaft (GR 3316/1-1) and institutional support to the HPI, which is provided by the Federal Ministry of Health and the City of Hamburg. The authors gratefully acknowledge Gwen Gage and the UT Austin School of Biological Sciences Graphics Dept. for help with constructing the figures.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abend JR, Uldrick T, Ziegelbauer JM. Regulation of TWEAKR expression by KSHV microRNA prevents TWEAK-induced apoptosis and inflammatory cytokine expression. J Virol. 2010 doi: 10.1128/JVI.00884-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson MG, Haasnoot PC, Xu N, Berenjian S, Berkhout B, Akusjarvi G. Suppression of RNA interference by adenovirus virus-associated RNA. J Virol. 2005;79:9556–9565. doi: 10.1128/JVI.79.15.9556-9565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio O, Razquin N, Zaratiegui M, Narvaiza I, Fortes P. Adenovirus virus-associated RNA is processed to functional interfering RNAs involved in virus production. J Virol. 2006;80:1376–1384. doi: 10.1128/JVI.80.3.1376-1384.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth S, Pfuhl T, Mamiani A, Ehses C, Roemer K, Kremmer E, Jaker C, Hock J, Meister G, Grasser FA. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2008;36:666–675. doi: 10.1093/nar/gkm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellare P, Ganem D. Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe. 2009;6:570–575. doi: 10.1016/j.chom.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bornkamm GW. Epstein-Barr virus and its role in the pathogenesis of Burkitt's lymphoma: an unresolved issue. Semin Cancer Biol. 2009;19:351–365. doi: 10.1016/j.semcancer.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Buck AH, Santoyo-Lopez J, Robertson KA, Kumar DS, Reczko M, Ghazal P. Discrete clusters of virus-encoded micrornas are associated with complementary strands of the genome and the 7.2-kilobase stable intron in murine cytomegalovirus. J Virol. 2007;81:13761–13770. doi: 10.1128/JVI.01290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Li G, Laimins LA, Cullen BR. Human papillomavirus genotype 31 does not express detectable microRNA levels during latent or productive virus replication. J Virol. 2006a;80:10890–10893. doi: 10.1128/JVI.01175-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci U S A. 2005;102:5570–5575. doi: 10.1073/pnas.0408192102. Epub 2005 Mar 5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006b;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantalupo P, Doering A, Sullivan CS, Pal A, Peden KW, Lewis AM, Pipas JM. Complete nucleotide sequence of polyomavirus SA12. J Virol. 2005;79:13094–13104. doi: 10.1128/JVI.79.20.13094-13104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll PA, Brazeau E, Lagunoff M. Kaposi's sarcoma-associated herpesvirus infection of blood endothelial cells induces lymphatic differentiation. Virology. 2004;328:7–18. doi: 10.1016/j.virol.2004.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandriani S, Xu Y, Ganem D. The lytic transcriptome of Kaposi's sarcoma-associated herpesvirus reveals extensive transcription of noncoding regions, including regions antisense to important genes. J Virol. 2010;84:7934–7942. doi: 10.1128/JVI.00645-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SJ, Chen GH, Chen YH, Liu CY, Chang KP, Chang YS, Chen HC. Characterization of Epstein-Barr virus miRNAome in nasopharyngeal carcinoma by deep sequencing. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy EY, Siu KL, Kok KH, Lung RW, Tsang CM, To KF, Kwong DL, Tsao SW, Jin DY. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med. 2008;205:2551–2560. doi: 10.1084/jem.20072581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clurman BE, Hayward WS. Multiple proto-oncogene activations in avian leukosis virus-induced lymphomas: evidence for stage-specific events. Mol Cell Biol. 1989;9:2657–2664. doi: 10.1128/mcb.9.6.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira DE, Ballon G, Cesarman E. NF-kappaB signaling modulation by EBV and KSHV. Trends Microbiol. 2010;18:248–257. doi: 10.1016/j.tim.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- Dolken L, Krmpotic A, Kothe S, Tuddenham L, Tanguy M, Marcinowski L, Ruzsics Z, Elefant N, Altuvia Y, Margalit H, Koszinowski UH, Jonjic S, Pfeffer S. Cytomegalovirus microRNAs Facilitate Persistent Virus Infection in Salivary Glands. PLoS Pathog. 2010;6:e1001150. doi: 10.1371/journal.ppat.1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolken L, Perot J, Cognat V, Alioua A, John M, Soutschek J, Ruzsics Z, Koszinowski U, Voinnet O, Pfeffer S. Mouse cytomegalovirus microRNAs dominate the cellular small RNA profile during lytic infection and show features of posttranscriptional regulation. J Virol. 2007;81:13771–13782. doi: 10.1128/JVI.01313-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari FB, Adams MD, Pagano JS. Unconventional processing of the 3' termini of the Epstein-Barr virus DNA polymerase mRNA. Proc Natl Acad Sci U S A. 1993;90:378–382. doi: 10.1073/pnas.90.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem D. KSHV and the pathogenesis of Kaposi sarcoma: listening to human biology and medicine. J Clin Invest. 2010;120:939–949. doi: 10.1172/JCI40567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto G, Rossi A, Rossi D, Kroening S, Bonatti S, Mallardo M. Epstein-Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-kappaB pathway. Nucleic Acids Res. 2008;36:6608–6619. doi: 10.1093/nar/gkn666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godshalk SE, Bhaduri-McIntosh S, Slack FJ. Epstein-Barr virus-mediated dysregulation of human microRNA expression. Cell Cycle. 2008;7:3595–3600. doi: 10.4161/cc.7.22.7120. [DOI] [PubMed] [Google Scholar]
- Gottwein E, Cullen BR. A human herpesvirus microRNA inhibits p21 expression and attenuates p21-mediated cell cycle arrest. J Virol. 2010;84:5229–5237. doi: 10.1128/JVI.00202-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT, Braich R, Manoharan M, Soutschek J, Ohler U, Cullen BR. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096–1099. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey F, Antoniewicz A, Allen E, Saugstad J, McShea A, Carrington JC, Nelson J. Identification and characterization of human cytomegalovirus-encoded microRNAs. J Virol. 2005;79:12095–12099. doi: 10.1128/JVI.79.18.12095-12099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey F, Meyers H, White EA, Spector DH, Nelson J. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 2007;3:e163. doi: 10.1371/journal.ppat.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey F, Nelson J. Identification and function of human cytomegalovirus microRNAs. J Clin Virol. 2008;41:186–191. doi: 10.1016/j.jcv.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey F, Tirabassi R, Meyers H, Wu G, McWeeney S, Hook L, Nelson JA. A viral microRNA down-regulates multiple cell cycle genes through mRNA 5'UTRs. PLoS Pathog. 2010;6:e1000967. doi: 10.1371/journal.ppat.1000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. Rna. 2006;12:733–750. doi: 10.1261/rna.2326106. Epub 2006 Mar 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther T, Grundhoff A. The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PLoS Pathog. 2010;6:e1000935. doi: 10.1371/journal.ppat.1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen A, Henderson S, Lagos D, Nikitenko L, Coulter E, Roberts S, Gratrix F, Plaisance K, Renne R, Bower M, Kellam P, Boshoff C. KSHV-encoded miRNAs target MAF to induce endothelial cell reprogramming. Genes Dev. 2010;24:195–205. doi: 10.1101/gad.553410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong YK, Foreman K, Shin JW, Hirakawa S, Curry CL, Sage DR, Libermann T, Dezube BJ, Fingeroth JD, Detmar M. Lymphatic reprogramming of blood vascular endothelium by Kaposi sarcoma-associated herpesvirus. Nat Genet. 2004;36:683–685. doi: 10.1038/ng1383. [DOI] [PubMed] [Google Scholar]
- Hussain M, Taft RJ, Asgari S. An insect virus-encoded microRNA regulates viral replication. J Virol. 2008;9:9. doi: 10.1128/JVI.01109-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Iizasa H, Wulff BE, Alla NR, Maragkakis M, Megraw M, Hatzigeorgiou A, Iwakiri D, Takada K, Wiedmer A, Showe L, Lieberman P, Nishikura K. Editing of Epstein-Barr virus-encoded BART6 microRNAs controls their dicer targeting and consequently affects viral latency. J Biol Chem. 2010;285:33358–33370. doi: 10.1074/jbc.M110.138362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. Epub 2003 Dec 2014. [DOI] [PubMed] [Google Scholar]
- Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R. A Pumilio-induced RNA structure switch in p27-3' UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol. 2010;12:1014–1020. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;15:15. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, Hatzigeorgiou A. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Kreuze JF, Savenkov EI, Cuellar W, Li X, Valkonen JP. Viral class 1 RNase III involved in suppression of RNA silencing. J Virol. 2005;79:7227–7238. doi: 10.1128/JVI.79.11.7227-7238.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lei X, Bai Z, Ye F, Xie J, Kim CG, Huang Y, Gao SJ. Regulation of NF-kappaB inhibitor IkappaBalpha and viral replication by a KSHV microRNA. Nat Cell Biol. 2010;12:193–199. doi: 10.1038/ncb2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. Epub 2005 Jan 2030. [DOI] [PubMed] [Google Scholar]
- Lin J, Cullen BR. Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J Virol. 2007;81:12218–12226. doi: 10.1128/JVI.01390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YT, Kincaid RP, Arasappan D, Dowd SE, Hunicke-Smith SP, Sullivan CS. Small RNA profiling reveals antisense transcription throughout the KSHV genome and novel small RNAs. Rna. 2010;16:1540–1558. doi: 10.1261/rna.1967910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish HF, Zhou B, Liu G, Chen CZ. Micromanagement of the immune system by microRNAs. Nat Rev Immunol. 2008;8:120–130. doi: 10.1038/nri2252. [DOI] [PubMed] [Google Scholar]
- Lu CC, Li Z, Chu CY, Feng J, Sun R, Rana TM. MicroRNAs encoded by Kaposi's sarcoma-associated herpesvirus regulate viral life cycle. EMBO Rep. 2010a;11:784–790. doi: 10.1038/embor.2010.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Stedman W, Yousef M, Renne R, Lieberman PM. Epigenetic regulation of Kaposi's sarcoma-associated herpesvirus latency by virus-encoded microRNAs that target Rta and the cellular Rbl2-DNMT pathway. J Virol. 2010b;84:2697–2706. doi: 10.1128/JVI.01997-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol. 2004;78:12868–12876. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure LV, Sullivan CS. Kaposi's sarcoma herpes virus taps into a host microRNA regulatory network. Cell Host Microbe. 2008;3:1–3. doi: 10.1016/j.chom.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Medina PP, Slack FJ. microRNAs and cancer: an overview. Cell Cycle. 2008;7:2485–2492. doi: 10.4161/cc.7.16.6453. [DOI] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Morgan R, Anderson A, Bernberg E, Kamboj S, Huang E, Lagasse G, Isaacs G, Parcells M, Meyers BC, Green PJ, Burnside J. Sequence conservation and differential expression of Marek's disease virus microRNAs. J Virol. 2008;82:12213–12220. doi: 10.1128/JVI.01722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E, Vanicek J, Robins H, Shenk T, Levine AJ. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc Natl Acad Sci U S A. 2008;105:5453–5458. doi: 10.1073/pnas.0711910105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muylkens B, Coupeau D, Dambrine G, Trapp S, Rasschaert D. Marek's disease virus microRNA designated Mdv1-pre-miR-M4 targets both cellular and viral genes. Arch Virol. 2010 doi: 10.1007/s00705-010-0777-y. [DOI] [PubMed] [Google Scholar]
- Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe. 2009;5:376–385. doi: 10.1016/j.chom.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Nair V, Zavolan M. Virus-encoded microRNAs: novel regulators of gene expression. Trends Microbiol. 2006;14:169–175. doi: 10.1016/j.tim.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. Epub 2005 Feb 2016. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- Polic B, Hengel H, Krmpotic A, Trgovcich J, Pavic I, Luccaronin P, Jonjic S, Koszinowski UH. Hierarchical and redundant lymphocyte subset control precludes cytomegalovirus replication during latent infection. J Exp Med. 1998;188:1047–1054. doi: 10.1084/jem.188.6.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polic B, Jonjic S, Pavic I, Crnkovic I, Zorica I, Hengel H, Lucin P, Koszinowski UH. Lack of MHC class I complex expression has no effect on spread and control of cytomegalovirus infection in vivo. J Gen Virol. 1996;77(Pt 2):217–225. doi: 10.1099/0022-1317-77-2-217. [DOI] [PubMed] [Google Scholar]
- Qin Z, Kearney P, Plaisance K, Parsons CH. Pivotal advance: Kaposi's sarcoma-associated herpesvirus (KSHV)-encoded microRNA specifically induce IL-6 and IL-10 secretion by macrophages and monocytes. J Leukoc Biol. 2010;87:25–34. doi: 10.1189/jlb.0409251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Riley KJ, Rabinowitz GS, Steitz JA. Comprehensive analysis of Rhesus lymphocryptovirus microRNA expression. J Virol. 2010;84:5148–5157. doi: 10.1128/JVI.00110-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouha H, Thurner C, Mandl CW. Functional microRNA generated from a cytoplasmic RNA virus. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samols MA, Hu J, Skalsky RL, Renne R. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J Virol. 2005;79:9301–9305. doi: 10.1128/JVI.79.14.9301-9305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samols MA, Skalsky RL, Maldonado AM, Riva A, Lopez MC, Baker HV, Renne R. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 2007;3:e65. doi: 10.1371/journal.ppat.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Seo GJ, Chen CJ, Sullivan CS. Merkel cell polyomavirus encodes a microRNA with the ability to autoregulate viral gene expression. Virology. 2009;383:183–187. doi: 10.1016/j.virol.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Seo GJ, Fink LH, O'Hara B, Atwood WJ, Sullivan CS. Evolutionarily conserved function of a viral microRNA. J Virol. 2008;82:9823–9828. doi: 10.1128/JVI.01144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto E, Moosmann A, Gromminger S, Walz N, Grundhoff A, Hammerschmidt W. Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro JS, Varble A, Pham AM, Tenoever BR. Noncanonical cytoplasmic processing of viral microRNAs. Rna. 2010;16:2068–2074. doi: 10.1261/rna.2303610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Singh CP, Bhavani A, Nagaraju J. Discovering microRNAs from Bombyx mori nucleopolyhedrosis virus. Virology. 2010;407:120–128. doi: 10.1016/j.virol.2010.07.033. [DOI] [PubMed] [Google Scholar]
- Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, Baker HV, Renne R. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol. 2007;81:12836–12845. doi: 10.1128/JVI.01804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, Horwitz E, Prokocimer Z, Prichard M, Hahn G, Goldman-Wohl D, Greenfield C, Yagel S, Hengel H, Altuvia Y, Margalit H, Mandelboim O. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–381. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N, Saleh N, Goldberg MD, Prichard M, Wolf DG, Mandelboim O. Analysis of human cytomegalovirus-encoded microRNA activity during infection. J Virol. 2009;83:10684–10693. doi: 10.1128/JVI.01292-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, Steer CJ. MicroRNAs as gatekeepers of apoptosis. J Cell Physiol. 2010;223:289–298. doi: 10.1002/jcp.22066. [DOI] [PubMed] [Google Scholar]
- Sullivan CS. New roles for large and small viral RNAs in evading host defences. Nat Rev Genet. 2008;20:20. doi: 10.1038/nrg2349. [DOI] [PubMed] [Google Scholar]
- Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- Sullivan CS, Sung CK, Pack CD, Grundhoff A, Lukacher AE, Benjamin TL, Ganem D. Murine Polyomavirus encodes a microRNA that cleaves early RNA transcripts but is not essential for experimental infection. Virology. 2009;7:7. doi: 10.1016/j.virol.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Bertke AS, Patel A, Wang K, Cohen JI, Krause PR. An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc Natl Acad Sci U S A. 2008;105:10931–10936. doi: 10.1073/pnas.0801845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng G, Papavasiliou FN. Shhh! Silencing by microRNA-155. Philos Trans R Soc Lond B Biol Sci. 2009;364:631–637. doi: 10.1098/rstb.2008.0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth Z, Maglinte DT, Lee SH, Lee HR, Wong LY, Brulois KF, Lee S, Buckley JD, Laird PW, Marquez VE, Jung JU. Epigenetic analysis of KSHV latent and lytic genomes. PLoS Pathog. 2010;6:e1001013. doi: 10.1371/journal.ppat.1001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach JL, Cullen BR. In-depth analysis of Kaposi's sarcoma-associated herpesvirus microRNA expression provides insights into the mammalian microRNA-processing machinery. J Virol. 2009;84:695–703. doi: 10.1128/JVI.02013-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;2:2. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach JL, Nagel MA, Cohrs RJ, Gilden DH, Cullen BR. Analysis of human alpha herpesvirus microRNA expression in latently infected human trigeminal ganglia. J Virol. 2009;83:10677–10683. doi: 10.1128/JVI.01185-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach JL, Yen HL, Poon LL, Cullen BR. Influenza A Virus Expresses High Levels of an Unusual Class of Small Viral Leader RNAs in Infected Cells. MBio. 2010;1 doi: 10.1128/mBio.00204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varble A, Chua MA, Perez JT, Manicassamy B, Garcia-Sastre A, tenOever BR. Engineered RNA viral synthesis of microRNAs. Proc Natl Acad Sci U S A. 2010;107:11519–11524. doi: 10.1073/pnas.1003115107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waidner LA, Morgan RW, Anderson AS, Bernberg EL, Kamboj S, Garcia M, Riblet SM, Ouyang M, Isaacs GK, Markis M, Meyers BC, Green PJ, Burnside J. MicroRNAs of Gallid and Meleagrid herpesviruses show generally conserved genomic locations and are virus-specific. Virology. 2009;388:128–136. doi: 10.1016/j.virol.2009.02.043. [DOI] [PubMed] [Google Scholar]
- Walz N, Christalla T, Tessmer U, Grundhoff A. A global analysis of evolutionary conservation among known and predicted gamma herpesvirus microRNAs. J Virol. 2010;84:716–728. doi: 10.1128/JVI.01302-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HW, Trotter MW, Lagos D, Bourboulia D, Henderson S, Makinen T, Elliman S, Flanagan AM, Alitalo K, Boshoff C. Kaposi sarcoma herpesvirus-induced cellular reprogramming contributes to the lymphatic endothelial gene expression in Kaposi sarcoma. Nat Genet. 2004;36:687–693. doi: 10.1038/ng1384. [DOI] [PubMed] [Google Scholar]
- Wen KW, Damania B. Kaposi sarcoma-associated herpesvirus (KSHV): molecular biology and oncogenesis. Cancer Lett. 2010;289:140–150. doi: 10.1016/j.canlet.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T, O'Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB, Ramos JC, Luz E, Pedroso C, Manrique M, Toomey NL, Brites C, Dittmer DP, Harrington WJ., Jr EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 2008;68:1436–1442. doi: 10.1158/0008-5472.CAN-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Xue C, Li J, Bi Y, Cao Y. MDV1-miR-M3 of Marek's Disease Virus suppresses Cisplatin-induced apoptosis by targeting SMAD2 of TGF-{beta} signal pathway. J Virol. 2010 doi: 10.1128/JVI.01392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, McBride J, Fewell C, Lacey M, Wang X, Lin Z, Cameron J, Flemington EK. MicroRNA-155 is an Epstein-Barr virus-induced gene that modulates Epstein-Barr virus-regulated gene expression pathways. J Virol. 2008;82:5295–5306. doi: 10.1128/JVI.02380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Harada JN, Brown HJ, Deng H, Song MJ, Wu TT, Kato-Stankiewicz J, Nelson CG, Vieira J, Tamanoi F, Chanda SK, Sun R. Systematic identification of cellular signals reactivating Kaposi sarcoma-associated herpesvirus. PLoS Pathog. 2007;3:e44. doi: 10.1371/journal.ppat.0030044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. Embo J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. Epub 2004 Nov 2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yao Y, Xu H, Lambeth L, Smith LP, Kgosana L, Wang X, Nair V. A functional MicroRNA-155 ortholog encoded by the oncogenic Marek's disease virus. J Virol. 2009;83:489–492. doi: 10.1128/JVI.01166-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JY, Pfuhl T, Motsch N, Barth S, Nicholls J, Grasser F, Meister G. Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J Virol. 2009;83:3333–3341. doi: 10.1128/JVI.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelbauer JM, Sullivan CS, Ganem D. Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat Genet. 2009;41:130–134. doi: 10.1038/ng.266. [DOI] [PMC free article] [PubMed] [Google Scholar]