Abstract
Background
The integrated functions of 11 Ser/Thr protein kinases (STPKs) and one phosphatase manipulate the phosphorylation levels of critical proteins in Mycobacterium tuberculosis. In this study, we show that the lone Ser/Thr phosphatase (PstP) is regulated through phosphorylation by STPKs.
Principal Findings
PstP is phosphorylated by PknA and PknB and phosphorylation is influenced by the presence of Zn2+-ions and inorganic phosphate (Pi). PstP is differentially phosphorylated on the cytosolic domain with Thr137, Thr141, Thr174 and Thr290 being the target residues of PknB while Thr137 and Thr174 are phosphorylated by PknA. The Mn2+-ion binding residues Asp38 and Asp229 are critical for the optimal activity of PstP and substitution of these residues affects its phosphorylation status. Native PstP and its phosphatase deficient mutant PstPc D38G are phosphorylated by PknA and PknB in E. coli and addition of Zn2+/Pi in the culture conditions affect the phosphorylation level of PstP. Interestingly, the phosphorylated phosphatase is more active than its unphosphorylated equivalent.
Conclusions and Significance
This study establishes the novel mechanisms for regulation of mycobacterial Ser/Thr phosphatase. The results indicate that STPKs and PstP may regulate the signaling through mutually dependent mechanisms. Consequently, PstP phosphorylation may play a critical role in regulating its own activity. Since, the equilibrium between phosphorylated and non-phosphorylated states of mycobacterial proteins is still unexplained, understanding the regulation of PstP may help in deciphering the signal transduction pathways mediated by STPKs and the reversibility of the phenomena.
Introduction
Mycobacterium tuberculosis has an array of proteins to ensure its existence during the course of infection. In order to thrive and maintain its homeostasis, the pathogen continuously influences its surroundings mainly through surface-located sensor proteins. Extracellular signals are communicated through the sensors to the cytosol leading to the appropriate cell responses. Apparently, a large number of pathogens employ reversible phosphorylation of proteins by kinases and phosphatases as a way of transmitting the signals from extracellular milieu which helps in their survival and pathogenicity [1]–[4]. Kinases carry out the phosphorylation by transferring the phosphate moiety on target proteins and phosphatases convert them back to the unphosphorylated state, either by dephosphorylating the substrate or by regulating the activity of kinases.
Apart from the well recognized two component systems targeting His/Asp residues in bacteria, Ser, Thr and Tyr residues are also the major targets for phosphorylation. M. tuberculosis is known to have 11 Ser/Thr protein kinases (STPKs PknA-L, except C), one tyrosine kinase (PtkA), one Ser/Thr phosphatase (PstP) and two tyrosine phosphatases (PtpA and PtpB) [5], [6]. Till date a large number of mycobacterial proteins are shown to be regulated through phosphorylation by STPKs [7]–[11]. Some of these substrates are also known to be dephosphorylated by PstP [9], [11]–[17]. PstP is a PP2C phosphatase (PPM family) that strictly requires Mn2+-ion for its activity [13]. It is a membrane localized enzyme with intracellular catalytic domain of 237 amino acids joined by a juxtamembrane region to the extracellular domain of 191 residues with a single transmembrane helix [18]. Using multi-wavelength anomalous diffraction studies, Pullen et al. determined the structure of the catalytic phosphatase domain of PstP [18]. PstP contains three metal-binding centers in its structure in contrast to two metal centers found in most of the PP2C phosphatases. Using atomic absorption spectroscopy and X-ray analysis, it has been shown that all the bound metal-ions are Mn2+. Similarities between Human Ser/Thr phosphatase PP2Cα and the mycobacterial enzyme have been explained on the basis of structural folds, metal binding and conserved residues [18]. Mutational analyses of PP2Cα have depicted the significance of certain conserved amino acid residues [19]. The corresponding residues in PstP are involved in binding to metal-ions and catalysis in addition to managing the binding and release of phosphate moiety. These residues in PP2Cα are critical for its activity [19] and thus, they are hypothesized to be important for PstP also.
The interesting feature of M. tuberculosis Ser/Thr signaling molecules is that both the essential STPKs, PknB (Rv0014c) and PknA (Rv0015c) and the only Ser/Thr phosphatase PstP (Rv0018c) are located in the same genomic cluster which is conserved in several mycobacterial species [6], [9], [20]. Transcriptional analysis in earlier studies revealed that PknA, PknB and PstP show similar expression profiles [20] and thus, implicate that strong regulation is required for their own functions as both the classes of enzymes functionally counteract each other. In this study, we show that the activity of PstP is modulated by phosphorylation. This is the first report on the regulation of any bacterial Ser/Thr phosphatase by post-translation modification. PstP was found to be phosphorylated differentially by PknA and PknB, both in vitro and in the surrogate host Escherichia coli. Additionally, we found that zinc ions (Zn2+) and inorganic phosphate (Pi) can inhibit the activity of PstP which in turn affects the phosphorylation status of both the kinases and phosphatase.
Materials and Methods
Bacterial strains and growth conditions
E. coli DH5α strain (Novagen) was used for cloning and BL21 (DE3) (Stratagene) was used for the expression of recombinant proteins. E. coli cells were grown and maintained with constant shaking (220 rpm) at 37°C in LB medium supplemented with 100 µg/ml ampicillin.
Gene manipulation
The genes coding for PknAc (rv0015c, representing the cytosolic region of 1-337aa) and PstP (rv0018c, PstP: 1-514aa) were PCR amplified using M. tuberculosis H37Rv genomic DNA. Resulting PCR products were digested with corresponding restriction enzymes and ligated into the vectors pProEx-HTc (Invitrogen) and/or pGEX-5X-3 (GE Healthcare Bio-Sciences) previously digested with the same enzymes. Htc-PknBc and Htc-PstPc were obtained as described earlier [9]. pGEX-PknBc was sub-cloned from Htc-PknBc using standard protocols under the same restriction sites. For cloning in dual-expression vector pETDuet-1 (Novagen), genes coding for PstPc or PstPc D38G were inserted in MCS1 having N-terminal His6-tag while kinases PknA and PknB (full length) were cloned in MCS2 with N-terminal MBP-tag (Maltose-binding protein tag upstream of the kinase). MBP-alone (without kinase) was taken as control vector having PstPc or PstPc D38G in MCS1. The protocols used for cloning in pETDuet-1 have been discussed earlier [21].
Mutagenesis of specific residues was carried out using the QuikChange XL site-directed mutagenesis kit (Stratagene) as per manufacturer's instructions. Mutants of PstP and PstPc were created as R20G, D38G and D229G using Htc-PstP and Htc-PstPc as templates. Htc-PstPc was utilized for the generation of Htc-PstPct5a and Htc-PstPct141e. Htc-pknBc was employed as template for generation of double mutant Htc-pknBct171/173d. The details of all the primers and clones are provided in tables 1 and 2, respectively. The integrity of all clones was confirmed by DNA sequencing (TCGA, New Delhi).
Table 1. Primers used in the study.
| Primer Name | Sequence Details (5′→3′) ** |
| PknBc T171/173D FP | CGGCAACAGCGTGGACCAGGACGCAGCAGTGATCG |
| PknBc T171/173D RP | CGATCACTGCTGCGTCCTGGTCCACGCTGTTGCCG |
| PknA FP | TGATCGAAGCCGGAATTCAGGGGGAACCATGA EcoR1 |
| PknAc RP | AGCACCCCCGCGGCCGCGAGCAGCGCTCACTGACCGGAC Not1 |
| PstPc D38G FP | CTATTGGCCCTGGCCGGCGGCATGGGTGGGCAT |
| PstPc D38G RP | ATGCCCACCCATGCCGCCGGCCAGGGCCAATAG |
| PstPc R20G FP | GATCGCGGCTTGGTAGGCGCCAACAACGAAGACTCGGTC |
| PstPc R20G RP | GACCGAGTCTTCGTTGTTGGCGCCTACCAAGCCGCGATC |
| PstPc D229G FP | GGCGGCGGCCCCGGCAACGTCACTGTCGTCGTC |
| PstPc D229G RP | GACGACGACAGTGACGTTGCCGGGGCCGCCGCC |
| PstPc T5A FP | GGAGAGTGGCGCGCGTGGCCCTGGTCCTGCGATAC |
| PstPc T5A RP | GTATCGCAGGACCAGGGCCACGCGCGCCACTCTCC |
| PstPc T141E FP | GACGACACGTTTGTCCAAGCGCTGGTCGACGAAGGCCG |
| PstPc T141E RP | CGGCCTTCGTCGACCAGCGCTTGGACAAACGTGTCGTC |
| pETDuet-PstP FP | CACC GCGGCCGCTCATATG GCGCGCGTGACCCTGG Not1 |
| pETDuet-PstPc RP | CGGTCACCAGTGCGGCCGCGAATGCTCACCGTCGGCC Not1 |
**Restriction sites/stop codon/mutated sequences have been underlined.
Table 2. Description of the plasmids used in this study.
| Plasmid construct | Description | Reference |
| pProEx-HTc | E. coli expression vector containing N-terminal His6-tag | Invitrogen |
| pProEx-HTc-PknBc | Expression of His6PknB1-331 (cytosolic domain) | [9] |
| pProEx-HTc-PknBc T171/173D | pProEx-HTc-PknBc with activation loop residues Thr171 and Thr173 mutated to Asp, phosphomimetic amino acid | This study |
| pProEx-HTc-PknAc | Expression of His6PknA1-337 (cytosolic domain) | This study |
| pProEx-HTc-PstPc | Expression of His6PstP1-300 (cytosolic domain) | [9] |
| pProEx-HTc-PstPc R20G | pProEx-HTc-PstPc with Arg20 mutated to Gly | This study |
| pProEx-HTc-PstPc D38G | pProEx-HTc-PstPc with Asp38 mutated to Gly | This study |
| pProEx-HTc-PstPc D229G | pProEx-HTc-PstPc with Asp229 mutated to Gly | This study |
| pProEx-HTc-PstPc T5A | pProEx-HTc-PstPc with Thr5 mutated to Ala | This study |
| pProEx-HTc-PstPc T141E | pProEx-HTc-PstPc with Thr141 mutated to Glu | This study |
| pGEX-5X-3 | E. coli expression vector containing N-terminal Glutathione S-Transferase tag | GE Healthcare |
| pGEX-5X-3-PknAc | Expression of GST-PknA1-337 (cytosolic domain) | This study |
| pGEX-5X-3-PknBc | Expression of GST-PknB1-331 (cytosolic domain) | This study |
| pETDuet1 | E. coli dual expression vector containing N-terminal His6-tag in MCS1 and C-terminal S-tag in MCS2 | Novagen |
| pETDuet1-PstPc D38G/MBP | Expression of His6-PstPc D38G in MCS1 with Myelin basic protein (MBP) in MCS2 | This study, [21] |
| pETDuet1-PstPc D38G/MBP-PknA | Expression of His6-PstPc D38G in MCS1 with MBP-tagged PknA in MCS2 | This study, [21] |
| pETDuet1-PstPc D38G/MBP-PknB | Expression of His6-PstPc D38G in MCS1 with MBP-tagged PknB in MCS2 | This study, [21] |
Protein expression and purification
Proteins were expressed and purified from E. coli as described previously [9]. The purified proteins were assessed by SDS-PAGE and concentrations were estimated by Bradford assay (Bio-Rad).
In vitro kinase assays and phosphoamino acid analysis
In vitro phosphorylation of PstPc or its mutants (0.5–3 µg) by PknAc (0.5–1 µg) or PknBc (1–3 µg) was carried out in kinase buffer (20 mM PIPES [pH 7.2], 5 mM MnCl2, 5 mM MgCl2) containing 2 µCi [γ-32P]ATP (BRIT, Hyderabad, India) followed by incubation at 25°C for 20 min. Reactions were terminated by 5X SDS sample buffer followed by boiling at 100°C for 5 min. Proteins were separated by 12% SDS-PAGE and analyzed by PhosphorImager (FLA 2000, Fuji). Zn2+ and Pi were added to the kinase assay reactions as per requirement of the assay. For the visualization of phosphorylation signal on cleaved proteins, removal of recombinant tags was achieved by addition of TEV protease (for His6-tagged PstP/PstPc and their mutants) in TEV buffer (Tris-Cl [pH 8.5], 5 mM EDTA, 300 mM NaCl and 1 mM DTT) after the kinase reaction followed by an additional incubation for 2 hr at 20°C. For phosphoamino acid analysis, PstPc D38G was phosphorylated by PknBc and PknAc and cleaved with TEV protease as mentioned above, separated by SDS-PAGE and electroblotted onto Immobilon PVDF membrane (Millipore). Phosphoamino acid analysis by two-dimensional thin layer electrophoresis was performed as described earlier [9], [22].
In vitro dephosphorylation and p-nitrophenol phosphate (pNPP) hydrolysis assays
PknBc and PknAc were autophosphorylated by in vitro kinase assays using [γ-32P]ATP. 1 µg of purified PstPc/PstPc D38G/PstPc R20G/PstPc D229G were added in four sets of reactions and incubated at 25°C for increasing time points up to 30 min to measure the dephosphorylation potential of PstPc and its mutants. For auto-dephosphorylation assays, PknBc and PknBc T171/173D (2 µg each) were autophosphorylated by in vitro kinase assays and exposed to dephosphorylation by PstPc and PstPc D38G (1 µg). Reactions were stopped by adding 5X SDS sample buffer and boiled for 5 min at 100°C. The samples were separated by 12% SDS-PAGE and phosphorylated bands were observed and analysed by PhosphorImager.
pNPP hydrolysis assay was performed as a measure of phosphatase activity. PstPc was added to a reaction mixture containing phosphatase assay buffer (50 mM Tris pH 8.0, 5 mM DTT, 4 mM MnCl2) and 10 mM pNPP in a 96-well plate and incubated at 37°C for indicated time points and absorbance was read at 405 nm (Microplate reader, Bio-Rad). To assay the relative activity of PstPc and its phosphatase-deficient variants, increasing concentrations of enzymes were added to the reaction mix and processed as above. Alkaline phosphatase (Roche) and PknBc were taken as positive and negative controls, respectively, for the pNPP hydrolysis assays. Variations of PstPc activity by addition of Zn2+ and Pi was assessed by adding ZnCl2 or sodium phosphate [pH 7.2] to the reaction mixture as above, to achieve the indicated final concentrations. pETDuet-1 purified PstPc and PstPc D38G, co-expressed with or without kinases, were employed for pNPP-assays to measure the effect of phosphorylation on their activities.
Metabolic labeling in E. coli
The procedure described by Kumar et al. was followed for metabolic labeling [23]. E. coli (BL21-DE3) transformants harbouring either pETDuet-PstPc/PstPcD38G-mbp or pETDuet-PstPc/PstPcD38G-mbpPknA or pETDuet-PstPc/PstPcD38G-mbpPknB were grown in 5 ml LB medium containing 100 µg/ml ampicillin to an O.D600 of ∼0.6. The cells were induced with 1 mM IPTG and further grown for 4 hr at 16°C. Cultures were harvested, washed with 5 ml of M9 medium [pH 7.0] without phosphate salts (for 1 L: NH4Cl-1 g, NaCl-0.5 g, 20% Glucose-10 ml, MgSO4.7H2O-1 ml, Thiamine-HCl-1 ml, CaCl2-1 ml). The cells were resuspended in 2 ml of M9 media supplemented with 1 mCi of [32P]orthophosphoric acid (BRIT, Hyderabad, India), 100 µg/ml ampicillin and 1 mM IPTG and further grown at 16°C for 4 hr. Under specific conditions, Sodium phosphate [pH 7.2] (2 mM) or ZnCl2 (4 mM) were added to M9 media and subsequent processing steps of metabolic labeling. The cells were harvested and lysed by sonication in the lysis buffer containing phosphate-buffered saline, 5% glycerol and protease inhibitor cocktail. The cell lysate was clarified and the lysates containing His6-fusion protein were incubated with lysis buffer equilibrated Ni2+-NTA affinity beads for 2 hr at 4°C. The beads were then thoroughly washed with lysis buffer containing 20 mM imidazole and resuspended in 5X SDS sample buffer followed by boiling for 15 min. The samples were resolved on SDS-PAGE followed by autoradiography.
Identification of phosphorylation sites in PstPc D38G
PknBc and PknAc were employed for in vitro kinase assay using 50 µM cold ATP and PstPc D38G. The samples were run on 12% SDS-PAGE, stained with Coomassie Brilliant Blue and de-stained. Bands corresponding to PstPc D38G were excised from the gel and washed with MilliQ water. The samples were processed for identification of phosphorylation sites by using Thermo-Finnagen LTQ electrospray instrument (Proteomics Core Facility, Children's Hospital, Boston). The detailed protocol of sample processing for identification of phosphorylation sites has been provided in File S1.
Generation of polyclonal antibodies for PstPc in rabbit and immunoblotting
Polyclonal antibodies against PstPc were generated in rabbit. To confirm the presence of PstPc/PstPc D38G in Ni2+-NTA pulled-out proteins after metabolic labeling by western blot analysis, the samples were resolved by SDS-PAGE along with positive (purified PstPc) and negative controls (GST-PknBc) and transferred onto nitrocellulose membrane (Bio-Rad). Standard procedure for immunoblotting was followed [9], [11]. The blots were developed using SuperSignalR West Pico Chemiluminescent Substrate kit (Pierce Protein Research Products) according to manufacturer's instructions.
Results
Identification of the residues critical for the activity of PstP
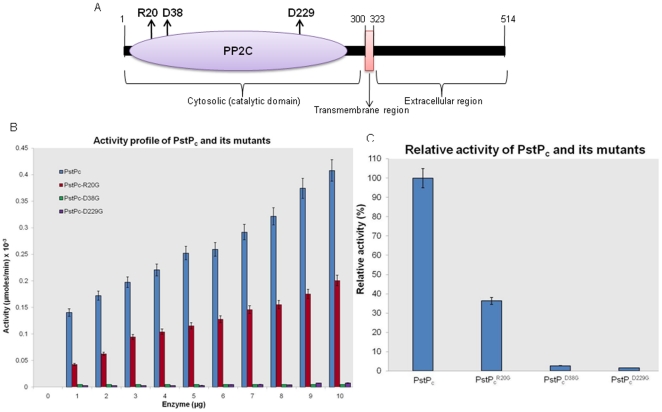
On the basis of structural data available for PstP and alignment with the residues important for Human PP2Cα activity [18], PstPc mutants were generated using site-directed mutagenesis. These residues include the Mn2+-ion binding sites-Asp38 and Asp229 and phosphate (Pi) binding residue-Arg20 (Figure 1A). In the resulting mutants, these sites were converted to Glycine (PstPc D38G, PstPc D229G and PstPc R20G). The activity of these mutants was compared using chromogenic substrate pNPP. To confirm the authenticity of the assay, increasing concentrations of alkaline phosphatase were utilized as a positive control while PknBc was used as negative control (Figure S1). The pNPP assay with increasing amounts of PstPc-mutants showed that the mutation of Asp38 and Asp229 to Gly resulted in >90% loss of the dephosphorylation activity of PstPc, while the PstPc R20G mutant lost about 60% of its activity (Figure 1B and 1C). Thus, Arg20, Asp38 and Asp229 were identified as the residues required for optimum activity of PstP. To confirm that the loss in activity was specifically due to mutagenesis of Asp38, Asp229 and Arg20, irrelevant residues (Thr5 and Thr141) in PstPc were mutagenized to generate PstPc T5A and PstPc T141E. The relative activities of these mutants were compared with the native enzyme through pNPP-assay (Figure S2). There were no significant changes observed in the mutants in comparison to PstPc, thus reinforcing the importance of Arg20, Asp38 and Asp229 residues.
Figure 1. Critical residues of PstP.
(A) Schematic representation of PstP with critical residues (Arg20, Asp38 and Asp229) being highlighted with upward arrows. (B) Activity profiles of PstPc and its mutants: Activity assays were performed by pNPP-hydrolysis mediated by PstPc, PstPc R20G, PstPc D38G and PstPc D229G. Increasing concentrations of proteins were taken with constant substrate concentration (10 mM pNPP) and incubated at 37°C for 30 mins. As shown in the graph, the mutants had lost phosphatase activity to different extents. Activity is calculated as a measure of µmoles of pNPP hydrolyzed per min. at a given enzyme concentration. (C) The relative activity of all the phosphatase variants (5 µg each, 30 min.) showed that PstPc D38G and PstPc D229G had lost >90% of activity while PstPc R20G lost ∼60% of the activity as compared to PstPc. The error bars indicate the SD of three individual experiments.
Phosphatase activity of PstPc and its mutants
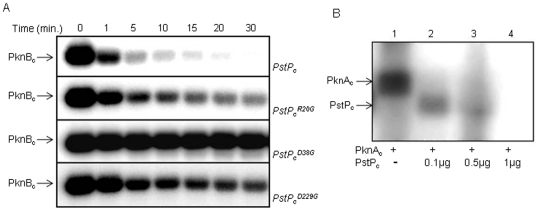
The dephosphorylation potential of PstPc and its mutants was also assessed by their ability to dephosphorylate PknBc in a time-dependent dephosphorylation (Figure 2A) and pNPP hydrolysis assays (Figure S3). PstPc R20G dephosphorylated the autophosphorylated PknBc to some extent, whereas substantial loss of phosphatase activity was observed with PstPc D38G and PstPc D229G (Figure 2A). The activity of PstPc D229G was relatively higher than that of PstPc D38G as opposed to the observation in pNPP-assays (Figures 1C and S3). Similar observations have been reported earlier where the activity of an enzyme, specifically Ser/Thr phosphatases, is shown to be dependent on the nature of substrate [24]–[26]. pNPP is an artificial substrate while PknB is a natural substrate of PstP, which may be recognized and subsequently dephosphorylated more optimally. Additionally, in this case, the activity of the phosphatase also depends on the activity of PknB, as discussed in later sections. The assays were also performed using autophosphorylated PknAc which showed similar results (data not shown). Surprisingly, in this assay, additional phosphorylated bands corresponding to the size of PstPc D38G were observed when incubated with kinase for longer time. No such bands were observed with PstPc, PstPc R20G and PstPc D229G at the given concentrations.
Figure 2. Dephosphorylation by PstPc and its mutants.
(A) Autoradiogram showing autophosphorylated PknBc, exposed to dephosphorylation by PstPc, PstPc R20G, PstPc D38G and PstPc D229G. Time-dependent dephosphorylation was performed with 1 µg of phosphatase after carrying out autophosphorylation of PknBc (2 µg) in an in vitro kinase assay. Noticeably, PstPc D38G was observed to be phosphorylated with increasing time points (3rd panel from the top). (B) Autoradiogram showing phosphorylation of PstPc by PknAc (1 µg). Increasing concentrations of PstPc were used to measure the extent of dephosphorylation. Unexpectedly, the phosphatase itself got phosphorylated at higher kinase to phosphatase ratio, though kinase was completely dephosphorylated. No phosphorylation was observed at higher PstPc concentrations.
To further assess this observation, PknAc or PknBc were incubated with increasing concentrations of PstPc. Interestingly, PstPc was phosphorylated by PknAc at higher kinase to phosphatase ratio (Figure 2B). An increase in the concentration of PstPc resulted in complete dephosphorylation of both the proteins. This serendipitous observation intrigued us to explore whether PstP is a target of Ser/Thr protein kinases. Due to strong dephosphorylation activity of PstP, it was difficult to achieve the phosphotransfer on native phosphatase. Therefore, further studies were carried out with the mutants of PstP that were deficient in phosphatase activity.
Phosphorylation of PstPc D38G, PstPc D229G and PstPc R20G
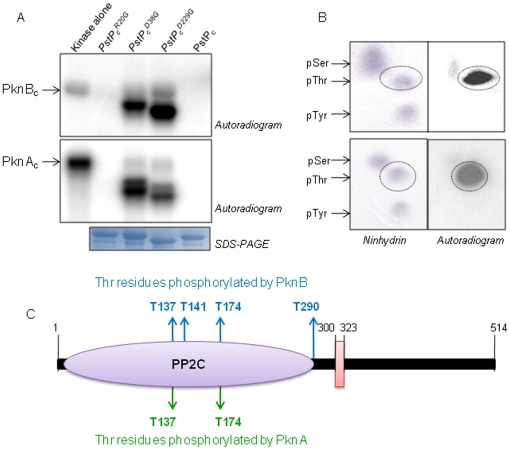
After identification of the residues critical for PstPc activity and measuring the activity of corresponding mutants, the phosphorylation status of PstPc mutants was studied. PknA and PknB were employed for the phosphorylation assays. PstPc D38G and PstPc D229G were efficiently phosphorylated by both PknAc and PknBc (Figure 3A), whereas faint signal on PstPc R20G was observed owing to its partial phosphatase activity. Phosphorylation of PstPc (at 3 µg concentration) was not observed by in vitro kinase assay as it completely dephosphorylated PknAc and PknBc, making them inactive (heat-inactive PstPc was found to be phosphorylated-data not shown). To confirm that the observed phosphorylation is on PstPc-mutants and not on the N-terminally attached His6-tag, TEV-protease cleavage of the tag was performed after the kinase assays. Phosphorylation was confirmed to be specifically localized on the cleaved substrate protein (data not shown). Additionally, the R20G, D38G and D229G mutants were also created in full length PstP construct and pNPP-hydrolysis assays and phosphorylation reactions were also confirmed using full length PstP and its mutants (data not shown).
Figure 3. Phosphorylation of PstPc and its mutants by PknA and PknB.
(A) Phosphorylation of PstPc and its mutants (3 µg each) by 2 µg PknBc (upper panel) and 0.5 µg PknAc (middle panel). PstPc D38G and PstPc D229G were efficiently phosphorylated by both the kinases due to loss of phosphatase activity. Phosphorylation on PstPc R20G mutant was low due to its partial phosphatase activity. The corresponding SDS-PAGE is shown (lowest panel) as a loading control. (B) Phosphoamino acid analysis by 2D-TLE illustrates that both PknAc (upper panel) and PknBc (lower panel) phosphorylates PstPc D38G on Thr residues. (C) Sites of phosphorylation of PknBc (blue) and PknAc (green) in PstPc D38G were identified by mass spectrometric analysis. PknBc phosphorylates PstPc D38G majorly on four Thr residues-Thr137, Thr141, Thr174 and Thr290 while two Thr residues were phosphorylated by PknAc-Thr137 and Thr174.
Phosphoamino acid analysis and identification of phosphorylation site(s) of PknA and PknB in PstPc D38G
Phosphoamino acid analysis by two-dimensional thin layer electrophoresis showed that both PknAc (Figure 3B, upper panel) and PknBc (Figure 3B, lower panel) phosphorylated PstPc D38G on Thr residues while no signal was observed on the spots corresponding to pSer and pTyr. For further experiments, PstPc D38G was utilized.
The sites of PknA and PknB phosphorylation on PstPc D38G were identified through mass-spectrometric analysis by Thermo-Finnagen LTQ electrospray Mass-Spectrometer, using in vitro phosphorylated protein. The results showed that four Thr residues were phosphorylated by PknB (Thr137, Thr141, Thr174 and Thr290) while PknA phosphorylated PstPc D38G on two residues (Thr137 and Thr174) (Figure 3C, supplementary file 2). Thus, PstPc D38G is differentially phosphorylated by PknA and PknB which may have important implications on the activity of PstP.
Validation of PstP phosphorylation in E. coli
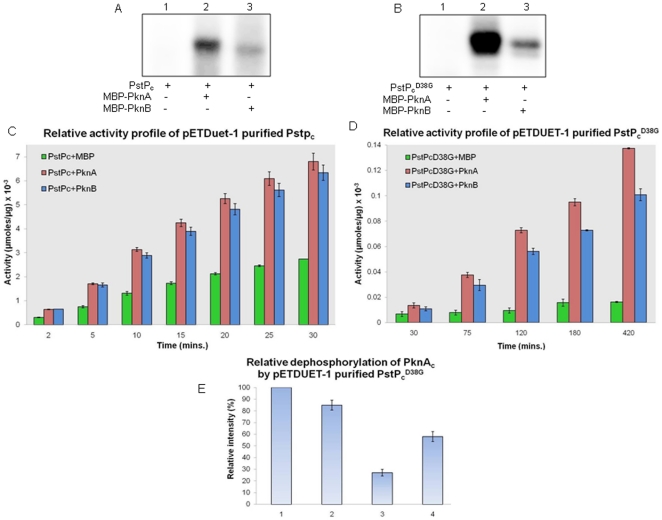
To further substantiate our results, the phosphorylation status of PstPc and PstPc D38G was examined specifically by PknA and PknB in E. coli using a dual expression system. PstPc and PstPc D38G were cloned in pETDuet1 expression vector along with either MBP alone or MBP-tagged PknA or PknB. E. coli BL21 (DE3) cells transformed with pETDuet1-PstPc/PstPcD38G-MBP or pETDuet1-PstPc/PstPcD38G-MBP-kinase (kinase, PknA or PknB) were metabolically labelled with [32P]orthophosphoric acid. Phosphorylation of PstPc and PstPc D38G could only be detected when PknA or PknB were co-expressed (Figures 4A and 4B), suggesting the phosphorylation of phosphatase by both the kinases in native conditions in E. coli. Western blot analysis of Ni2+-NTA purified samples using rabbit anti-PstPc antibodies confirmed the metabolically labelled protein to be PstPc (data not shown).
Figure 4. Co-expression analysis of STPKs and PstPc/PstPc D38G.
(A) Metabolic labeling of PstPc: PstPc co-expressed with MBP-PknA (lane 2) or MBP-PknB (lane 3) gets phosphorylated in E. coli under native conditions while PstPc co-expressed with MBP alone (lane 1) was not phosphorylated. (B) Metabolic labeling of PstPc D38G: PstPc D38G co-expressed with MBP-PknA (lane 2) or MBP-PknB (lane 3) gets phosphorylated in E. coli while PstPc D38G co-expressed with MBP alone (lane 1) was not phosphorylated. As expected, the intensity of phosphorylation on PstPc D38G was comparatively higher than that of PstPc. (C) Relative activity profile of pETDuet1 purified PstPc and (D) PstPc D38G: pNPP assays were performed with PstPc and PstPc D38G (1 µg each) purified from pETDuet1 co-expressing MBP or MBP-PknA/PknB. The dephosphorylation potential of phosphorylated PstPc and PstPc D38G (co-expressed with either kinase) is higher than that of unphosphorylated protein. For PstPc D38G, activity was evaluated over long time points due to its low dephosphorylation activity. Activity is calculated as a measure of µmoles of pNPP hydrolyzed per µg of protein at a given time. The error bars indicate the SD of three individual experiments. (E) Relative dephosphorylation of PknAc by pETDuet-1 purified PstPc D38G: Autophosphorylated PknAc was incubated for 30 mins with unphosphorylated and phosphorylated PstPc D38G and the extent of dephosphorylation was assessed by in vitro dephosphorylation assays. The image obtained after autoradiography was analyzed by ImageGauge software (Fuji) and relative intensity of phosphorylation was measured: (1) PknAc alone, (2) PknAc+MBP-PstPc D38G, (3) PknAc+PstPc D38G phosphorylated by PknA and (4) PknAc+PstPc D38G phosphorylated by PknB. As shown, the PknA-phosphorylated PstPc D38G dephosphorylated the kinase to a greater extent in comparison to the unphosphorylated PstPc D38G. The error bars represent the SD of the three individual experiments. The corresponding autoradiogram is shown in Figure S4.
Activity assays of pETDuet1-purified PstPc and PstPc D38G
The activity profiles of PstPc and PstPc D38G co-expressed with and without PknA/PknB, were evaluated. According to the pNPP assays, the activity of phosphorylated PstPc (co-expressed with PknA or PknB) was higher than that of unphosphorylated phosphatase (co-expressed with MBP alone) (Figure 4C). The phenomenon was also confirmed by measuring the activity of PstPc D38G. As already discussed, PstPc D38G had retained about 10% of the dephosphorylation activity as a result of which, it was phosphorylated efficiently by kinases. The relative activity of phosphorylated PstPc D38G with PknA/PknB and unphosphorylated protein was measured for 420 min. Interestingly, the activity of phosphorylated PstPc D38G was remarkably higher than that of unphosphorylated protein, thus the similar profile as that of PstPc was observed (Figure 4D). Also, the activity of PknA phosphorylated phosphatase was even more than the protein phosphorylated by PknB. Noticeably, the increase in phosphatase activity after phosphorylation may also account for the observed increase in the activity of PstPc D229G in the time-dependent dephosphorylation assays (Figure 2A).
The dephosphorylation of in vitro autophosphorylated PknAc was assessed by PstPc D38G+MBP, PstPc D38G+MBP-PknA and PstPc D38G+MBP-PknB. As expected, due to higher activity of phosphorylated PstPc D38G, intensity of phosphorylation on PknAc was low as compared to the reaction containing unphosphorylated PstPc D38G+MBP (Figures 4E and S4). Also, since PknA-phosphorylated PstPc D38G was more active than PknB-phosphorylated PstPc D38G (Figure 4D), the extent of dephosphorylation was more in lane 3 as compared to lane 4.
Auto-dephosphorylation of PstPc
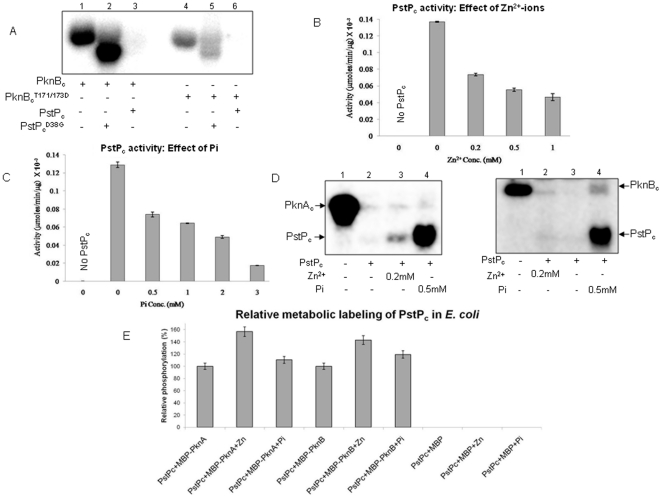
Next, we tried to understand whether the inability of PstPc to be effectively phosphorylated was due to its dephosphorylation activity on the kinases resulting in their inactivation or it was due to auto-dephosphorylation. Consequently, phosphomimetic mutants of PknBc were generated for the Thr residues of activation loop in catalytic domain [12], forming PknBc T171/173D which cannot be dephosphorylated by PstPc on Thr171 and Thr173. As reported by Boitel et al., PknB does not lose phosphorylation signals after mutagenesis of Thr171 and Thr173. Through a series of careful analysis of single and double mutants of PknB, it has been shown that PknB can be additionally phosphorylated on Ser166 and/or Ser169 residues [12]. Thus, we utilized PknBc and PknBc T171/173D, that were autophosphorylated in an in vitro kinase assay using [γ-32P]ATP, before incubation with PstPc. Phosphorylation of PstPc was still not observed with constitutively active PknBc T171/173D, as confirmed by phosphotransfer observed on PstPc D38G (Figure 5A). This suggests that PstPc can dephosphorylate itself. Additionally, PknBc T171/173D was completely dephosphorylated in presence of PstPc, suggesting that PstP could also dephosphorylate the surplus sites Ser166/Ser169.
Figure 5. Factors affecting PstP activity.
(A) Auto-dephosphorylation of PstPc: Autoradiogram showing phosphorylation by PknBc. PstPc and PstPc D38G (3 µg each) were used for in vitro phosphorylation assay by PknBc and PknBc T171/173D (2 µg each). Since PknBc T171/173D cannot be dephosphorylated by PstPc, lack of signal signifies auto-dephosphorylation of phosphatase. PstPc D38G was used as positive control to show that PknBc T171/173D is active. Regulation of PstPc activity: pNPP assay showing the effect on activity of PstPc (1 µg) by (B) Zn2+ and (C) Pi. pNPP assay was carried out for 30 mins and activity was calculated as a measure of µmoles of pNPP hydrolyzed per min per µg of protein. The error bars show SD of three independent experiments. (D) Phosphorylation of PstPc: Autoradiogram showing the phosphorylation of PstPc (1 µg) by GST-PknAc (left panel) and GST-PknBc (right panel) in presence of 0.2 mM Zn2+ and 0.5 mM Pi. Since His6-tagged STPKs were not resolved properly from PstPc on SDS-PAGE (Figure S5), the assay was also performed with GST-tagged kinases having higher molecular weights. (E) Metabolic labeling of PstPc by PknA and PknB in E. coli in presence of Zn2+ and Pi: Phosphorylation level of PstPc was observed to be increased when Zn2+ (4 mM) and Pi (2 mM) were added during the culture conditions and subsequent processing steps. The autoradiograms obtained after SDS-PAGE were analyzed by ImageGauge software and intensity of the band corresponding to PstPc phosphorylation without any added factor was taken as 100%. Relative phosphorylation is depicted in the bar graph.
Identification of the factors affecting the activity of PstP
The phosphorylation of PstP suggested that additional factors may be involved in the cellular milieu that can regulate and control the phosphatase activity, preceding its phosphorylation. In general, phosphatases are known to be affected by a number of factors like metal-cations, Pi, creatine phosphate (CP) and ATP/ADP ratio. PstPc activity assay was carried out in the presence of selected factors. Interestingly, activity of PstPc was reduced in the presence of Zn2+ and Pi, as assessed by pNPP assay. Reduction of almost 50% activity was observed at 0.2 mM Zn2+ (Figure 5B) and 0.5 mM Pi (Figure 5C). Maximum inhibition of PstPc was observed at 1 mM Zn2+ and 4 mM Pi. Inhibition by Zn2+ at >1 mM was not calculable due to protein precipitation in the reaction mixture.
Phosphorylation of PstPc in the presence of Zn2+ and Pi
The inhibition of PstPc in the presence of Zn2+ and Pi provided a condition that could favour the phosphorylation of PstPc by STPKs. PstPc was indeed phosphorylated by PknAc and PknBc in presence of 0.2 mM Zn2+ or 0.5 mM Pi (Figure 5D), under in vitro conditions. Since the phosphorylated bands of His6-tagged PknAc/PknBc and PstPc were not able to resolve on SDS-PAGE (Figure S5), the assay was performed with GST-tagged kinases and similar results were obtained. To further assess the effects of Zn2+ and Pi, metabolic labeling of PstPc by co-expressed kinases PknA and PknB was performed in E. coli in the presence of Zn2+ (4 mM) and Pi (2 mM) (Figure 5E). Phosphorylation of PstPc was indeed enhanced in the presence of Zn2+ by ∼40%-50%. The enhancement in phosphorylation in the presence of Pi was not as prominent (∼10%–20%), possibly due to competition of phosphate ions with [32P]orthophosphoric acid. Nevertheless, as a proof of principle, Zn2+ and Pi were identified as the novel regulators which can inhibit the activity of PstPc and facilitate its phosphorylation.
Discussion
The coordinated regulation of Ser/Thr protein kinases and phosphatases is essential for maintaining the appropriate equilibrium of protein phosphorylation. Membrane associated kinases and phosphatases are known or hypothesized to be regulated by external stimulus. It is of great relevance to decipher the regulatory mechanisms especially in the systems like M. tuberculosis where one Ser/Thr phosphatase PstP is accountable for the effects caused by 11 STPKs. In general, the processes involved in regulating the phosphatases include some external signals, variation in pH [27], cellular concentrations of ATP, ADP, Pi (or their ratios) [28], [29], cytosolic cations like Mn2+, Zn2+, Mg2+, Ca2+ [13], [27], [29]–[31] and post-translation modifications (phosphorylation, methylation) [28], [30], [32]–[39]. Present study demonstrates an example of PknA and PknB mediated regulation of PstP through inter-dependent phosphorylation-dephosphorylation reactions. Regulation of phosphatases by phosphorylation is a critical step for cell signaling pathways. It is also associated with feedback phenomena in case where phosphatases are phosphorylated by the kinases that are in turn dephosphorylated by the same phosphatase. Certain examples illustrate the phosphorylation of PP2C phosphatases such as rat Mg2+-dependent protein phosphatase α (MPPα) by casein kinase II [39], Soybean kinase associated protein phosphatase (Soybean KAPP) [37], Oryza sativa KAPP [40], but these have not been detailed in terms of feedback regulation.
PstP has conserved domain architecture of PP2C-phosphatases (PPM family). PPM family phosphatases play an imperative role in a number of systems described earlier [41]–[48]. Except a few PP2C-phosphatases like Human PP2Cα [49] and Arabidopsis KAPP [50], not much is known about other members of this family. For PstP, we have previously shown that PknA and PknB are the targets for dephosphorylation by PstP and detailed the basic biochemical requirements of this enzyme along with its membrane localization [13]. In a later study, Pullen et al. resolved the crystal structure of PstP catalytic domain and described the most important features of this molecule having characteristic PP2C-fold along with three-metal binding centers that associate with Mn2+ [18]. The discovery of third-metal centre was a unique feature of PstP as other PP2C phosphatases were found to have two metal-binding centres. In the recent studies, the PP2C-phosphatases of Streptococcus agalactiae and Thermosynechococcus elongatus have been shown to have a similar third-metal binding centre [51], [52]. The third metal ion center in PstP is proposed to be involved in structural perturbations leading to altered phosphoprotein recognition profiles.
In this study, three conserved residues were selected for generation of site-directed mutants in PstPc, on the basis of similarity with Human phosphatase PP2Cα [18]. Arg20 (PP2Cα Arg33) is responsible for hydrolysis of phosphate moiety from pSer/pThr residues in target proteins. Asp38 (PP2Cα Asp60) and Asp229 (PP2Cα Asp282) constitute a part of Mn2+-metal centers and coordinate with the two critical Mn2+. Mutations of Asp38 and Asp229 affected the activity of PstP rendering it active to minimal level, though R20G mutant retained about 40% activity. Thus, the residues that are involved in Mn2+-ion binding and hydrolysis of phosphate are deciphered to be critical for its activity. Accordingly, the extent of phosphorylation of each mutant was dependent on the remaining dephosphorylation activity, so that PstPc D38G and PstPc D229G were efficiently phosphorylated by PknA and PknB.
Association with metals is crucial for PP2C phosphatases and any perturbation with inherently associated metals may lead to altered functional profile. The minimum requirement for PstPc activity is the presence of Mn2+ [13]. For PP2C-class of phosphatases, divalent ions other than Mn2+/Mg2+ can inhibit their activity by competitively replacing the Mn2+ in the core enzyme structure [27] and Zn2+ are the most potent regulators, having comparable ionic radii with that of Mn2+. PstPc was partially inactive in the presence of 0.2 mM ZnCl2 and displayed lower activity on increasing the Zn2+-ion concentration upto 2 mM, as observed by pNPP assays. In vitro kinase assays with PknAc and PknBc in presence of Zn2+ resulted in phosphorylation of PstPc. Also, there was increase in phosphorylation of PstPc during metabolic labeling by PknA and PknB in the presence of Zn2+ added in the E. coli culture. These results indicate that in mycobacterial cell, if cytosolic Zn2+ concentration increases, it may inhibit PstP perhaps leading to its phosphorylation. In an elaborative elemental analysis, Wagner et al. have reported that during infection, intravacuolar Zn2+-ion concentration increases from 0.037 mM to 0.46 mM in macrophages infected with M. tuberculosis [53]. Although there is no report of concomitant increase in mycobacterial Zn2+-ion concentration, it can only be speculated that if these changes in vacuolar ionic concentrations alter the mycobacterial ionic profile, a condition may develop where the enzymes that respond to Zn2+ (like PstP) can be activated or deactivated.
End-product inhibition of enzymes is a well established phenomenon to prevent the accumulation of a particular metabolite. In case of reversible reactions, end-product accumulation can change the direction of the reaction. Similarly, Pi is known to inhibit a number of phosphatases [27], [42], [49] and in present study, PstPc mediated pNPP hydrolysis is inhibited by Pi. To confirm that this effect is not limited to pNPP, in vitro kinase assays and metabolic labeling in E. coli showed PstPc to be phosphorylated by PknA and PknB in presence of Pi because of its inhibition. Pi content is indicative of nutrient availability and energy status of the cell. In general, high Pi is associated with energy-starved conditions, when all the ATP is depleted and metabolite homeostasis is in unbalanced state. Such conditions usually arise during late-log and stationary phases in culture conditions.
Metabolic labeling by [32P]orthophosphoric acid in the presence of co-expressed STPK (PknA or PknB) in E. coli lead to the specific phosphorylation of PstPc and PstPc D38G. Co-expression in pETDuet-1 has previously been utilized extensively to assess the interaction of mycobacterial STPKs with their cognate substrates in the surrogate host E. coli [21], [23]. Such dual-expression systems are increasingly becoming useful for analysis of protein-protein interactions specifically for challenging systems like mycobacteria [54]. Activity assays of the pETDuet-1 purified PstPc and PstPc D38G revealed the higher activity of PknA-phosphorylated phosphatase as compared to the PknB-phosphorylated protein. Prominent variations in the activity of phosphorylated and unphosphorylated PstPc D38G were observed with phosphorylated protein being proficient to hydrolyze pNPP to a greater extent (∼15-fold higher) in contrast to the unphosphorylated protein. The difference in the activities of phosphorylated and unphosphorylated PstPc was not as prominent as that of PstPc D38G (∼2–3 fold higher). These differences may be attributed to the fact that PstPc may get auto-dephosphorylated to a greater extent than PstPc D38G during expression and purification procedures. Higher activity of phosphorylated phosphatase is suggestive of reverse regulation of signaling cascade emanating from the kinases. In the constitutively active state, STPKs perform their regular functions and phosphorylate the target substrates following the stimulus. This may ultimately lead to the phosphorylation of PstP. The resulting increase in the activity of phosphatase may itself act as a control mechanism for kinases, eventually impeding the continued effect of that particular stimulus. The overall process has to be dynamic due to auto-dephosphorylation of PstP, eventually ceasing the effect of signaling cascade. In the conditions of high Zn2+ or high Pi content of the cell, PstP may not be active and will allow the kinase to work at its maximal activity. The proposed phosphorylation of PstP in such conditions may act as a mechanism to overcome the inhibition of PstP, hence balancing the cellular signaling pathways.
Supporting Information
p NPP-assay. To confirm the authenticity of pNPP assay, increasing amounts of alkaline phosphatase (0-100 ng) was used a positive control and PknBc (0–5 µg) was used as a negative control. The assay was performed for 30 mins at 37°C and the activity is calculated as µmoles of pNPP hydrolyzed per min at a given amount of enzyme used. As clearly evident, alkaline phosphatase showed very high activity while no such activity was detected in PknBc.
(TIF)
Effect of mutations on the activity of PstPc. To show that the loss in activity of PstPc was specifically due to mutations of Arg20, Asp38 and Asp229, PstPc was mutagenized on irrelevant residues Thr5 and Thr141 to Ala and Glu, respectively and pNPP hydrolysis was performed for 30 mins at 37°C. Activity of PstPc was taken as 100% and relative activity was calculated. As evident from the bar graph, there were no significant changes in the activity of the mutants PstPc T5A and PstPc T141E as compared to PstPc.
(TIF)
Time-dependent p NPP-assay. pNPP-hydrolysis was performed in a time-dependent manner for 30 mins using PstPc, PstPc R20G, PstPc D38G and PstPc D229G variants (2 µg each) at 37°C. Alkaline phosphatase (2 ng) was used a positive control and PknBc (5 µg) was used as a negative control. Activity was calculated as nmoles of pNPP hydrolyzed per µg of enzyme used at a given time and depicted in logarithmic scale. Nevertheless, the results are essentially similar as that of time-dependent dephosphorylation of PknBc (Figure 2A).
(TIF)
In vitro dephosphorylation activity of pETDuet-1 purified PstPcD38G. Autophosphorylated PknAc was incubated with unphosphorylated and phosphorylated PstPc D38G. As shown in the autoradiogram, the PknA-phosphorylated PstPc D38G dephosphorylated the kinase to a greater extent in comparison to the unphosphorylated PstPc D38G. The image was also analyzed by ImageGauge software and corresponding values are depicted by bar-graph (Figure 4E).
(TIF)
Phosphorylation of PstPc. Autoradiogram showing the phosphorylation of PstPc (1 µg) by His6-tagged STPKs PknAc (upper panel) and PknBc (lower panel) in presence of 0.2 mM Zn2+ and 0.5 mM Pi. Due to overlapping molecular weights of PknAc and PknBc with PstPc, the bands were not separated properly. Still, the phosphotransfer on PstPc was evident in presence of Zn2+ and Pi by both the kinases. The reaction was also performed with GST-tagged STPKs to clearly depict the reaction (Figure 5D).
(TIF)
Detailed protocol of sample processing for identification of phosphorylation sites.
(DOC)
Acknowledgments
We thank Zachary Weldon (Proteomics Core Facility, Children's hospital, Boston) for providing assistance in using Mass-spectrometric analysis for identification of phosphorylation sites.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Financial support to the work was provided by Council of Scientific and Industrial Research (NWP-0038). AS is a Senior Research Fellow of University Grants Commission, India. GA and MG are Senior Research Fellows of Council of Scientific and Industrial Research, India. SU is a Junior Research Fellow of Council of Scientific and Industrial Research, India. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Echenique J, Kadioglu A, Romao S, Andrew PW, Trombe MC. Protein serine/threonine kinase StkP positively controls virulence and competence in Streptococcus pneumoniae. Infect Immun. 2004;72:2434–2437. doi: 10.1128/IAI.72.4.2434-2437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galyov EE, Hakansson S, Forsberg A, Wolf-Watz H. A secreted protein kinase of Yersinia pseudotuberculosis is an indispensable virulence determinant. Nature. 1993;361:730–732. doi: 10.1038/361730a0. [DOI] [PubMed] [Google Scholar]
- 3.Juris SJ, Rudolph AE, Huddler D, Orth K, Dixon JE. A distinctive role for the Yersinia protein kinase: actin binding, kinase activation, and cytoskeleton disruption. Proc Natl Acad Sci U S A. 2000;97:9431–9436. doi: 10.1073/pnas.170281997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, Li C, Yang H, Mushegian A, Jin S. A novel serine/threonine protein kinase homologue of Pseudomonas aeruginosa is specifically inducible within the host infection site and is required for full virulence in neutropenic mice. J Bacteriol. 1998;180:6764–6768. doi: 10.1128/jb.180.24.6764-6768.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bach H, Wong D, Av-Gay Y. Mycobacterium tuberculosis PtkA is a novel protein tyrosine kinase whose substrate is PtpA. Biochem J. 2009;420:155–160. doi: 10.1042/BJ20090478. [DOI] [PubMed] [Google Scholar]
- 6.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 7.Chao J, Wong D, Zheng X, Poirier V, Bach H, et al. Biochim Biophys Acta. Vol. 1804. 620-627; 2010. Protein kinase and phosphatase signaling in Mycobacterium tuberculosis physiology and pathogenesis. pp. 620–627. [DOI] [PubMed] [Google Scholar]
- 8.Chao JD, Papavinasasundaram KG, Zheng X, Chavez-Steenbock A, Wang X, et al. Convergence of Ser/Thr and two-component signaling to coordinate expression of the dormancy regulon in Mycobacterium tuberculosis. J Biol Chem. 2010;285:29239–29246. doi: 10.1074/jbc.M110.132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta M, Sajid A, Arora G, Tandon V, Singh Y. Forkhead-associated domain-containing protein Rv0019c and polyketide-associated protein PapA5, from substrates of serine/threonine protein kinase PknB to interacting proteins of Mycobacterium tuberculosis. J Biol Chem. 2009;284:34723–34734. doi: 10.1074/jbc.M109.058834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prisic S, Dankwa S, Schwartz D, Chou MF, Locasale JW, et al. Extensive phosphorylation with overlapping specificity by Mycobacterium tuberculosis serine/threonine protein kinases. Proc Natl Acad Sci U S A. 2010;107:7521–7526. doi: 10.1073/pnas.0913482107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arora G, Sajid A, Gupta M, Bhaduri A, Kumar P, et al. Understanding the role of PknJ in Mycobacterium tuberculosis: biochemical characterization and identification of novel substrate pyruvate kinase A. PLoS One. 2010;5:e10772. doi: 10.1371/journal.pone.0010772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boitel B, Ortiz-Lombardia M, Duran R, Pompeo F, Cole ST, et al. PknB kinase activity is regulated by phosphorylation in two Thr residues and dephosphorylation by PstP, the cognate phospho-Ser/Thr phosphatase, in Mycobacterium tuberculosis. Mol Microbiol. 2003;49:1493–1508. doi: 10.1046/j.1365-2958.2003.03657.x. [DOI] [PubMed] [Google Scholar]
- 13.Chopra P, Singh B, Singh R, Vohra R, Koul A, et al. Phosphoprotein phosphatase of Mycobacterium tuberculosis dephosphorylates serine-threonine kinases PknA and PknB. Biochem Biophys Res Commun. 2003;311:112–120. doi: 10.1016/j.bbrc.2003.09.173. [DOI] [PubMed] [Google Scholar]
- 14.Greenstein AE, MacGurn JA, Baer CE, Falick AM, Cox JS, et al. M. tuberculosis Ser/Thr protein kinase D phosphorylates an anti-anti-sigma factor homolog. PLoS Pathog. 2007;3:e49. doi: 10.1371/journal.ppat.0030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Hare HM, Duran R, Cervenansky C, Bellinzoni M, Wehenkel AM, et al. Regulation of glutamate metabolism by protein kinases in mycobacteria. Mol Microbiol. 2008;70:1408–1423. doi: 10.1111/j.1365-2958.2008.06489.x. [DOI] [PubMed] [Google Scholar]
- 16.Sharma K, Gupta M, Pathak M, Gupta N, Koul A, et al. Transcriptional control of the mycobacterial embCAB operon by PknH through a regulatory protein, EmbR, in vivo. J Bacteriol. 2006;188:2936–2944. doi: 10.1128/JB.188.8.2936-2944.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sureka K, Hossain T, Mukherjee P, Chatterjee P, Datta P, et al. Novel role of phosphorylation-dependent interaction between FtsZ and FipA in mycobacterial cell division. PLoS One. 2010;5:e8590. doi: 10.1371/journal.pone.0008590. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Pullen KE, Ng HL, Sung PY, Good MC, Smith SM, et al. An alternate conformation and a third metal in PstP/Ppp, the M. tuberculosis PP2C-Family Ser/Thr protein phosphatase. Structure. 2004;12:1947–1954. doi: 10.1016/j.str.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 19.Jackson MD, Fjeld CC, Denu JM. Probing the function of conserved residues in the serine/threonine phosphatase PP2Calpha. Biochemistry. 2003;42:8513–8521. doi: 10.1021/bi034074+. [DOI] [PubMed] [Google Scholar]
- 20.Kang CM, Abbott DW, Park ST, Dascher CC, Cantley LC, et al. The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 2005;19:1692–1704. doi: 10.1101/gad.1311105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan S, Nagarajan SN, Parikh A, Samantaray S, Singh A, et al. Phosphorylation of enoyl-acyl carrier protein reductase InhA impacts mycobacterial growth and survival. J Biol Chem. 2010;285:37860–37871. doi: 10.1074/jbc.M110.143131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyle WJ, van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- 23.Kumar P, Kumar D, Parikh A, Rananaware D, Gupta M, et al. The Mycobacterium tuberculosis protein kinase K modulates activation of transcription from the promoter of mycobacterial monooxygenase operon through phosphorylation of the transcriptional regulator VirS. J Biol Chem. 2009;284:11090–11099. doi: 10.1074/jbc.M808705200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dahche H, Abdullah A, Ben PM, Kennelly PJ. A PPM-family protein phosphatase from the thermoacidophile Thermoplasma volcanium hydrolyzes protein-bound phosphotyrosine. Extremophiles. 2009;13:371–377. doi: 10.1007/s00792-008-0211-5. [DOI] [PubMed] [Google Scholar]
- 25.Perrino BA, Wilson AJ, Ellison P, Clapp LH. Substrate selectivity and sensitivity to inhibition by FK506 and cyclosporin A of calcineurin heterodimers composed of the alpha or beta catalytic subunit. Eur J Biochem. 2002;269:3540–3548. doi: 10.1046/j.1432-1033.2002.03040.x. [DOI] [PubMed] [Google Scholar]
- 26.Sugiura T, Noguchi Y. Substrate-dependent metal preference of PPM1H, a cancer-associated protein phosphatase 2C: comparison with other family members. Biometals. 2009;22:469–477. doi: 10.1007/s10534-009-9204-9. [DOI] [PubMed] [Google Scholar]
- 27.Fjeld CC, Denu JM. Kinetic analysis of human serine/threonine protein phosphatase 2Calpha. J Biol Chem. 1999;274:20336–20343. doi: 10.1074/jbc.274.29.20336. [DOI] [PubMed] [Google Scholar]
- 28.Chen J, Martin BL, Brautigan DL. Regulation of protein serine-threonine phosphatase type-2A by tyrosine phosphorylation. Science. 1992;257:1261–1264. doi: 10.1126/science.1325671. [DOI] [PubMed] [Google Scholar]
- 29.Zhao S, Zhu Q, Somerville RL. The sigma(70) transcription factor TyrR has zinc-stimulated phosphatase activity that is inhibited by ATP and tyrosine. J Bacteriol. 2000;182:1053–1061. doi: 10.1128/jb.182.4.1053-1061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 31.Taylor WP, Widlanski TS. Charged with meaning: the structure and mechanism of phosphoprotein phosphatases. Chem Biol. 1995;2:713–718. doi: 10.1016/1074-5521(95)90098-5. [DOI] [PubMed] [Google Scholar]
- 32.Ahn JH, McAvoy T, Rakhilin SV, Nishi A, Greengard P, et al. Protein kinase A activates protein phosphatase 2A by phosphorylation of the B56delta subunit. Proc Natl Acad Sci U S A. 2007;104:2979–2984. doi: 10.1073/pnas.0611532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Awano K, Amano K, Nagaura Y, Kanno S, Echigo S, et al. Phosphorylation of protein phosphatase 2Czeta by c-Jun NH2-terminal kinase at Ser92 attenuates its phosphatase activity. Biochemistry. 2008;47:7248–7255. doi: 10.1021/bi800067p. [DOI] [PubMed] [Google Scholar]
- 34.Barford D, Das AK, Egloff MP. The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu Rev Biophys Biomol Struct. 1998;27:133–164. doi: 10.1146/annurev.biophys.27.1.133. [DOI] [PubMed] [Google Scholar]
- 35.Bukczynska P, Klingler-Hoffmann M, Mitchelhill KI, Lam MH, Ciccomancini M, et al. The T-cell protein tyrosine phosphatase is phosphorylated on Ser-304 by cyclin-dependent protein kinases in mitosis. Biochem J. 2004;380:939–949. doi: 10.1042/BJ20031780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doehn U, Gammeltoft S, Shen SH, Jensen CJ. p90 ribosomal S6 kinase 2 is associated with and dephosphorylated by protein phosphatase 2Cdelta. Biochem J. 2004;382:425–431. doi: 10.1042/BJ20040948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyahara A, Hirani TA, Oakes M, Kereszt A, Kobe B, et al. Soybean nodule autoregulation receptor kinase phosphorylates two kinase-associated protein phosphatases in vitro. J Biol Chem. 2008;283:25381–25391. doi: 10.1074/jbc.M800400200. [DOI] [PubMed] [Google Scholar]
- 38.Virshup DM, Shenolikar S. From promiscuity to precision: protein phosphatases get a makeover. Mol Cell. 2009;33:537–545. doi: 10.1016/j.molcel.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi T, Kanno S, Terasawa T, Murakami T, Ohnishi M, et al. Phosphorylation of Mg(2+)-dependent protein phosphatase alpha (type 2C alpha) by casein kinase II. Biochem Biophys Res Commun. 1993;195:484–489. doi: 10.1006/bbrc.1993.2069. [DOI] [PubMed] [Google Scholar]
- 40.van der Knaap E, Song WY, Ruan DL, Sauter M, Ronald PC, et al. Expression of a gibberellin-induced leucine-rich repeat receptor-like protein kinase in deepwater rice and its interaction with kinase-associated protein phosphatase. Plant Physiol. 1999;120:559–570. doi: 10.1104/pp.120.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adler E, Donella-Deana A, Arigoni F, Pinna LA, Stragler P. Structural relationship between a bacterial developmental protein and eukaryotic PP2C protein phosphatases. Mol Microbiol. 1997;23:57–62. doi: 10.1046/j.1365-2958.1997.1801552.x. [DOI] [PubMed] [Google Scholar]
- 42.Halbedel S, Busse J, Schmidl SR, Stulke J. Regulatory protein phosphorylation in Mycoplasma pneumoniae. A PP2C-type phosphatase serves to dephosphorylate HPr(Ser-P). J Biol Chem. 2006;281:26253–26259. doi: 10.1074/jbc.M605010200. [DOI] [PubMed] [Google Scholar]
- 43.Jan G, Delorme V, Saksouk N, Abrivard M, Gonzalez V, et al. A Toxoplasma type 2C serine-threonine phosphatase is involved in parasite growth in the mammalian host cell. Microbes Infect. 2009;11:935–945. doi: 10.1016/j.micinf.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 44.Jang J, Wang L, Jeanjean R, Zhang CC. PrpJ, a PP2C-type protein phosphatase located on the plasma membrane, is involved in heterocyst maturation in the cyanobacterium Anabaena sp. PCC 7120. Mol Microbiol. 2007;64:347–358. doi: 10.1111/j.1365-2958.2007.05654.x. [DOI] [PubMed] [Google Scholar]
- 45.Jiang L, Yang J, Fan F, Zhang D, Wang X. The Type 2C protein phosphatase FgPtc1p of the plant fungal pathogen Fusarium graminearum is involved in lithium toxicity and virulence. Mol Plant Pathol. 2010;11:277–282. doi: 10.1111/j.1364-3703.2009.00598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lammers T, Lavi S. Role of type 2C protein phosphatases in growth regulation and in cellular stress signaling. Crit Rev Biochem Mol Biol. 2007;42:437–461. doi: 10.1080/10409230701693342. [DOI] [PubMed] [Google Scholar]
- 47.Travis SM, Berger HA, Welsh MJ. Protein phosphatase 2C dephosphorylates and inactivates cystic fibrosis transmembrane conductance regulator. Proc Natl Acad Sci U S A. 1997;94:11055–11060. doi: 10.1073/pnas.94.20.11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, et al. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci U S A. 2009;106:17588–17593. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Das AK, Helps NR, Cohen PT, Barford D. Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 A resolution. EMBO J. 1996;15:6798–6809. [PMC free article] [PubMed] [Google Scholar]
- 50.Stone JM, Collinge MA, Smith RD, Horn MA, Walker JC. Interaction of a protein phosphatase with an Arabidopsis serine-threonine receptor kinase. Science. 1994;266:793–795. doi: 10.1126/science.7973632. [DOI] [PubMed] [Google Scholar]
- 51.Rantanen MK, Lehtio L, Rajagopal L, Rubens CE, Goldman A. Structure of Streptococcus agalactiae serine/threonine phosphatase. The subdomain conformation is coupled to the binding of a third metal ion. FEBS J. 2007;274:3128–3137. doi: 10.1111/j.1742-4658.2007.05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schlicker C, Fokina O, Kloft N, Grune T, Becker S, et al. Structural analysis of the PP2C phosphatase tPphA from Thermosynechococcus elongatus: a flexible flap subdomain controls access to the catalytic site. J Mol Biol. 2008;376:570–581. doi: 10.1016/j.jmb.2007.11.097. [DOI] [PubMed] [Google Scholar]
- 53.Wagner D, Maser J, Lai B, Cai Z, Barry CE, III, et al. Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell's endosomal system. J Immunol. 2005;174:1491–1500. doi: 10.4049/jimmunol.174.3.1491. [DOI] [PubMed] [Google Scholar]
- 54.Molle V, Leiba J, Zanella-Cleon I, Becchi M, Kremer L. An improved method to unravel phosphoacceptors in Ser/Thr protein kinase-phosphorylated substrates. Proteomics. 2010;10:3910–3915. doi: 10.1002/pmic.201000316. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
p NPP-assay. To confirm the authenticity of pNPP assay, increasing amounts of alkaline phosphatase (0-100 ng) was used a positive control and PknBc (0–5 µg) was used as a negative control. The assay was performed for 30 mins at 37°C and the activity is calculated as µmoles of pNPP hydrolyzed per min at a given amount of enzyme used. As clearly evident, alkaline phosphatase showed very high activity while no such activity was detected in PknBc.
(TIF)
Effect of mutations on the activity of PstPc. To show that the loss in activity of PstPc was specifically due to mutations of Arg20, Asp38 and Asp229, PstPc was mutagenized on irrelevant residues Thr5 and Thr141 to Ala and Glu, respectively and pNPP hydrolysis was performed for 30 mins at 37°C. Activity of PstPc was taken as 100% and relative activity was calculated. As evident from the bar graph, there were no significant changes in the activity of the mutants PstPc T5A and PstPc T141E as compared to PstPc.
(TIF)
Time-dependent p NPP-assay. pNPP-hydrolysis was performed in a time-dependent manner for 30 mins using PstPc, PstPc R20G, PstPc D38G and PstPc D229G variants (2 µg each) at 37°C. Alkaline phosphatase (2 ng) was used a positive control and PknBc (5 µg) was used as a negative control. Activity was calculated as nmoles of pNPP hydrolyzed per µg of enzyme used at a given time and depicted in logarithmic scale. Nevertheless, the results are essentially similar as that of time-dependent dephosphorylation of PknBc (Figure 2A).
(TIF)
In vitro dephosphorylation activity of pETDuet-1 purified PstPcD38G. Autophosphorylated PknAc was incubated with unphosphorylated and phosphorylated PstPc D38G. As shown in the autoradiogram, the PknA-phosphorylated PstPc D38G dephosphorylated the kinase to a greater extent in comparison to the unphosphorylated PstPc D38G. The image was also analyzed by ImageGauge software and corresponding values are depicted by bar-graph (Figure 4E).
(TIF)
Phosphorylation of PstPc. Autoradiogram showing the phosphorylation of PstPc (1 µg) by His6-tagged STPKs PknAc (upper panel) and PknBc (lower panel) in presence of 0.2 mM Zn2+ and 0.5 mM Pi. Due to overlapping molecular weights of PknAc and PknBc with PstPc, the bands were not separated properly. Still, the phosphotransfer on PstPc was evident in presence of Zn2+ and Pi by both the kinases. The reaction was also performed with GST-tagged STPKs to clearly depict the reaction (Figure 5D).
(TIF)
Detailed protocol of sample processing for identification of phosphorylation sites.
(DOC)