Abstract
Environmental factors (e.g., BaP) have been pointed out as one of the etiologies of pancreatic cancer. However, very limited experimental assays are available to identify pancreatic specific environmental mutagens or susceptibility genes. In this study, we have developed a simple in vitro cell culture model system that can be used to study the molecular and biochemical aspects of carcinogenesis in a near-normal immortalized pancreatic ductal epithelial cell lines. In order to demonstrate that xenobiotic stress response is intact in these cells we employed standard molecular biology techniques. For examples, luciferase reporter and/or real-time quantitative PCR assays were used to determine stress-induced CYP1A1 and CYP1B1 gene expression. Western blotting and immunocytochemistry assays were used to demonstrate that TCDD or BaP could activate AhR signaling. For exploring the carcinogenesis mechanism, we incubated cells with [3H]BaP and determined BaP-DNA binding activity by measuring its radioactivity. BaP-DNA adduct formation was further confirmed by [32P]-postlabeling assay. Finally, we demonstrated the effects of endogenous AhR or BRCA1 in BaP-DNA adduct accumulation in our cell system: As results, no apparent BaP-DNA adduct accumulation by [32P]-postlabeling assay was found in either control-siRNA or AhR-siRNA pretreated cells. On the other hand, a significant increase of BaP-DNA adduct accumulation was found in BRCA1 knockdown cells. In conclusion, we suggest that this in vitro model may provide the feasibility for future studies on the molecular basis of pancreatic ductal cell carcinogenesis caused by dietary mutagens.
Keywords: AhR, CYP1A1, pancreatic cancer, benzo(a)pyrene, TCDD, BRCA1
1. Introduction
Pancreatic cancer has the highest lethality among human malignancies. In 2008, estimated deaths from pancreatic cancers (34,290) were approximately equal to its incidence (37,680) in the United States (Jemal et al., 2008). Most pancreatic cancer patients have been diagnosed at unrespectable stages (Kim and Saif, 2007). Thus, they usually receive palliative chemotherapy (Kang and Saif, 2008), which show poor results due to drug resistance (Kim and Saif, 2007; Kang and Saif, 2008). The cause of pancreatic cancer is not clearly demonstrated as other cancers. Up to 10 % of patients have family history of pancreatic cancer (Koorstra et al., 2008). A subset of these patients harbors germline mutations harboring in K-ras (Almoguera et al., 1988), CDKN2A/p16 (Cyclin-dependent kinase inhibitor 2A) (Caldas et al., 1994), p53 (Redston et al., 1994), and BRCA2 (Breast Cancer Type 2 susceptibility protein) (Goggins et al., 1996). However, the vast majority of onsets in pancreatic cancer has been attributed to environmental factors such as age, cigarette smoking (Silverman et al., 1994; Lin et al., 2002), diet pattern and obesity (Michaud et al., 2001; Michaud et al., 2002), and diabetes mellitus type II (Huxley et al., 2005).
So far, smoking and dietary mutagens are well characterized and consistently implicated in epidemiological studies (Silverman et al., 1994). Lowenfels and Maisonneuve (Lowenfels and Maisonneuve, 2006) estimated that smoking causes about 25 % of all pancreatic cancer. Cigarette smoke contains lots of carcinogenic chemicals including dioxins (e.g., polychlorinated dibenzodioxins (PCDDs) and Benzo(a)pyrene (BaP)), which are present in a wide range of materials including plastics, resins, or bleaches (Agency for Toxic Substances and Disease Registry, 1998). The most toxic dioxin, 2,3,7,8-tetrachlorodibenzodioxin (TCDD), became well known as a contaminant of Agent Orange (2,4,5-Trichlorophenoxyacetic acid) used in the Vietnam War. In addition, high consumption of smoked meat is a source of carcinogenic heterocyclic amines (HCAs) and polycyclic aromatic hydrocarbons (PAHs) (Skog, 1993; Sugimura et al., 1994).
Presumably, humans have evolved to be able to detoxify a wide range of xenobiotic chemicals. Biotransformation and elimination of xenobiotics are processed through the set of metabolic pathways such as phase I, II and III (Xu et al., 2005). In phase I, enzymes such as cytochrome P450 oxidases (CYPs) introduce reactive or polar groups into xenobiotics. These modified compounds are then conjugated to polar compounds in phase II and excreted by phase III enzymes (Denison and Nagy, 2003). Aryl hydrocarbon Receptor (AhR) is a main transcription factor that regulates Phase I gene expression. It resides in cytosol and normally inactive, bound to several co-chaperones. Upon AhR binding to chemicals such as TCDD and BaP, the chaperones dissociate resulting in dimerization of AhR with AhR nuclear translocator (ARNT) (Hankinson, 1995). The AhR/ARNT heterodimeric complex, then translocates into the nucleus, where it interacts with DNA by binding to recognition sequences, referred to xenobiotic-responsive element (XRE), in the promoter region of AhR responsive genes. The activation of AhR leads to expression of various genes encoding Phase I enzymes such as CYP1A1 (cytochrome P450, family 1, subfamily A, polypeptide 1), and CYP1B1, and Phase II enzymes such as NQO1 (NAD(P)H dehydrogenase, quinone 1), and UGT1A2 (UDP glucuronosyltransferase 1 family, polypeptide A2) (Hankinson, 1995).
AhR is expressed ubiquitously in human and rodent systemic organs including the pancreas (Dolwick et al., 1993, Yamamoto et al., 2004). There are several lines of evidence showing that overexpression of AhR promotes tumorigenesis. Transgenic mice, which express constitutively active AhR, spontaneously develop hepatocarcinoma (Moennikes et al., 2004) and stomach cancer (Andersson et al., 2002). Whereas AhR-knockout has increased resistance to carcinogens (Shimada et al., 2002) and knockdown of AhRR, AhR repressor, promotes tumor cell growth (Zudaire et al., 2008). Therefore, identification of environmental risk factors and understanding the mechanism of AhR activation in the pancreas is essential for elucidating etiology of pancreatic cancer.
Immortalized but not transformed human pancreatic ductal epithelial (HPDE) cell lines were developed by expressing human papilloma virus (HPV)-16 E6E7 genes in normal human pancreatic epithelial cells as an in vitro model system (Furukawa et al., 1996). Although these cells have been altered by viral gene products, subsequently isolated clones such as HPDE6-C7 and HPDE6-C11 showed near normal genotypic and phenotypic characteristics of normal human pancreatic cells including anchorage-dependent growth requirement and nontumorigenic in immune-deficient mice (Ouyang et al., 2000). Moreover, tumorigenicity of HPDE6-C7 cells in SCID mice need to further transformation by oncogenic K-ras protein (Qian et al., 2005).
In this study, we are reporting these two non-transformed human pancreatic cell lines as in vitro models that are useful tools in identifying genes that are susceptible to environmental stress. In addition, these cell lines will be useful in identifying environmental factors and susceptible genes in normal pancreas.
2. Materials and Methods
2.1. Cell culture and chemicals
Human pancreatic ductal epithelial (HPDE) cells, HPDE6-C7 and HPDE6-C11 from Dr. Tsao (Furukawa et al., 1996), were cultured in keratinocyte serum-free (KSF) medium supplemented by an epidermal growth factor and bovine pituitary extract (Invitrogen) and 1X Antibiotic-Antimycotic (Invitrogen). 3-Methylcholanthrene (3MC), indole-3-carbinol (I3C) and BaP were purchased from Sigma and dissolved in dimethyl sulfoxide (DMSO). TCDD was obtained from Ultra Scientific (N. Kingstown, RI). Radio-labeled [3H]BaP was purchased from American Radiolabeled Chemicals, Inc (St. Louis, MO) and [γ-32P] ATP was obtained from Perkin Elmer, Inc (Waltham, MA).
2.2. Reporter gene assay
Cells (5×104 cells/well) were seeded in 24-well plates and transfected overnight with reporter plasmids, p(XRE.1A1)-Luc (125 ng/well) (Kang et al., 2006(a)), with or without human AhRR expression plasmid (pCDNA3-AhRR Δexon) (Karchner et al., 2009) or fish AhRR expression plasmid (pCDNA3-FAhRR) (Watanabe et al., 2001) using Lipofectamine Plus (Invitrogen). Next, cells were further incubated for 24 hr with various chemicals as indicated and lysed in a reporter lysis buffer (Promega, Madison, WI). Luciferase activity was measured using a luminometer and the luminescence signals were normalized for relative transfection efficiency by measuring β-galactosidase activity of a co-transfected reporter plasmid (Kang et al., 2006(a); Kang et al., 2006(b)).
2.3. Real-time PCR
HPDE6 cells incubated with 10 nM of TCDD or 5 μM of BaP for indicated times were harvested and RNAs were purified with Trizol solution (Invitrogen). Reverse transcription (RT) was performed using Superscript II reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. Quantitative real-time PCR was performed as previously described (Kang et al., 2006(a)). Reactions were performed in tetraplicate using the 1x TaqMan Universal PCR Master Mix (Roche Applied Science), on an Applied Biosystems-Prism Sequence Detector System 7700 and analyzed with SDS software. The mRNA level of GAPDH was also determined for each RNA sample and was used for normalization. The following sequences were used for primer: 5′-ctt gga cct ctt tgg agc tg-3′ (forward) and 5′-cga agg aag agt gtc gga ag-3′ (reverse) for CYP1A1 (NM_000499, +933 ~ +1144); 5′-cac caa ggc tga gac agt ga-3′ (forward) and 5′-gat gac gac tgg gcc tac at-3′ (reverse) for CYP1B1 (NM_000104, +3837 ~ +4000); 5′-gta tga caa cga att tgg cta cag -3′ (forward) and 5′-agc aca ggg tac ttt att gat ggt -3′ (reverse) for GAPDH.
2.4. Western blotting
Cells were cultured for 24 hr, treated with TCDD or BaP, lysed and centrifuged for total protein analysis. Cytosolic/nuclear fraction and Standard Western blots were prepared and analyzed as described previously (Kang et al., 2006(b)). Primary antibodies used in this experiment are as follows: rabbit polyclonal anti-BRCA1 (C-20, Santa Cruz Biotechnology, Inc), rabbit polyclonal anti-AhR (H-211, Santa Cruz Biotechnology, Inc), rabbit polyclonal anti-CYP1A1 (Affinity BioReagents, Golden, CO), rabbit polyclonal anti-Lamin B1 (Abcam, Cambridge, MA), mouse monoclonal anti-β-tubulin (Sigma), mouse monoclonal anti-β-actin (Sigma). Anti-mouse and -rabbit IgG–peroxidase antibody produced in goat (Sigma) were used for secondary antibody and ECL solution (Santa Cruz Biotechnology, Inc) was used for detection.
2.5. Immunocytochemistry assay
Immunofluorescence staining was used to identify the changes of cellular localization of AhR. Cells cultured on coverslips were incubated with 10 nM TCDD for indicated times then fixed with 4 % paraformaldehyde for 15 min. Cells were permeabilized with incubation of 0.1 % Triton X-100 for 5 min and rinsed three times with PBS containing 0.5 % BSA. Then they were incubated with rabbit anti-AhR antibody (H-211, Santa Cruz Biotechnology, Inc) for 3 hr and washed with PBS three times and further incubated with Alexa-fluor 488-conjugated goat anti-rabbit IgG (Molecular Probes, Eugene, OR) for 1 hr. Fluorescence images were taken with a fluorescence microscope (Carl Zeiss, Berlin, Germany).
2.6. Ethoxyresorufin-O-deethylation activity
CYP1A1 enzymatic activity was measured using a CYP1A1 ethoxyresorufin-O-deethylation (EROD) activity kit (IKZUS Environment, Italy) according to the procedures provided by manufacturer. Briefly, HPDE6 cells treated with various doses of BaP and TCDD for 24 hr, and then cells were washed twice with PBS and incubated with reaction buffers, containing 5 μM of 7-ethoxyresorufin and 10 μM of dicumarol for 30 min at 37 °C. Fluorescence was measured every 10 min for 60 min at 37 °C with an Ultra 384 fluorometry (Tecan, Männedorf, Switzerland) using 535 nm excitation and 590 nm emission filter as previously described (Burke and Mayer, 1974; Kang et al., 2008(b)).
2.7. [3H]BaP-DNA binding assay
Cells (HPDE6) were transfected with 100 nM of each siRNA (Dharmacon) using Lipofectamine 2000 (Invitrogen) for 72 hr (Bae et al., 2004). The sequence of each siRNAs are as follows: control, 5′-GAC GAG CGG CAC GUG CAC A-3′; AhR, 5′-UAC UCC CAC CUC AGU UGG C-3′; BRCA1, 5′-UGA UAA AGC UCC AGC AGG A -3′. Then cells were treated with [3H]BaP for 24 hr. Genomic DNA was isolated using Wizard SV Genomic DNA purification system (Promega, Madison, WI). The purity and amount of genomic DNA were measured using spectrophotometer at absorbance 260 nm and 280 nm. The radioactivity of [3H]BaP-DNA adduct in equal amount of DNA was counted using Beckman Coulter liquid scintillation counter LS6500 (Fullerton, CA). Each radioactive value was normalized with [3H]BaP non-treated (background) value, and represented as pmole of [3H]BaP per mg of DNA.
2.8. [32P]-Postlabeling assay
Cells (HPDE6) transfected with siRNA (control-siRNA, AhR-siRNA, or BRCA1-siRNA) for 72 hr were incubated with BaP (5 μM) for 24 hr. Isolated genomic DNA (10 μg) was incubated with a digestion mixture [10 units/ml micrococcal nuclease, 1.1 unit/ml spleen phosphodiesterase, 25 mM CaCl2, and 75 mM sodium succinate (pH 6.0)] at 37°C for 4 hr. After digestion, 50 mM ammonium formate (pH 7.1) was added. The SPE column (Varian, Inc., Harbor City, CA) was equilibrated with methanol. After the sample was loaded onto the column, it was washed with 50 mM ammonium formate (pH 7.1). The DNA-adducts was eluted with 50 % methanol, and were completely dried using SpeedVac. Then nuclease P1 mixture [2 μg/μl nuclease P1, 0.15 mM zinc chloride, 62.5 mM sodium acetate (pH 5.0)] was added. After 1 hr incubation at 37 °C, 500 mM Tris base was added and reaction mixture was dried. The adducts were labeled with [γ-32P]-ATP in the mixture [[γ-32P]ATP (10 μCi/μl), T4 polynucleotide kinase, and apyrase] at 37 °C for 30 min. The labeled adduct was spotted on TLC sheet and developed as follows: The BaP-DNA adduct developed in; D1 (bottom to top) in 1.7 M NaH2PO4 (pH 5.8) for overnight; D3 (bottom to top) in 3.4 M lithium formate, 6.4 M urea (pH 3.5) for 4 hr; D4 (left to right) in 0.8 M NaH2PO4, 0.5 M Tris-HCl, 7.5 M urea (pH 3.5) for 4 hr (Pan et al., 2006). After development, the TLC was dried, exposed the X-ray film.
3. Results
3.1. Various xenobiotic stressors induce Xenobiotic stress-responsible genes, CYP1A1 and CYP1B1
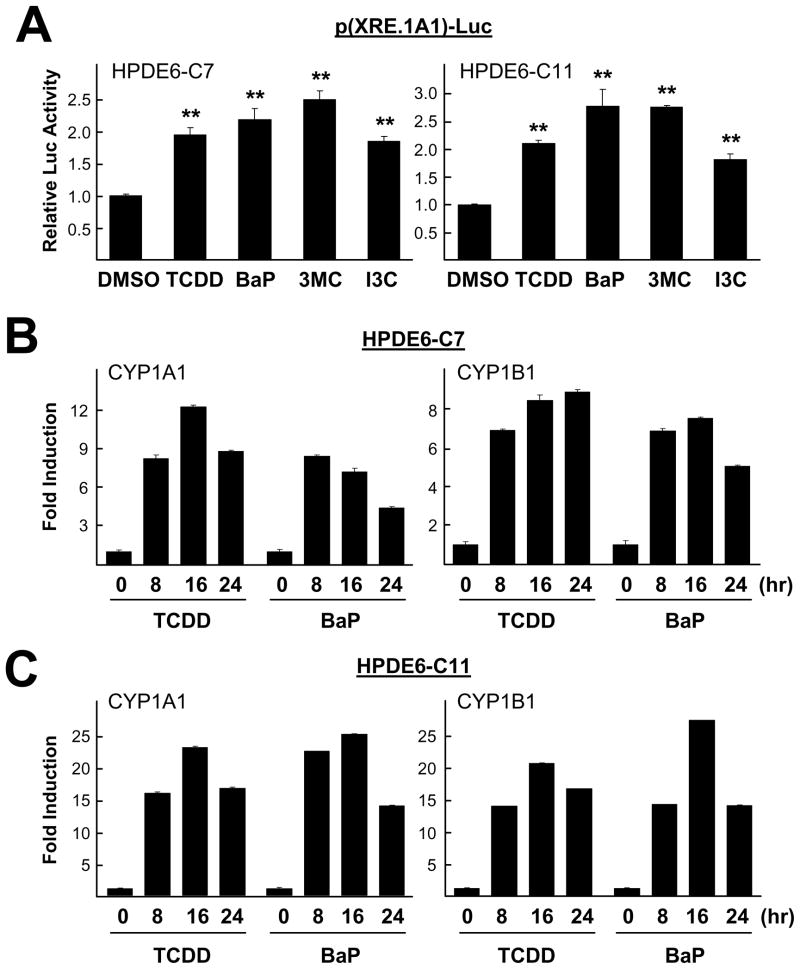
In order to investigate whether immortalized near-normal pancreatic cells respond to various xenobiotic mutagens, we examined expression of xenobiotic stress inducible genes in both HPDE6-C7 and HPDE6-C11 cells. First, cells transfected with p(XRE.1A1)-Luc reporter overnight were treated with xenobitics including 3MC, BaP and TCDD and a chemopreventive agent (I3C), respectively, for 24 hr. All these chemicals are known to stimulate AhR signaling in other tissues or cell types (e.g., breast cells). Luciferase reporter assay results showed that TCDD (10 nM), BaP (5 μM), 3MC (1 μM), and I3C (50 μM) significantly increased p(XRE.1A1)-Luc reporter activity in both cell lines, compared with DMSO-treated samples (as a vehicle control) (Fig. 1A). Total RNAs were extracted from cells treated with either 10 nM TCDD or 5 μM BaP for 0, 8, 16 and 24 hr and were used for real-time PCR as described in Materials and Methods. Both TCDD and BaP significantly increased the expression of CYP1A1 and CYP1B1 mRNA in both HPDE6-C7 (Fig. 1B) and HPDE6-C11 (Fig. 1C) cells.
Fig. 1.
Various xenobiotic stresses induced CYP1A1 gene expression in pancreatic cell lines. (A) Cells (HPDE6-C7 and HPDE6-C11) transfected with p(XRE.1A1)-Luc for overnight were treated with various AhR ligands (10 nM TCDD, 5 μM BaP, 1 μM 3MC, and 50 μM I3C) for 24 hr were harvested to measure luciferase activity. All experiments were performed three times. The two-tailed Student t test was used for statistical analysis and the symbol (**, P < 0.01) in the figure indicates statistically significant differences. Total RNA was extracted from (B) HPDE6-C7 and (C) HPDE6-C11 cells treated with 10 nM TCDD or 5 μM BaP for indicated times. Expression level of CYP1A1 and CYP1B1 mRNA were compared using real-time PCR.
3.2. Xenobiotics induce AhR activation and nuclear translocation within short time exposure
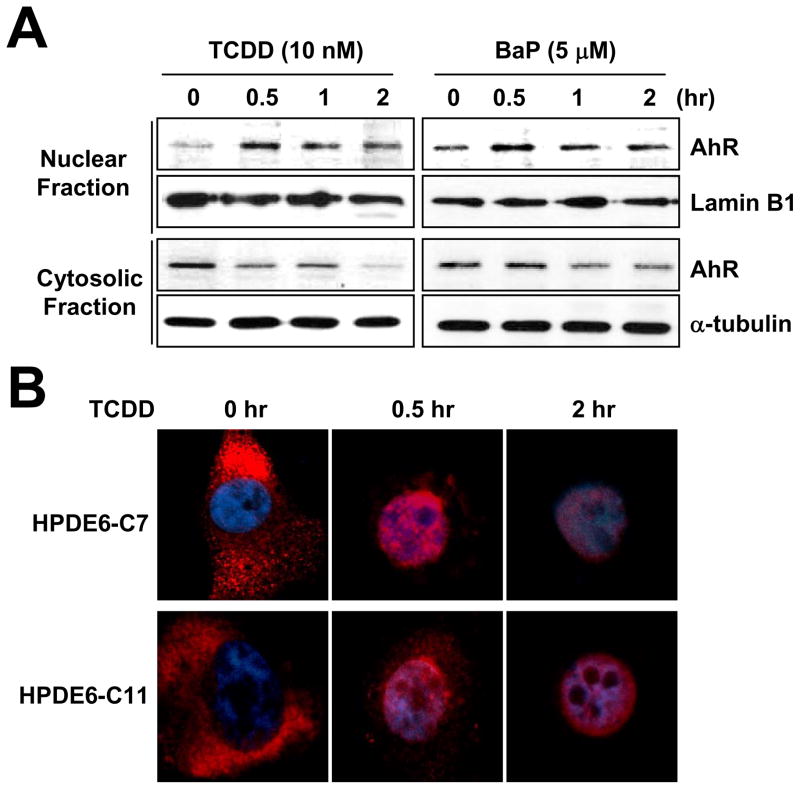
Since activated AhR translocates from the cytosol into the nucleus under xenobiotic stress (Hankinson, 1995), we investigated the localization of AhR following treatment of either TCDD or BaP. Western blotting analysis of fractionated lysates from HPDE6-C7 cells revealed that cytosolic AhR rapidly disappeared and nuclear AhR accumulated upon TCDD treatment. Similarly, incubation of 5 μM BaP also stimulated the AhR nuclear translocalization within 30 min (Fig. 2A). We also found that the cytosolic AhR rapidly translocalized into nucleus upon TCDD treatment, which was determined by an immunocytochemistry assay (Fig. 2B).
Fig. 2.
PAH induced nuclear accumulation of AhR. (A) HPDE6-C7 cells were incubated with 10 nM TCDD or 5 μM BaP for indicated times then they were harvested and fractionated into nuclear and cytosolic fraction. Lamin B1 for nuclear fraction control and β-tubulin for cytosolic fraction control were used. (B) Pancreatic normal cell lines were treated with 10 nM TCDD for indicated times then they were fixed and stained with anti-AhR antibody (red) and DAPI (blue).
3.3. TCDD and BaP induced CYP1A1 enzymatic activity in immortalized pancreatic cell lines
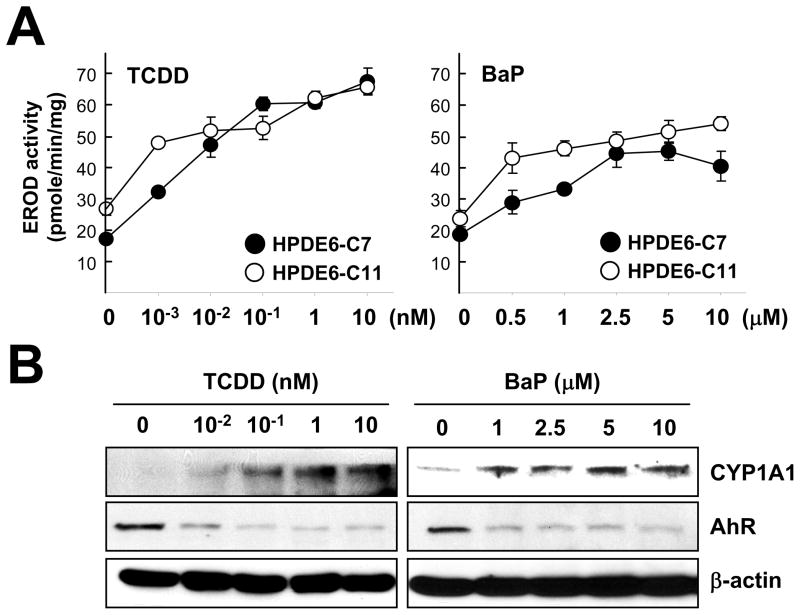
To determine whether the mRNA induction of CYP1A1 by TCDD or BaP led to increasing enzyme activity, we measured its activity (EROD activity) in cells treated with different dose of TCDD or BaP (Kang et al., 2008(b)). We found that either TCDD or BaP increased the EROD activity in a dose-dependent manner (Fig. 3A) in both HPDE6-C7 and HPDE6-C11 cell lines. Western blotting further confirmed that elevated EROD enzyme activity was correlated with increased protein amounts of CYP1A1 in HPDE6-C7 cells (Fig. 3B). Decreased amounts of AhR protein were observed after treatment of TCDD or BaP, which may be due to the AhR activation-coupled proteasome-mediated protein degradation (Davarinos and Pollenz, 1999).
Fig. 3.
Exposure to xenobiotic stress increased CYP1A1 activity. (A) Pancreatic cells were incubated with indicated concentration of TCDD and BaP for 24 hr then EROD activity was measured as described in materials and methods. (B) HPDE6-C7 cells exposed to TCDD and BaP for 24 hr were harvested and protein levels were measured with standard Western blotting using anti-CYP1A1 and anti-AhR antibodies. β-Actin was used as a loading and transfer control.
3.4. AhR inhibitors, AhRR and FAhRR, reduced AhR activity
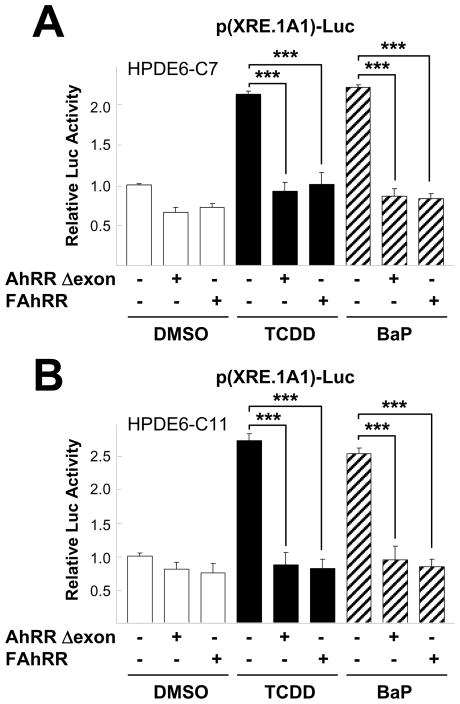
As AhR activity is regulated by its repressor, AhRR (AhR repressor), we investigated whether AhRR reduce AhR activity in non-transformaed pancreatic cell lines. Reporter gene assays showed that incubation of TCDD or BaP significantly induced p(XRE.1A1)-Luc reporter activity in both pCDNA transfected HPDE6-C7 (Fig. 4A) and HPDE6-C11 cells (Fig. 4B). As expected, when cells were co-transfected with AhRR Δexon or FAhRR, the reporter activity following TCDD and BaP treatment was significantly repressed in these cell lines (Fig. 4).
Fig. 4.
AhRR reduced TCDD or BaP-stimulated p(XRE.1A1)-Luc reporter activity. Pancreatic cells, HPDE6-C7 (A) and HPDE6-C11 (B), transfected with pCDNA3, AhRR Δexon, or FAhRR as well as p(XRE.1A1)-Luc for overnight were further incubated with 10 nM TCDD or 5 μM BaP for 24 hr then harvested to measure luciferase activity. All experiments were performed three times. The two-tailed Student t test was used for statistical analysis and the symbol (***, P < 0.001) in the figure indicates statistically significant differences.
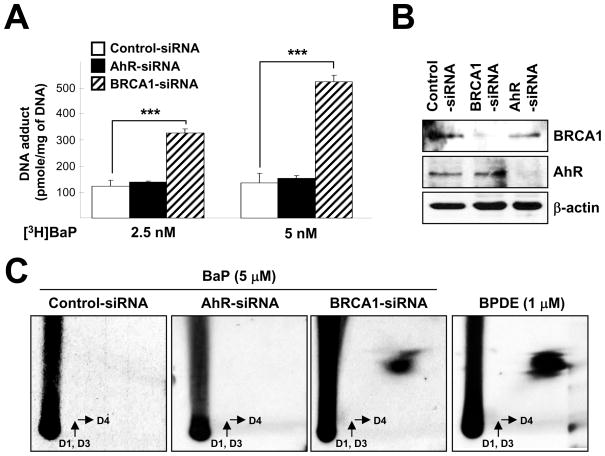
3.5. [3H]BaP-DNA adducts accumulation is AhR-independent
Isotope labeled-[3H]BaP has been used to determine the amount of DNA-bound BaP as previously described (Kang and Lee, 2005). In this study, cells pretreated with siRNA (control vs AhR) for 72 hr were incubated with 2.5 or 5 nM of [3H]BaP for an additional 24 hr before their genomic DNA were isolated. We found that AhR-siRNA did not significantly affect BaP-DNA binding activity (e.g., DNA adduct formation) in HPDE6-C7 cell (Fig. 5A). [32P]-postlabeling assay also showed that there is no detectable BPDE level in both control- and AhR-siRNA pretreated cells (Fig. 5C). The knockdown efficiency was confirmed by Western blotting (Fig. 5B).
Fig. 5.
BaP-DNA adduct accumulation in pancreatic ductal cells. (A) HPDE6-C7 cells were transfected with control, AhR, and BRCA1-specific siRNA for 72 hr and further incubated with 2.5 or 5 nM [3H]BaP for 24 hr. Then DNA incorporated [3H]BaP were measured from purified genomic DNA using a liquid scintillation counter. (B) Efficiency of knockdown was measured by standard Western blotting. (C) HPDE6-C7 cells were transfected with control, AhR, and BRCA1-specific siRNA for 72 hr and further incubated with 5 μM BaP for 24 h. Then post-labeled with [γ-32P]ATP and [32P] labeled DNA adduct was detected as described in material and methods. 1 μM of BPDE treated HPDE6-C7 cells were used as positive control for DNA-BPDE adduct spot. ***, p < 0.001
3.6. BRCA1 knockdown increases [3H]BaP-DNA adduct accumulation
Mutations of BRCA1 or loss of BRCA1 expression can confer an increased risk for breast and ovarian cancers (Miki et al., 1994). However, increased risk for other human cancer types such as pancreas or stomach in BRCA1 defective patients has also been reported (Friedenson, 2005; Lynch et al., 2005; Mai et al., 2009). Thus, we investigated the effects of BRCA1 knockdown in BaP-DNA adduct accumulation in our model system. Cells pretreated with siRNA (control vs BRCA1) for 72 hr were incubated with various doses of [3H]BaP for 24 hr. Interestingly, BaP-DNA adduct formation was markedly increased in BRCA1 knockdown compared to control or AhR knockdown (Fig. 5A). We further confirmed the increased BaP-DNA adduct formation by [32P]-postlabeling method (Fig. 5C). In general, [32P]-postlabeling method results in much greater sensitivity of detection than [3H] incorporation method (Phillips and Arlt, 2007). Western blot analysis showed the level of endogenous BRCA1 in BRCA1-siRNA-transfected HPDE6 cells (Fig. 5B).
4. Discussion
In this study, we demonstrated that the two immortalized near-normal pancreatic ductal cell lines, HPDE6-C7 and HPDE6-C11, have a functional AhR signaling pathway, which responds to the environmental carcinogens such as BaP and TCDD. In addition, incubation of BaP and TCDD substantially increased CYP1A1 expression (mRNA and protein) and its enzymatic activity in the immortalized pancreatic cells. In in vitro, CYP1A1 expression has been extensively studied and its induction has been considered as pro-metabolic activation (Whitlock, 1999). However, a recent study on Cyp1a1 knockout mouse using BaP as a xenobiotic agent suggested that Cyp1a1’s function in detoxification can be more important than in metabolic activation (Uno et al., 2004).
There are few experimental reports that elucidate the cytotoxicities of dietary or environmental mutagens in the pancreas, which may be due to lack of an appropriate experimental model. In the rat model of National Toxicology Program study, TCDD and other mutagens induced acinar-cell vacuolation and inflammation occurred at a high incidence (National Toxicology Program, 2006), however, no biochemical mechanism has been obtained from that study. Thus, our approach using newly developed in vitro cell culture model may provide an effective and simple tool to study the molecular and biochemical aspect of carcinogenesis in normal pancreatic ductal cells. In this experiment, we used both radio-labeled [3H]BaP and [32P]-postlabeling assay to measure the DNA adduct formation in an in vitro culture system. Measuring DNA adduct after BaP incubation can be applied to determining toxicity of other dietary mutagens in vitro. In order to test whether the accumulation of BaP-DNA adduct in HPDE6-C7 cell line is derived by AhR signaling pathway, cells pretreated with AhR-siRNA were challenged with radio-labeled BaP. We found that either the presence or absence of endogenous AhR did not significantly affect the BaP-DNA adduct accumulation in our in vitro system. Although previous studies show that carcinogenicity of BaP was lost in the absence of AhR (Shimizu et al., 2000), the level of BaP-DNA adduct depends on multiple steps, such as metabolic activation by phase I enzymes, detoxification of BaP metabolites by both phase I and phase II enzymes, and the rate of repair of BaP–DNA adducts (Arlt et al., 2008; Uno et al., 2004). Several CYPs, which are transcriptionally modulated by AhR, are involved in activation of BaP and DNA adduct formation. However, constitutive expression of these CYPs in several tissues of AhR null mice was reported (Kondraganti et al., 2003; Shimizu et al., 2000). Therefore, our results suggest that further transcriptional and/or translational regulation of phase I and II enzymes may exist in human pancreatic cells during BaP metabolism. Actually, multiple nuclear receptors are involved in transcriptional regulation of xenobiotic metabolizing enzymes (Nakata et al., 2006).
To demonstrate that our assay system, including manipulation of gene expression levels (knockdown of target genes) plus [3H]BaP-DNA binding assay, can be used to identify environmental stress susceptible genes, we employed BRCA1 as a model. Loss of heterozygosity (LOH) at the BRCA1 locus or its mutations has been reported in pancreatic cancer (Al-Sukhni et al., 2008). A recent study shows decreased expression level of BRCA1 in multiple chronic pancreatis and pancreatic cancer tissues (Beger et al., 2004). However, whether or how the alteration in BRCA1 affects pancreatic carcinogenesis has not been well documented. BRCA1 has been demonstrated to have function in DNA damage repair of double strand breaks, ubiquitination and transcriptional regulation (Starita and Parvin, 2003). Our group has previously reported that BRCA1 modulates a range of expression of genes induced by various environmental stressors including xenobiotics or oxidative stressors (Bae et al., 2004; Kang et al., 2006(a); Kang et al., 2006(b); Kang et al., 2008(a); Kang et al., 2008(b)). In this study, we demonstrated increased accumulation of the BaP-DNA adducts in the BRCA1-siRNA treated cells, which implicates that loss of BRCA1 or its expression may confer sensitivity to environmental stresses such as BaP. However, since BPDE-induced DNA damage is reported to be repaired by nucleotide excision repair (NER) (Motykiewicz et al., 2002), and BRCA1 has a function in DNA repair, specifically, the NER pathway following UV irradiation (Hartman and Ford, 2002), we cannot exclude the possible involvement of the impaired NER pathway by BRCA1-knockdown in increasing BaP-DNA adducts in our cell culture system. Nevertheless, our recent study by pulse-chase experiments showed that BaP-DNA adducts accumulation was removed at the similar rate in both control-siRNA and BRCA1-siRNA transfected human normal mammary cells (paper in preparation). Therefore, our data implicate that the function of BRCA1 in detoxification by regulation of Phase I and Phase II gene expression could be more important in BaP-induced DNA adduct accumulation and protection in these cells (paper in preparation). Further investigation is needed to understand the role of BRCA1 function in pancreatic carcinogenesis. Taken together, our in vitro system can be useful for further understanding of pancreatic carcinogenesis and for revealing the gene-environment interaction.
Acknowledgments
We appreciate BioMedText, Inc./Dr. Rashmi Nemade for helpful discussions and editing. We also appreciate to Dr. Ming-Sound Tsao at Ontario Cancer Institute/Princess Margaret Hospital, who kindly provided us HPDE6-C7 and HPDE6-C11 cell lines. Dr. Bae has been partially supported by National Institute of Health (1R03CA152530), by Susan G Komen for the Cure (FAS0703858), and by R31-10069 (WCU program) through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology.
Abbreviation
- CYP1A1
cytochrome P450, family 1, subfamily A, polypeptide 1
- CYP1B1
cytochrome P450, family 1, subfamily B, polypeptide 1
- AhR
aryl hydrocarbon receptor
- TCDD
2,3,7,8-Tetrachlorodibenzodioxin
- BaP
Benzo(a)pyrene
- AhRR
aryl-hydrocarbon receptor repressor
- BRCA1
breast cancer susceptibility gene 1
Footnotes
Conflict of Interest
There is no conflict of interest for all authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agency for Toxic Substances and Disease Registry, Division of Toxicology/Toxicology Information Branch. Toxicological Profile for Chlorinated Dibenzo-p-Dioxins. Public Health Service, U.S. Department of Health and Human Services; Atlanta, GA: 1998. [PubMed] [Google Scholar]
- Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53:549–554. doi: 10.1016/0092-8674(88)90571-5. [DOI] [PubMed] [Google Scholar]
- Al-Sukhni W, Rothenmund H, Eppel Borgida A, Zogopoulos G, O’Shea AM, Pollett A, Gallinger S. Germline BRCA1 mutations predispose to pancreatic adenocarcinoma. Hum Genet. 2008;124:271–278. doi: 10.1007/s00439-008-0554-0. [DOI] [PubMed] [Google Scholar]
- Andersson P, McGuire J, Rubio C, Gradin K, Whitelaw ML, Pettersson S, Hanberg A, Poellinger L. A constitutively active dioxin/aryl hydrocarbon receptor induces stomach tumors. Proc Natl Acad Sci USA. 2002;99:9990–9995. doi: 10.1073/pnas.152706299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt VM, Stiborová M, Henderson CJ, Thiemann M, Frei E, Aimová D, Singh R, Gamboa da Costa G, Schmitz OJ, Farmer PB, Wolf CR, Phillips DH. Metabolic activation of benzo[a]pyrene in vitro by hepatic cytochrome P450 contrasts with detoxification in vivo: experiments with hepatic cytochrome P450 reductase null mice. Carcinogenesis. 2008;29:656–665. doi: 10.1093/carcin/bgn002. [DOI] [PubMed] [Google Scholar]
- Bae I, Fan S, Meng Q, Rih JK, Kim HJ, Kang HJ, Xu J, Goldberg ID, Jaiswal AK, Rosen EM. BRCA1 induces antioxidant gene expression and resistance to oxidative stress. Cancer Res. 2004;64:7893–7909. doi: 10.1158/0008-5472.CAN-04-1119. [DOI] [PubMed] [Google Scholar]
- Beger C, Ramadani M, Meyer S, Leder G, Krüger M, Welte K, Gansauge F, Beger HG. Down-regulation of BRCA1 in chronic pancreatitis and sporadic pancreatic adenocarcinoma. Clin Cancer Res. 2004;10:3780–3787. doi: 10.1158/1078-0432.CCR-0992-3. [DOI] [PubMed] [Google Scholar]
- Burke MD, Mayer RT. Ethoxyresorufin: direct fluorimetric assay of a microsomal O-dealkylation, which is preferentially inducible by 3-methylcholanthrene. Drug Metab Dispos. 1974;2:583–588. [PubMed] [Google Scholar]
- Caldas C, Hahn SA, da Costa LT, Redston MS, Schutte M, Seymour AB, Weinstein CL, Hruban RH, Yeo CJ, Kern SE. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat Genet. 1994;8:27–32. doi: 10.1038/ng0994-27. [DOI] [PubMed] [Google Scholar]
- Davarinos NA, Pollenz RS. Aryl hydrocarbon receptor imported into the nucleus following ligand binding is rapidly degraded via the cytosplasmic proteasome following nuclear export. J Biol Chem. 1999;274:28708–28715. doi: 10.1074/jbc.274.40.28708. [DOI] [PubMed] [Google Scholar]
- Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol. 2003;43:309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- Dolwick KM, Schmidt JV, Carver LA, Swanson HI, Bradfield CA. Cloning and expression of a human Ah receptor cDNA. Mol Pharmacol. 1993;44:911–917. [PubMed] [Google Scholar]
- Friedenson B. BRCA1 and BRCA2 pathways and the risk of cancers other than breast or ovarian. MedGenMed. 2005;7:60. [PMC free article] [PubMed] [Google Scholar]
- Furukawa T, Duguid WP, Rosenberg L, Viallet J, Galloway DA, Tsao MS. Long term culture and immortalization of epithelial cells from normal adult human pancreatic ducts by transfection with the E6E7 gene of human papilloma virus 16. Am J Pathol. 1996;148:1763–1770. [PMC free article] [PubMed] [Google Scholar]
- Goggins M, Schutte M, Lu J, Weinstein CL, Petersen GM, Yeo CJ, Jackson CE, Lynch HT, Hruban RH, Kern SE. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res. 1996;56:5360–5364. [PubMed] [Google Scholar]
- Hankinson O. The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- Hartman AR, Ford JM. BRCA1 induces DNA damage recognition factors and enhances nucleotide excision repair. Nat Genet. 2002;32:180–184. doi: 10.1038/ng953. [DOI] [PubMed] [Google Scholar]
- Huxley R, Ansary-Moghaddam A, Berrington de González A, Barzi F, Woodward M. Type-II diabetes and pancreatic cancer: a meta-analysis of 36 studies. Br J Cancer. 2005;92:2076–2083. doi: 10.1038/sj.bjc.6602619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer Statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kim HJ, Cho CH, Hu Y, Li R, Bae I. BRCA1 transcriptional activity is enhanced by interactions between its AD1 domain and AhR. Cancer Chemother Pharmacol. 2008a;62:965–975. doi: 10.1007/s00280-008-0686-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kim HJ, Kim SK, Barouki R, Cho CH, Khanna KK, Rosen EM, Bae I. BRCA1 modulates xenobiotic stressinducible gene expression by interacting with ARNT in human breast cancer cells. J Biol Chem. 2006a;281:14654–14662. doi: 10.1074/jbc.M601613200. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kim HJ, Kwon SH, Kang BD, Eling TE, Lee SH, Bae I. BRCA1 modulates sensitivity to 5F-203 by regulating xenobiotic stress-inducible protein levels and EROD activity. Cancer Chemother Pharmacol. 2008b;62:689–697. doi: 10.1007/s00280-007-0657-7. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kim HJ, Rih JK, Mattson TL, Kim KW, Cho CH, Isaacs JS, Bae I. BRCA1 plays a role in the hypoxic response by regulating HIF-1alpha stability and by modulating vascular endothelial growth factor expression. J Biol Chem. 2006b;281:13047–13056. doi: 10.1074/jbc.M513033200. [DOI] [PubMed] [Google Scholar]
- Kang SC, Lee BM. Effect of estrogen receptor (ER) on benzo[a]pyrene-DNA adduct formation in human breast cancer cells. J Toxicol Environ Health A. 2005;68:1833–1840. doi: 10.1080/15287390500182883. [DOI] [PubMed] [Google Scholar]
- Kang SP, Saif MW. Pharmacogenomics and pancreatic cancer treatment. Optimizing current therapy and individualizing future therapy. JOP. 2008;9:251–266. [PubMed] [Google Scholar]
- Karchner SI, Jenny MJ, Tarrant AM, Evans BR, Kang HJ, Bae I, Sherr DH, Hahn ME. The active form of human aryl hydrocarbon receptor (AHR) repressor lacks exon 8, and its Pro 185 and Ala 185 variants repress both AHR and hypoxia-inducible factor. Mol Cell Biol. 2009;29:3465–3477. doi: 10.1128/MCB.00206-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R, Saif MW. Is there an optimal neoadjuvant therapy for locally advanced pancreatic cancer? JOP. 2007;8:279–288. [PubMed] [Google Scholar]
- Kondraganti SR, Fernandez-Salguero P, Gonzalez FJ, Ramos KS, Jiang W, Moorthy B. Polycyclic aromatic hydrocarbon-inducible DNA adducts: evidence by 32P-postlabeling and use of knockout mice for Ah receptor-independent mechanisms of metabolic activation in vivo. Int J Cancer. 2003;103:5–11. doi: 10.1002/ijc.10784. [DOI] [PubMed] [Google Scholar]
- Koorstra JB, Hustinx SR, Offerhaus GJ, Maitra A. Pancreatic Carcinogenesis. Pancreatology. 2008;8:110–125. doi: 10.1159/000123838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Tamakoshi A, Kawamura T, Inaba Y, Kikuchi S, Motohashi Y, Kurosawa M JACC Study Group, Japan Collaborative Cohort. A prospective cohort study of cigarette smoking and pancreatic cancer in Japan. Cancer Causes Control. 2002;13:249–254. doi: 10.1023/a:1015052710213. [DOI] [PubMed] [Google Scholar]
- Lowenfels AB, Maisonneuve P. Epidemiology and risk factors for pancreatic cancer. Best Pract Res Clin Gastroenterol. 2006;20:197–209. doi: 10.1016/j.bpg.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Lynch HT, Deters CA, Snyder CL, Lynch JF, Villeneuve P, Silberstein J, Martin H, Narod SA, Brand RE. BRCA1 and pancreatic cancer: pedigree findings and their causal relationships. Cancer Genet Cytogenet. 2005;158:119–125. doi: 10.1016/j.cancergencyto.2004.01.032. [DOI] [PubMed] [Google Scholar]
- Mai PL, Chatterjee N, Hartge P, Tucker M, Brody L, Struewing JP, Wacholder S. Potential excess mortality in BRCA1/2 mutation carriers beyond breast, ovarian, prostate, and pancreatic cancers, and melanoma. PLoS One. 2009;4:e4812. doi: 10.1371/journal.pone.0004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud DS, Giovannucci E, Willett WC, Colditz GA, Stampfer MJ, Fuchs CS. Physical activity, obesity, height, and the risk of pancreatic cancer. JAMA. 2001;286:921–929. doi: 10.1001/jama.286.8.921. [DOI] [PubMed] [Google Scholar]
- Michaud DS, Liu S, Giovannucci E, Willett WC, Colditz GA, Fuchs CS. Dietary sugar, glycemic load, and pancreatic cancer risk in a prospective study. J Natl Cancer Inst. 2002;94:1293–1300. doi: 10.1093/jnci/94.17.1293. [DOI] [PubMed] [Google Scholar]
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- Moennikes O, Loeppen S, Buchmann A, Andersson P, Ittrich C, Poellinger L, Schwarz M. A constitutively active dioxin/aryl hydrocarbon receptor promotes hepatocarcinogenesis in mice. Cancer Res. 2004;64:4707–4710. doi: 10.1158/0008-5472.CAN-03-0875. [DOI] [PubMed] [Google Scholar]
- Motykiewicz G, Faraglia B, Wang LW, Terry MB, Senie RT, Santella RM. Removal of benzo(a)pyrene diol epoxide (BPDE)-DNA adducts as a measure of DNA repair capacity in lymphoblastoid cell lines from sisters discordant for breast cancer. Environ Mol Mutagen. 2002;40:93–100. doi: 10.1002/em.10095. [DOI] [PubMed] [Google Scholar]
- Nakata K, Tanaka Y, Nakano T, Adachi T, Tanaka H, Kaminuma T, Ishikawa T. Nuclear receptor-mediated transcriptional regulation in phase I, II, and III xenobiotic metabolizing systems. Drug Metab Pharmacokinet. 2006;21:437–457. doi: 10.2133/dmpk.21.437. [DOI] [PubMed] [Google Scholar]
- National Toxicology Program. NTP Toxicology and Carcinogenesis Studies of a Mixture of 2,3,7,8-Tetrachlorodibenzo-p-Dioxin (TCDD) (CAS No. 1746–01–6), 2,3,4,7,8-Pentachlorodibenzofuran (PeCDF) (CAS No 57117–31–4), and 3,3′,4,4′,5-Pentachlorobiphenyl (PCB 126) (CAS No 57465–28–8) in Female Harlan Sprague-Dawley Rats (Gavage Studies) Natl Toxicol Program Tech Rep Ser. 2006;526:1–180. [PubMed] [Google Scholar]
- Ouyang H, Mou L, Luk C, Liu N, Karaskova J, Squire J, Tsao MS. Immortal human pancreatic duct epithelial cell lines with near normal genotype and phenotype. Am J Pathol. 2000;157:1623–1631. doi: 10.1016/S0002-9440(10)64800-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Davis W, Trushin N, Amin S, Nath RG, Salem N, Jr, Chung FL. A solid-phase extraction/high-performance liquid chromatography-based 32P-postlabeling method for detection of cyclic 1,N(2)-propanodeoxyguanosine adducts derived from enals. Anal Biochem. 2006;348:15–23. doi: 10.1016/j.ab.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Phillips DH, Arlt VM. The 32P-postlabeling assay for DNA adducts. Nat Protocols. 2007;2:2772–2781. doi: 10.1038/nprot.2007.394. [DOI] [PubMed] [Google Scholar]
- Qian J, Niu J, Li M, Chiao PJ, Tsao MS. In vitro modeling of human pancreatic duct epithelial cell transformation defines gene expression changes induced by K-ras oncogenic activation in pancreatic carcinogenesis. Cancer Res. 2005;65:5045–5053. doi: 10.1158/0008-5472.CAN-04-3208. [DOI] [PubMed] [Google Scholar]
- Redston MS, Caldas C, Seymour AB, Hruban RH, da Costa L, Yeo CJ, Kern SE. p53 mutations in pancreatic carcinoma and evidence of common involvement of homocopolymer tracts in DNA microdeletions. Cancer Res. 1994;54:3025–3033. [PubMed] [Google Scholar]
- Shimada T, Inoue K, Suzuki Y, Kawai T, Azuma E, Nakajima T, Shindo M, Kurose K, Sugie A, Yamagishi Y, Fujii-Kuriyama Y, Hashimoto M. Arylhydrocarbon receptor-dependent induction of liver and lung cytochromes P450 1A1, 1A2, and 1B1 by polycyclic aromatic hydrocarbons and polychlorinated biphenyls in genetically engineered C57BL/6J mice. Carcinogenesis. 2002;23:1199–1207. doi: 10.1093/carcin/23.7.1199. [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Nakatsuru Y, Ichinose M, Takahashi Y, Kume H, Mimura J, Fujii-Kuriyama Y, Ishikawa T. Benzo[a]pyrene carcinogenicity is lost in mice lacking the aryl hydrocarbon receptor. Proc Natl Acad Sci U S A. 2000;97:779–782. doi: 10.1073/pnas.97.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman DT, Dunn JA, Hoover RN, Schiffman M, Lillemoe KD, Schoenberg JB, Brown LM, Greenberg RS, Hayes RB, Swanson GM. Cigarette smoking and pancreas cancer: a case-control study based on direct interviews. J Natl Cancer Inst. 1994;86:1510–1516. doi: 10.1093/jnci/86.20.1510. [DOI] [PubMed] [Google Scholar]
- Skog K. Cooking procedures and food mutagens: a literature review. Food Chem Toxicol. 1993;31:655–675. doi: 10.1016/0278-6915(93)90049-5. [DOI] [PubMed] [Google Scholar]
- Starita LM, Parvin JD. The multiple nuclear functions of BRCA1: transcription, ubiquitination and DNA repair. Curr Opin Cell Biol. 2003;15:345–350. doi: 10.1016/s0955-0674(03)00042-5. [DOI] [PubMed] [Google Scholar]
- Sugimura T, Nagao M, Wakabayashi K. Heterocyclic amines in cooked foods: candidates for causation of common cancers. J Natl Cancer Inst. 1994;86:2–4. doi: 10.1093/jnci/86.1.2. [DOI] [PubMed] [Google Scholar]
- Uno S, Dalton TP, Derkenne S, Curran CP, Miller ML, Shertzer HG, Nebert DW. Oral exposure to benzo[a]pyrene in the mouse: detoxication by inducible cytochrome P450 is more important than metabolic activation. Mol Pharmacol. 2004;65:1225–1237. doi: 10.1124/mol.65.5.1225. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Imoto I, Kosugi Y, Fukuda Y, Mimura J, Fujii Y, Isaka K, Takayama M, Sato A, Inazawa J. Human arylhydrocarbon receptor repressor (AHRR) gene: genomic structure and analysis of polymorphism in endometriosis. J Hum Genet. 2001;46:342–346. doi: 10.1007/s100380170070. [DOI] [PubMed] [Google Scholar]
- Whitlock JP., Jr Induction of cytochrome P4501A1. Annu Rev Pharmacol Toxicol. 1999;39:103–125. doi: 10.1146/annurev.pharmtox.39.1.103. [DOI] [PubMed] [Google Scholar]
- Xu C, Li CY, Kong AT. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch Pharm Res. 2005;28:249–268. doi: 10.1007/BF02977789. [DOI] [PubMed] [Google Scholar]
- Yamamoto J, Ihara K, Nakayama H, Hikino S, Satoh K, Kubo N, Iida T, Fujii Y, Hara T. Characteristic expression of aryl hydrocarbon receptor repressor gene in human tissues: organ-specific distribution and variable induction patterns in mononuclear cells. Life Sci. 2004;74:1039–1049. doi: 10.1016/j.lfs.2003.07.022. [DOI] [PubMed] [Google Scholar]
- Zudaire E, Cuesta N, Murty V, Woodson K, Adams L, Gonzalez N, Martínez A, Narayan G, Kirsch I, Franklin W, Hirsch F, Birrer M, Cuttitta F. The aryl hydrocarbon receptor repressor is a putative tumor suppressor gene in multiple human cancers. J Clin Invest. 2008;118:640–650. doi: 10.1172/JCI30024. [DOI] [PMC free article] [PubMed] [Google Scholar]