Abstract
Bestrophin 3 (Best3), a member of bestrophin Cl channel family, is a CaClcGMP channel candidate in vascular smooth muscle cells. The mechanism for its activation remains unclear. In previous studies, we reported that an autoinhibitory domain (356IPSFLGS362) existed in Best3 C-terminus and when the autoinhibitory domain was mutated, the Best3 channel was dramatically activated. In this study, we further dissected the roles of the C-terminal sequence in Best3 activation. We found that there were eight basic amino acids downstream of the AI domain within the region (384–397), which were also involved in Best3 activation. Mutations of these basic amino acids significantly activated Best3 as a Cl channel. Led by the assumption that the basic amino acids may be involved in the Best3 C-terminal membrane association through binding to membranous phospholipids, we discovered that PI3Kα inhibitor IV could strongly activate Best3.
Keywords: anion channel, chloride channel, bestrophin 3, channel activation, patch clamp, membrane protein, channel mutation
INTRODUCTION
Because bestrophin 1 (Best1) has been determined as a causal gene for the inherited Best vitelliform macular dystrophy,1,2 the biologic functions of the bestrophin family have attracted broad attention.3,4 Human bestrophins except bestrophin 3 (Best3) were able to express robust Cl and HCO –3− currents when they were transfected into heterologous cells.5–7 Best3 in heterologous cells induced small Cl currents if stimulations were in a physiological range.8,9 Recently, the Best3 gene has been suggested to encode cGMP-sensitive Ca-activated Cl channels (CaClcGMP) in vascular smooth muscle cells.10 This is exciting because the activity of CaCl channels has long been thought to regulate the tone of vascular smooth myocytes.11 In addition, because Best3 transcripts were robustly expressed in heart tissue,12 it is assumed that Best3 may play some important roles in cardiovascular function. The assumption is supported by the observation that Best3 proteins showed a ubiquitous distribution in both atrial and ventricular myocytes, including the sarcolemmal membrane.13
Although Best3 has been considered as a CaClcGMP channel candidate, its activation mechanism remains elusive. We previously investigated why the Best3 channel remained in low activity in heterologous cells. A 53 amino acid sequence (amino acid 354–404) was determined to be important for regulation of the channel activation.9,14 An autoinhibitory (AI) domain in the sequence composed of seven amino acids (356IPSFLGS362) was identified to be responsible for the low activity of the channel.
In this study, we further elucidate the role of C-terminal sequence in Best3 activation. We studied another critical regulatory domain (amino acids 384–397 downstream of the AI domain), which includes a cluster of eight basic amino acids. These basic residues were also involved in the inhibition of Best3 activity because their mutations significantly activated the channel. We previously found that mBest3 C-terminus could strongly associate with cellular membranes. When the 53 amino acids (354–404) in its C-terminus was deleted, the membrane associability disappeared while the mBest3–channel was activated robustly.14 We assumed that the positively charged basic amino acids may be involved in association with the negatively charged membranous phospholipids and inhibited the channel. Led by the assumption, we discovered that PI3Kα inhibitor IV was capable of strongly activating heterologously expressed Best3.
MATERIALS AND METHODS
Generation of Bestrophin Mutations and Heterologous Expression
Mouse bestrophin 3 (mBest3, mouse VMD2L3) cDNA in pcDNA3.1 vector8,9 was mutated site-specifically using a polymerase chain reaction-based mutagenesis kit (Quick-change; Stratagene, La Jolla, CA) as described previously.15 The cDNA expression vectors were cotransfected with pEGFP (Invitrogen) into HEK293 cells (ATCC, Manassas, VA) using FuGene-6 transfection reagent (Roche, Indianapolis, IN). The transfected cells were identified by the pEGFP-expressed green fluorescence. The transfection efficiency was 20% to 30%. Approximately 0.3 μg of plasmid was used to transfect HEK293 cells on one 35-mm culture dish. Cells were trypsinized and placed on the coverslips for electrophysiological recording 2 days after transfection.
Electrophysiology
Recordings were performed using whole-cell configuration of the patch clamp at 22°C to 24°C.15 Voltage steps and ramp protocols were used as indicated in the figure legends with a holding potential of 0 mV. Standard pipette solution (approximately 4.5 μM free Ca) contained (in mM) 146 CsCl, 2 MgCl2, 5 (Ca2+)-EGTA, 10 sucrose, 8 HEPES, pH 7.3.15 Standard extracellular solution contained (in mM) 140 NaCl, 4 KCl, 2 CaCl2, 1 MgCl2, 10 glucose, 10 HEPES, pH 7.3. This combination of solutions set Erev for Cl– currents to zero, whereas cation currents carried by Na+ or Cs+ had very positive or negative Erev, respectively.15 To verify the currents were carried by Cl–, cells were exposed to a solution (mM) containing 100 Na2SO4, 1 CaCl2, 10 HEPES, pH 7.3. Because SO4–2 is largely impermeable through bestrophin channels, this solution blocks outward whole-cell Cl– currents. Currents were normalized with cellular membrane capacitance (from 10 to 15 pF for HEK293 cells) for current densities (pA/pF).
Statistical Analysis
The Student t test was used for statistical analysis. The amplitude of whole-cell currents at the end of a 100-mV pulse was measured and is shown as mean ± standard error.
Solubility of Bestrophin C-termini
mBet3 wild-type, mutated, or deleted C-terminal cDNA subcloned in pcDNA3.1 vector8 was cotransfected with pEGFP into HEK293 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). The transfection efficiency was approximately 60% with approximately 3 μg DNA per 10-cm dish. The transfected cells were harvested 2 days after transfection and sonicated briefly in phosphate-buffered saline containing 1 mM DTT, 1 mM PMSF, and 1/200 (volume) protease inhibitor set III (Calbiochem, San Diego, CA) and ultracentrifuged at 42,500 × g (Beckman, Brea, CA; Optima TLX ultracentrifuge, TLS 55 rotor). The supernatant was collected as a soluble fraction (S). The pellet was washed with phosphate-buffered saline, dissolved in a nonionic detergent lysis buffer (mM) (1% Triton X-100, 150 NaCl, 0.5 EGTA, 10% Glycerol, 1 DTT, 10 HEPES, pH 7.3, 1 PMSF, 1/200 Protease inhibitor cocktail III), and centrifuged at 16,000 × g. The supernatant contained the Triton-soluble membrane proteins (M). Protein concentrations were measured with the BCA Protein Assay kit (Pierce, Rockford, IL). Western blotting was performed with equivalent amounts (10 μg/lane) of protein from each sample. mBest3 C-termini were detected with an anti-mBest3 antibody (05619)14 followed by horseradish peroxidase-conjugated goat anti-rabbit IgG and chemiluminescence.
Materials
PI3Kα inhibitor IV was purchased from Calbiochem (San Diego, CA). The compound was dissolved in DMSO as stock solutions (10 mM) and kept at –20°C before use.
RESULTS
A Cluster of Basic Amino Acids in C-terminus Is Involved in Best3 Activation
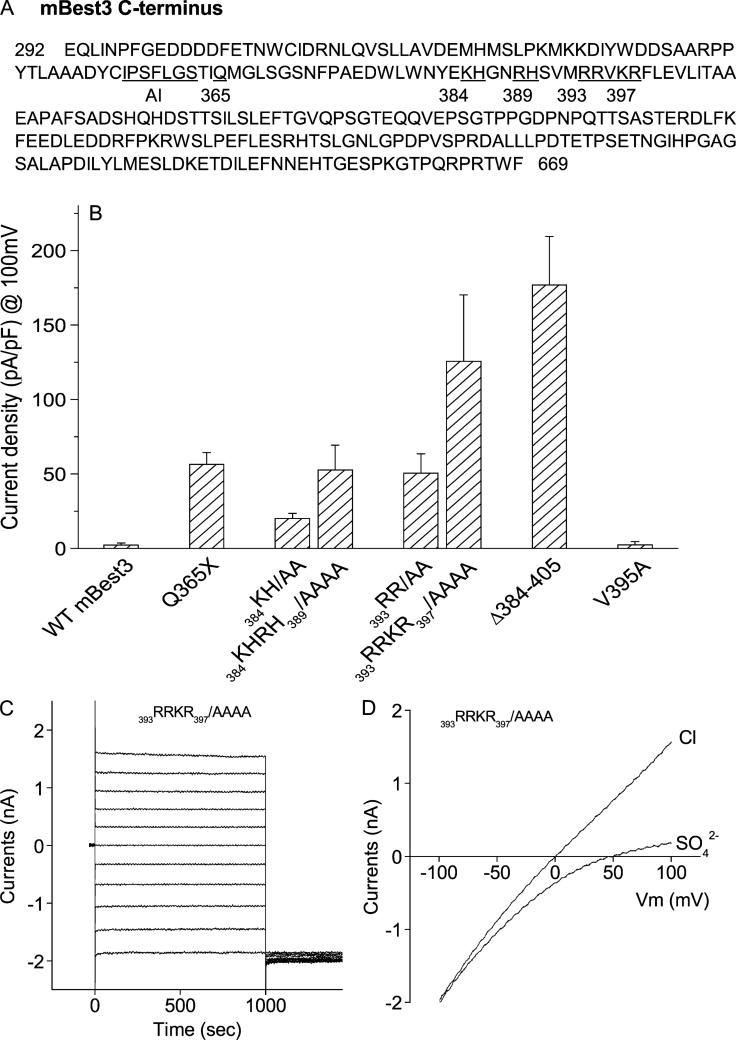
In a previous publication, we reported that an AI domain (356IPSFLGS362) in C-terminus was involved in activation of bestrophin 3 (Best3). Mutation of any amino acids in the AI domain activated the Cl channel.14 Further examining the amino acids downstream of the AI domain, we noticed that there were eight basic amino acids clustering from amino acid 384 to 397: K384, H385, R388, H389, R393, R394, K396, and R397 (Fig. 1A). Because these basic residues are not conserved among bestrophins,8,9 we hypothesized that they might play specific roles in the Best3 activation. To test the hypothesis, we first introduced a stop codon at position of 365 (Q365X) between the AI domain and the basic residues (Fig. 1A). This stop codon introduction left the AI domain complete but removed all the sequence downstream of it. Previously, we showed that mBest3 remained inactive when a stop codon was introduced at P405 downstream of AI domain (P405X).8,9 If any residue in the AI domain of P405X mutant was mutated, the mutant channel was dramatically activated, indicating that the mutant channel kept inactive when the AI domain was intact. Thus, here, mBest3 was supposed to remain inactive when the Q365X mutation was introduced. However, unexpectedly, Q365X mutation moderately activated Best3 (Fig. 1B). The result suggests that the residues downstream of the AI domain also play an inhibitory role. The eight basic amino acids were hypothesized to contribute to the inhibitory role. We mutated these basic residues to examine their roles in the channel activation. As expected, mutation of these amino acids did activate Best3 (Fig. 1B). Interestingly, the degree of activation is proportional to the number of basic residues mutated, ie, the more basic residues were mutated, the higher channel activity mBest3 gained. The current magnitude of 384KHRH389/AAAA mutant was larger than that of 384KH/AA mutant and 393RRKR397/AAAA larger than 393RR/AA. If the current amplitudes induced by 384KHRH389/AAAA mutant and 393RRKR397/AAAA mutant were added up, the total amplitude was close to that of the Δ384–405 deletion mutant where all eight basic residues were removed. A mutation of the nonbasic residue, V395A, between 393RR and KR397 did not activate Best3, indicating that the roles of the basic residue mutations were specific. The results suggest that the inactivating role of the region from 384 to 405 is contributed by the eight amino acids. The current waves behaved time- and voltage-independently among all mutations shown in Figure 1B. A representative of current traces from 393RRKR397/AAAA was presented (Fig. 1C). Ramp current traces showed Vrev near zero and were blocked by SO4–2, indicating that the currents were carried by Cl anions (Fig. 1D). Our results suggest that Best3 inactivation is completed by two separate domains in its C-terminus: the AI domain and the basic amino acids domain. They work together to exert an inhibitory effect on Best3 activation. Mutations occurring in either domain can activate the channel. At present, it remains unknown how the two domains regulate the Best3 activation.
FIGURE 1.
Mutations of basic amino acids in C-terminus activated mBest3. (A) Positions of mutated residues (both numbered and underlined) downstream of the autoinhibitory domain (AI) in mBest3 C-terminus. (B) Stimulatory effect of basic amino acid mutations residing from amino acids 384 to 397 on the mBest3 channel activation. Mutants were transiently transfected into HEK cells, which were voltage-clamped with high [Ca]i included in the pipette. Step voltages in 20-mV increments or ramp voltages were applied between −100 mV and 100 mV with 1-second duration. The Cl currents were recorded in whole-cell configuration. Internal and external solutions with symmetric [Cl–]were described in the ”Methods.” Average amplitudes of Cl– currents at the end of the +100-mV trace are shown. All single or combined mutations except V395A significantly activated mBest3 channel compared with wild-type (WT) mBest3 (P < 0.01, n = 4–7). (C–D) Representatives of current traces produced by 393RRKR397/AAAA mutant in HEK cells stimulated by step (C) or ramp voltages (D).
mBest3 C-terminus Associates With the Cellular Membrane Surface
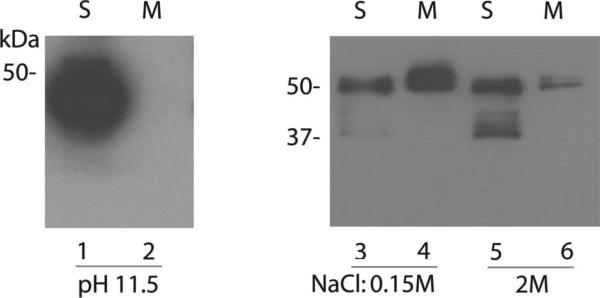
Previously we reported that the Best3 C-terminal sequence from 354 to 404 was involved in C-terminal membrane association.14 We assume that the inhibition of Best3 activation is based on the association in which these basic amino acids may participate. To study the mechanism for the association, we performed the following experiments to clarify whether the C-terminus associates with the membrane surface or integrates with the lipid bilayers. First, the membrane fractions from mBest3-C terminus-transfected HEK cells were treated with 0.1M Na2CO3 (pH 11.5) solution. As a result, the treatment totally made the C terminus dissociate from the membrane (Fig. 2, lane 2), suggesting that the C-terminus only associated with the membrane surface and was not an integral membrane protein.16 To delineate how strongly the C-terminus interacts with the membrane through their interfaces, the membrane fractions from mBest3-C terminus-transfected HEK cells were treated with a solution of high ionic strength (2 M NaCl). The solution significantly dissociated the C-terminus from the membrane (Fig. 2, lane 6). Because the treatment did not completely dissociate the C-terminus from the membrane, we assume that the membrane association is determined by two aspects. One is the ionic binding strength of positively charged amino acids to the negatively charged molecules (eg, phospholipids) inside the membrane or proteins embedded or associated with the membrane. Another is hydrophobic binding between Best3 C-terminus and membranes. With the MPEx program for hydropathy analysis, we have shown that the mBest3 sequence, 355CIPSFLGSTIQMGLSGSNF373 including the AI domain, is more hydrophobic than the flanking sequences.14 These hydrophobic amino acids may form hydrophobic binding with the membranes, which cannot be simply dissociated with solutions of high ionic strength (Fig. 2, lane 6).
FIGURE 2.
Association between Best3 C-terminus and membrane was disrupted by alkaline and high ionic strength solutions. Wild type of mBest3 C-terminus was transfected into HEK293 cells, which were sonicated 2 days after transfection with different buffers: 0.1 M Na2CO3, pH 11.5 (lanes 1 and 2); and 0.15 or 2 M NaCl, pH 7.3 (lanes 3–6). Cellular proteins in soluble (S) and membrane (M) fractions were extracted for Western blotting (see ”Methods” for details). Immunoblots were probed with anti-mBest3 antibody. The bands with molecular mass at approximately 50 kDa were expected mBest3 C-terminal proteins. The bands with smaller molecular masses (approximately 37 kDa) in lanes 3 and 5 (S) were probably the protease-degraded proteins.
A PI3 Kinase Inhibitor Activates Best3
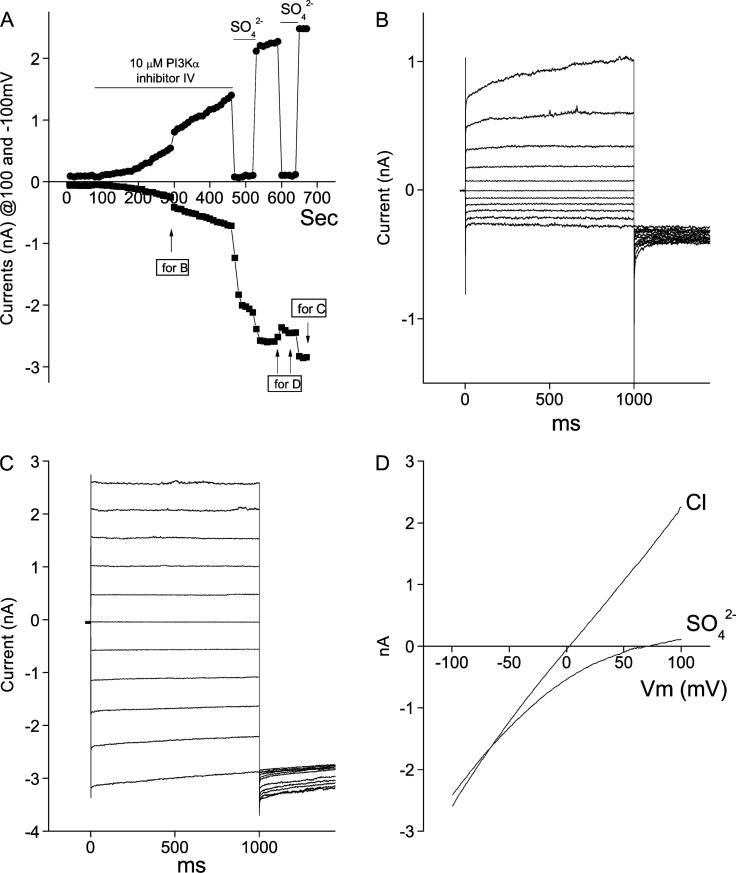
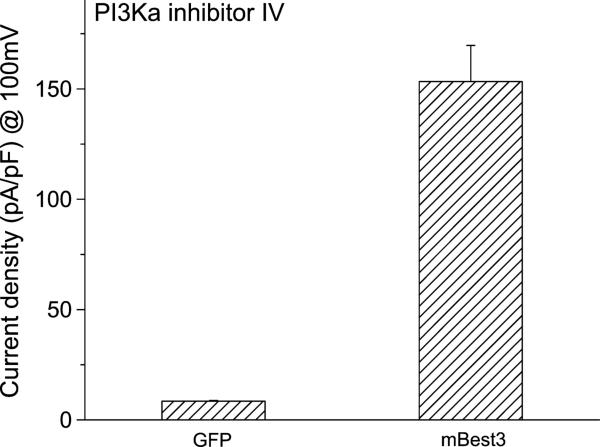
Because Best3 has been considered to be a candidate for the endogenous CaClcGMP channel,10 we believe that finding a Best3 activator will benefit the channel research and clinical treatment of some cardiovascular diseases. If our assumption is correct that the basic amino acids may bind the phospholipids in the cellular lipid bilayers, a PI3 kinase inhibitor, which can reduce the amount of phospholipids in the membrane and subsequently weaken the C-terminal membrane association, is supposed to be able to activate the channel. As assumed, we found that PI3Kα inhibitor IV, a PI3 kinase inhibitor at 10 μM, dramatically induced Cl currents in Best3-transfected HEK cells (Figs. 3A and 4). Because the PI3K inhibitor did not induce Cl current from GFP-transfected HEK cells, its stimulatory effect on mBest3 activation was considered to be specific (Fig. 4). The induction process was slow and irreversible (Fig. 3A). The induced Cl currents continuously developed although the PI3K inhibitor was washed out. The magnitude of whole-cell Cl currents reached peak in approximately 10 minutes. Before washing, the activated currents were outwardly rectifying and time-dependent (Fig. 3A–B). However, when the PI3K inhibitor was washed out, the currents behaved in a time- and voltage-independent manner (Figs. 3A and C), indicating that the PI3K inhibitor suppressed the induced inward currents. In Figure 3D, the ramp currents show the Vrev near zero. This indicates that the currents were carried by Cl anions, which is well blocked by impermeable SO42–.
FIGURE 3.
PI3Kα inhibitor IV slowly activated mBest3 in a representative HEK cell. The transfected cells were stimulated with ramp and step voltage protocols (see ”Methods” and Fig.1 legend). A, Time course of mBest3 activation by PI3Kα inhibitor IV. The inhibitor (10 μM) applied to the bath solution slowly induced Cl currents recorded with ramp voltages. Step voltages were applied twice in the middle (B) and the end (C) of recordings to observe the current wave forms. The induced currents were blocked by SO4–2 to eliminate the possibility of leaky currents. Note that the outward currents were completely blocked by extracellular SO4–2, whereas inward currents were not. B, First-step voltage stimulation in A in the presence of the inhibitor (B). Currents behaved as outwardly rectifying. C, Step voltage stimulation in A in absence of the inhibitor (C). The currents were time- and voltage-independent when the inhibitor had been washed out. D, Ramp currents that were blocked by SO4–2 showed the linear pattern with Vrev close to zero.
FIGURE 4.
PI3Kα inhibitor IV and Best3 activation. Average amplitudes of mBest3 currents activated by PI3Kα inhibitor IV. Wild-type mBest3 in pcDNA3.1 vector was cotransfected into HEK cells with GFP or cells were transfected with GFP only. Green fluorescent cells were selected for whole-cell recordings with ramp voltage stimulations (refer to Fig.1 legend). The PI3K inhibitor (10 μM) applied to the bath dramatically stimulated Cl currents from mBest3-transfected (n = 7) but not from GFP-only transfected HEK cells (n = 4). The currents obtained with 100-mV stimulation, 10 minutes after the PI3K inhibitor application to two groups, were averaged for statistical analysis (P < 0.01).
DISCUSSION
We assume that the mechanism of the PI3K inhibitor in activation of Best3 is related with the membranous phospholipids metabolism. Cellular membrane contains a large amount of phospholipids whose phosphate bases make the cytosolic surface of membrane negatively charged. The positively charged basic amino acids in Best3 C-terminus tend to attract the negative charges of phospholipids. The attraction leads to the binding of C-terminus and the membrane. A PI3 kinase inhibitor,17 which reduces the membranous phospholipids synthesis (eg, PIP3 and PIP2) and then theoretically lowers the amount of negative charges in the membrane, may weaken the interaction between the basic amino acids (from 384 to 397) and the membrane, which somehow activates Best3. When voltage dependence of a channel is changed by a reagent, it usually indicates that the reagent probably acts on the channel pore. Because PI3Kα inhibitor IV made the activated Best3 currents outwardly rectifying and time-dependent, the inhibitor likely activates Best3 through interacting with the channel pore structure. Therefore, we assumed that the inhibitor may affect Best3 activation through two aspects: one is to inhibit PI3 kinase and another is to bind to the channel and change Best3 pore structure.
Although the CaClcGMP channels may be represented by the Best3 gene, it is believed now that Best3, instead of a pore-forming protein, may be a regulator of the Anoctamin 1 or 2, which has been recently suggested as the gene encoding the pore-forming subunit of the native CaCl channel.18 Both Best3 and Anoctamin are expressed in cardiovascular tissues. However, how bestrophins interact with and regulate endogenous CaCl channels in native cardiac myocytes or vascular smooth muscle cells remains to be explored.19
Because Best3 is closely related with CaClcGMP channels in vascular smooth muscle cells,10 it will be interesting to see whether the Best3 activation by the PI3K inhibitor also occurs in the vascular tissue. Potential function of Best3 in cardiovascular system has been suggested.10,13 Enhancement of the endogenous CaClcGMP currents by the PI3K inhibitor will strengthen the suggestion that Best3 represents the CaClcGMP channel gene product. CaCl channel activation was regarded to be closely associated with arterial smooth muscle relaxation.11 If the PI3K inhibitor does work as an endogenous CaClcGMP channel activator, it will help develop a clinical medication for hypertension treatment.
Acknowledgments
This study is supported by Chinese National Natural Science Foundation (30840004) and American Heart Association Scientist Development Grant (0430204N) to ZQ.
Footnotes
The authors report no conflicts of interest.
REFERENCES
- 1.Petrukhin K, Koisti MJ, Bakall B, et al. Identification of the gene responsible for Best macular dystrophy. Nat Genet. 1998;19:241–247. doi: 10.1038/915. [DOI] [PubMed] [Google Scholar]
- 2.Marquardt A, Stohr H, Passmore LA, et al. Mutations in a novel gene, VMD2, encoding a protein of unknown properties cause juvenile-onset vitelliform macular dystrophy (Best's disease). Hum Mol Genet. 1998;7:1517–1525. doi: 10.1093/hmg/7.9.1517. [DOI] [PubMed] [Google Scholar]
- 3.Hartzell C, Qu Z, Putzier I, et al. Looking chloride channels straight in the eye: bestrophins, lipofuscinosis, and retinal degeneration. Physiology. 2005;20:292–302. doi: 10.1152/physiol.00021.2005. [DOI] [PubMed] [Google Scholar]
- 4.Hartzell HC, Qu Z, Yu K, et al. Molecular physiology of bestrophins: multifunctional membrane proteins linked to best disease and other retinopathies. Physiol Rev. 2008;88:639–672. doi: 10.1152/physrev.00022.2007. [DOI] [PubMed] [Google Scholar]
- 5.Sun H, Tsunenari T, Yau KW, et al. The vitelliform macular dystrophy protein defines a new family of chloride channels. Proc Natl Acad Sci U S A. 2002;99:4008–4013. doi: 10.1073/pnas.052692999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsunenari T, Sun H, Williams J, et al. Structure–function analysis of the bestrophin family of anion channels. J Biol Chem. 2003;278:41114–41125. doi: 10.1074/jbc.M306150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qu Z, Hartzell HC. Bestrophin Cl– channels are highly permeable to HCO3–. Am J Physiol Cell Physiol. 2008;294:C1371–C1377. doi: 10.1152/ajpcell.00398.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qu Z, Cui Y, Hartzell C. Corrigendum to ‘A short motif in the C–terminus of mouse bestrophin 3 inhibits its activation as a Cl channel.’. FEBS Lett. 2006;580:2141–2146. doi: 10.1016/j.febslet.2006.03.025. [DOI] [PubMed] [Google Scholar]; FEBS Lett. 2007;581:580. [Google Scholar]
- 9.Qu Z, Cui Y, Hartzell HC. A short motif in the C-terminus of mouse bestrophin 4 inhibits its activation as a Cl channel. FEBS Lett. 2006;580:2141–2146. doi: 10.1016/j.febslet.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 10.Matchkov VV, Larsen P, Bouzinova EV, et al. Bestrophin-3 (vitelliform macular dystrophy 2-like 3 protein) is essential for the cGMP-dependent calcium-activated chloride conductance in vascular smooth muscle cells. Circ Res. 2008;103:864–872. doi: 10.1161/CIRCRESAHA.108.178517. [DOI] [PubMed] [Google Scholar]
- 11.Large WA, Wang Q. Characteristics and physiological role of the Ca2+-activate Cl– conductance in smooth muscle. Am J Physiol. 1996;271:C435–C454. doi: 10.1152/ajpcell.1996.271.2.C435. [DOI] [PubMed] [Google Scholar]
- 12.Kramer F, Stohr H, Weber BH. Cloning and characterization of the murine Vmd2 RFP-TM gene family. Cytogenet Genome Res. 2004;105:107–114. doi: 10.1159/000078016. [DOI] [PubMed] [Google Scholar]
- 13.O'Driscoll KE, Hatton WJ, Burkin HR, et al. Expression, localization, and functional properties of bestrophin 3 channel isolated from mouse heart. Am J Physiol Cell Physiol. 2008;295:C1610–C1624. doi: 10.1152/ajpcell.00461.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qu Z, Yu K, Cui Y, et al. Activation of bestrophin Cl– channels is regulated by C-terminal domains. J Biol Chem. 2007;282:17460–17467. doi: 10.1074/jbc.M701043200. [DOI] [PubMed] [Google Scholar]
- 15.Qu Z, Fischmeister R, Hartzell C. Mouse bestrophin-2 is a bona fide Cl(–) channel: identification of a residue important in anion binding and conduction. J Gen Physiol. 2004;123:327–340. doi: 10.1085/jgp.200409031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wishart MJ, Andrews PC, Nichols R, et al. Identification and cloning of GP-3 from rat pancreatic acinar zymogen granules as a glycosylated membrane-associated lipase. J Biol Chem. 1993;268:10303–10311. [PubMed] [Google Scholar]
- 17.Rameh LE, Cantley LC. The role of phosphoinositide 3-kinase lipid products in cell function. J Biol Chem. 1999;274:8347–8350. doi: 10.1074/jbc.274.13.8347. [DOI] [PubMed] [Google Scholar]
- 18.Hartzell HC, Yu K, Xiao Q, et al. Anoctamin/TMEM16 family members are Ca2+-activated Cl– channels. J Physiol. 2009;587:2127–2139. doi: 10.1113/jphysiol.2008.163709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duan D. Phenomics of cardiac chloride channels: the systematic study of chloride channel function in the heart. J Physiol. 2009;587:2163–2177. doi: 10.1113/jphysiol.2008.165860. [DOI] [PMC free article] [PubMed] [Google Scholar]