Abstract
The C3H/HeJBir mouse model of intestinal inflammation was used for isolation of a Gram-positive, rod-shaped, non-spore-forming bacterium (B7T) from caecal suspensions. On the basis of partial 16S rRNA gene sequence analysis, strain B7T was a member of the class Actinobacteria, family Coriobacteriaceae, and was related closely to Enterorhabdus mucosicola Mt1B8T (97.6 %). The major fatty acid of strain B7T was C16 : 0 (19.1 %) and the respiratory quinones were mono- and dimethylated. Cells were aerotolerant, but grew only under anoxic conditions. Strain B7T did not convert the isoflavone daidzein and was resistant to cefotaxime. The results of DNA–DNA hybridization experiments and additional physiological and biochemical tests allowed the genotypic and phenotypic differentiation of strain B7T from the type strain of E. mucosicola. Therefore, strain B7T represents a novel species, for which the name Enterorhabdus caecimuris sp. nov. is proposed. The type strain is B7T (=DSM 21839T =CCUG 56815T).
The family Coriobacteriaceae currently comprises 13 genera, four of which have been described recently, and includes Adlercreutzia equolifaciens (Maruo et al., 2008), the type species of which was isolated from human faeces; Asaccharobacter celatus (Minamida et al., 2008), isolated from a rat caecum; Enterorhabdus mucosicola (Clavel et al., 2009), isolated from the inflamed ileal mucosa of a mouse; and Gordonibacter pamelaeae (Würdemann et al., 2009), isolated from a patient with Crohn's disease.
During the course of experiments focused on flagellated bacteria and their implication in intestinal inflammation (Duck et al., 2007), strain B7T was isolated from the caecum of a C3H/HeJBir mouse, a mouse substrain prone to spontaneous colitis (Sundberg et al., 1994), after 3 days growth at 37 °C on ATCC medium 602 E. Additional information on strain isolation and 16S rRNA gene sequencing has been published elsewhere (Duck et al., 2007). Unless otherwise stated, all experiments for the description of strain B7T were carried out as described previously (Clavel et al., 2009). Bacteroides vulgatus was used as a positive control for the determination of growth with bile salts (no. 48305; Fluka). Cellular fatty acids, respiratory quinones, peptidoglycan and whole-cell sugars were analysed by the DSMZ, Braunschweig, Germany, according to standard procedures (Sasser, 1990; Cashion et al., 1977; De Ley et al., 1970; Huß et al., 1983; Mesbah et al., 1989; Rhuland et al., 1955; Staneck & Roberts, 1974; Tamaoka & Komagata, 1984; Visuvanathan et al., 1989; Whiton et al., 1985).
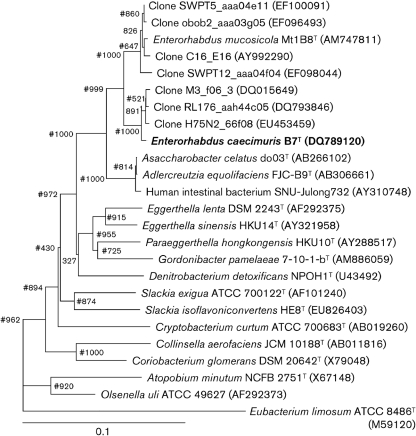
16S rRNA gene sequences from strain B7T (determined as described previously; Duck et al., 2007) and GenBank were aligned using BioEdit version 7.0.5.3 (Hall, 1999) and a rooted tree was constructed using the neighbour-joining method with clustal x version 1.8. Bootstrap values were calculated on the basis of 1000 resamplings. The maximum-parsimony method was used to confirm the topology of the phylogeny. Fig. 1 gives a current phylogenetic overview of the family Coriobacteriaceae and the position of strain B7T. Sequence similarity values were obtained with the DNA distance matrix function in the BioEdit software. The 16S rRNA gene sequence of strain B7T (1336 bp) was related most closely (>99 %) to sequences originating from as-yet-uncultured mouse intestinal bacteria (Ley et al., 2005, 2006) and E. mucosicola Mt1B8T (97.6 %). Lower similarities were found to sequences from Asaccharobacter celatus do03T (93.4 %), Adlercreutzia equolifaciens FJC-B9T (93.3 %) and two Eggerthella strains (<90 %). The resolution of 16S rRNA gene sequence analysis does not allow the identification of closely related species. However, it consistently depicts phylogenetic relationships from the level of domains to moderately related species (Stackebrandt & Goebel, 1994). It has been proposed that a genus could be defined as containing species that have 95 % 16S rRNA gene sequence similarity to each other (Rossello-Mora & Amann, 2001). We suggest that strain B7T does not belong to either of the genera Adlercreutzia or Asaccharobacter, as the strain has 16S rRNA gene sequence similarity values with members of these genera of <94 %.
Fig. 1.
Phylogenetic position of strain B7T within the family Coriobacteriaceae, based on a neighbour-joining analysis of 16S rRNA gene sequences (1338 bp). Bootstrap values based on 1000 resamplings are shown at branch nodes. Hash signs indicate that the corresponding nodes were also recovered in the tree generated using the maximum-parsimony method. Eubacterium limosum ATCC 8486T, a member of the phylum Firmicutes, was used as an outgroup. Bar, 10 substitutions per 100 nucleotide positions.
The gyrase B gene of strain B7T was amplified as described previously (Santos & Ochman, 2004). Amplicons (1500 bp) were purified using agarose gel electrophoresis and the Wizard SV Gel and PCR Clean-Up System (Promega). Purified products were sequenced using the primers gyrBBNDN1 (5′-CCGTCCACGTCGGCRTCNGYCAT-3′) and gyrBBAUP2 (5′-GCGGAAGCGGCCNGSNATGTA-3′). The gyrase B gene sequence of strain B7T shared 95.7 and 79.3 % similarity with sequences from E. mucosicola Mt1B8T (GenBank accession no. EU594341) and Eggerthella lenta DSM 2243T (EU594342; 524 bp), respectively. Strain B7T exhibited low DNA–DNA relatedness to E. mucosicola DSM 19490T (28.0±2.0 %, two experiments), which supported the fact that these two bacteria belong to different species. The DNA G+C content of strain B7T (64.5 mol%) was comparable to those reported in the literature for its phylogenetic neighbours.
The results of phenotypic and chemotaxonomic analyses are given in the species description and in Table 1. The fatty acid profile of strain B7T was similar to that of E. mucosicola DSM 19490T. The diamino acid in the peptidoglycan was identified as meso-diaminopimelic acid, which so far has been reported only for the peptidoglycan type A1γ and three variations of peptidoglycan type A4γ. The quinones were monomethylmenaquinone-6 (60 %) and dimethylmenaquinone-6 (40 %).
Table 1.
Characteristics that differentiate strain B7T and E. mucosicola DSM 19490T
Strains: 1, B7T; 2, E. mucosicola DSM 19490T. Data were taken from this study. DMA, Dimethylacetal; DMMK, dimethylmenaquinone; MMK, monomethylmenaquinone; r, resistant (MIC >32 μg ml−1); nd, not determined.
| Characteristic | 1 | 2 |
|---|---|---|
| Diaminopimelic acid | meso | ll |
| Major menaquinones (%) | ||
| MMK-6 | 60 | 100 |
| DMMK-6 | 40 | 0 |
| Whole-cell sugars | ||
| Glucose | + | − |
| Enzyme activities* | ||
| Aminopeptidase | − | + |
| Glutamic acid decarboxylase | + | − |
| Isoflavone conversion | ||
| Daidzein | − | + |
| Genistein | nd | + |
| Antibiotic MIC (μg ml−1)† | ||
| Cefotaxime | r (>32) | 1.250±0.112 |
| Ciprofloxacin | 0.305±0.035 | r (>32) |
| Clarithromycin | <0.016 | <0.016 |
| Clindamycin | 0.105±0.010 | <0.016 |
| Erythromycin | <0.016 | 0.048±0.007 |
| Metronidazole | 0.016±0.000 | 0.034±0.004 |
| Oxacillin | r (36.000±5.750) | 4.667±0.667 |
| Tetracycline | 0.120±0.005 | 0.115±0.007 |
| Tobramycin | 4.333±0.558 | 2.667±0.211 |
| Vancomycin | 1.500±0.129 | 1.333±0.105 |
| Cellular fatty acids | ||
| iso-C12 : 0 | 0.36 | 0.50 |
| C12 : 0 | 1.21 | 0.72 |
| iso-C13 : 0 | − | 0.15 |
| anteiso-C13 : 0 | 0.43 | 0.49 |
| C13 : 1c12 | 0.29 | 0.17 |
| iso-C14 : 0 | 3.83 | 2.48 |
| C14 : 0 | 15.08 | 12.11 |
| C14 : 0 DMA | 0.68 | 0.41 |
| iso-C15 : 0 | 1.20 | 1.60 |
| anteiso-C15 : 0 | 2.82 | 2.29 |
| C15 : 0 | 1.77 | 1.60 |
| C15 : 0 DMA | 0.40 | 0.42 |
| C16 : 0 ALDE | 2.28 | 2.31 |
| C16 : 0 | 19.14 | 18.06 |
| iso-C16 : 0 | 0.52 | 0.42 |
| C16 : 0 DMA | 9.62 | 11.56 |
| C16 : 1c9 | 0.94 | 0.93 |
| anteiso-C17 : 0 | 0.82 | 0.75 |
| anteiso-C17 : 0 DMA | − | 0.19 |
| C17 : 0 | 0.87 | − |
| C17 : 0 DMA | − | 0.20 |
| C17 : 1c8 | 1.16 | 0.52 |
| C17 : 1c9 | − | 0.41 |
| C18 : 0 DMA | 2.40 | 3.73 |
| C18 : 0 | 7.99 | 9.35 |
| C18 : 1c9 | 15.25 | 16.68 |
| C18 : 1c9 DMA | 4.94 | 4.75 |
| C18 : 1c11 DMA | 0.59 | 0.56 |
| C18 : 1c11/t9/t6 | 4.16 | 3.83 |
| C18 : 2c9,12 | 1.24 | 1.30 |
*Data obtained with Rapid ID32A identification system for anaerobes (bioMérieux).
†Values are expressed as mean±sd of six replicates (three independent experiments with duplicates).
For polar lipid analysis, batch cultures (1.5 l) of strain B7T and E. mucosicola DSM 19490T were grown under anoxic conditions for 48 h in GYBHIc [brain–heart infusion broth (no. 211059; BD) supplemented with (l−1) 4 g glucose, 4 g yeast extract and 0.05 % (w/v) cysteine] and harvested by centrifugation [5525 g for 10 min at room temperature in 500 ml containers using a 4K15C centrifuge (Sigma)]. Pellets were resuspended in filter-sterilized PBS [(l distilled water)−1: 8.60 g NaCl, 0.87 g Na2HPO4, 0.40 g KH2PO4; pH 7.2] and supernatants were centrifuged again as above. Resuspended pellets were pooled in 50 ml Falcon tubes and centrifuged as above for 15 min. Supernatants were discarded first by inverting the tubes and subsequently pipetting the remaining liquid after the tubes had been left to stand for 30 s. Samples were stored at −80 °C prior to shipping on dry ice. Polar lipid analysis was done by the Identification Service of the DSMZ and Dr B. J. Tindall (Braunschweig, Germany). The polar lipid pattern of strain B7T differed from that of E. mucosicola DSM 19490T (Supplementary Fig. S1, available in IJSEM Online). The major polar lipids were diphosphatidylglycerol, phosphatidylglycerol, one unknown phospholipid, two unknown glycolipids and one unidentified lipid.
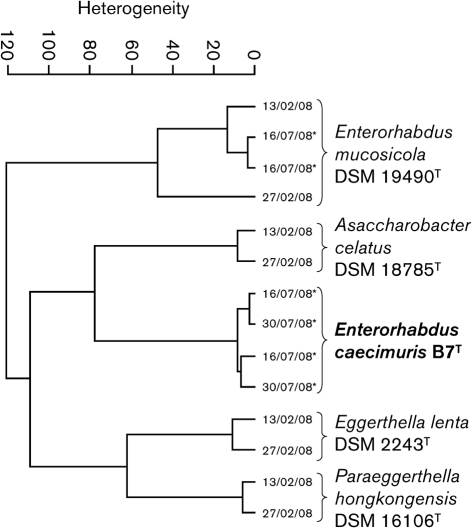
Fourier-transform infrared spectroscopy (FT-IRS) was used to further differentiate strain B7T and E. mucosicola DSM 19490T. FT-IRS relies on the absorption of infrared radiation by cell components and results in fingerprint-like spectra that reflect the cellular chemical composition and allow the identification of closely related bacteria (Kirschner et al., 2001; Wenning et al., 2008). Clusters were calculated using Ward's algorithm and vector-normalized first derivatives of the spectra (Savitzky–Golay algorithm) in the ranges 3000–2800 and 1800–700 cm−1 (Fig. 2). Measurements of duplicate cultures of E. mucosicola DSM 19490T at two time points clustered together and demonstrated the reproducibility of the technique. For each strain, spectra from independent cultures were more similar to one another than to those from other species and attested to the robustness of the observed spectral variations between taxa. Interestingly, the spectra from strain B7T were less closely related to those from E. mucosicola DSM 19490T than to those from more distant phylogenetic neighbours, providing evidence at the whole-cell biochemical level that there were differences between these two organisms. Thus, although it is not useful for taxonomic purposes, FT-IRS can be used for the rapid identification of members of the family Coriobacteriaceae if the dataset is extended to other members of the family.
Fig. 2.
Cluster analysis of FT-IRS spectra of strain B7T and closely related strains. Data were taken from this study and Clavel et al. (2009). Dates indicate date of measurement. Asterisks indicate results from this study.
The sensitivity of strain B7T was tested towards ten antimicrobial agents as described previously (Clavel et al., 2009). The MICs are presented in Table 1. Strain B7T was resistant to cefotaxime, a broad-spectrum antibiotic interfering with cell-wall synthesis, and oxacillin, a narrow-spectrum β-lactam antibiotic, but highly sensitive to clarithromycin, erythromycin and metronidazole. Strain B7T grew in the presence of 2 % (w/v) NaCl and at pH 6.0–9.0, but not in the presence of 0.5 % (w/v) bile salts. At pH 6.9, cysteine was not required for growth. In contrast to E. mucosicola DSM 19490T, strain B7T did not convert a variety of amino acid derivatives and was positive for glutamic acid decarboxylase (Rapid ID32A; bioMérieux). Strain B7T was also positive for arginine dihydrolase.
Because E. mucosicola DSM 19490T produces equol from the isoflavone daidzein (Matthies et al., 2008), we investigated daidzein conversion in strain B7T. Stock solutions (20 mM) of daidzein (no. D7802; Sigma-Aldrich) and (R,S)-equol (no. ALX-385-032; Axxora) were prepared in DMSO and stored at −20 °C. The daidzein stock solution was filter-sterilized prior to storage (Millex-LG PTFE membrane, 0.22 μm; Millipore). E. mucosicola DSM 19490T was used as a positive control; separate negative controls for daidzein and bacteria were included. Each bacterium was tested in duplicate at 37 °C under anoxic conditions (100 % N2) in GYBHIc. The initial concentration of daidzein was approximately 120 μM. Samples were taken over time with a syringe and stored at −20 °C. Supernatants (14 000 g for 5 min; 200 μl) were diluted fivefold in a mixture of the eluents (30 %, v/v, B in A; see below) and 50 μl samples were used for reversed-phase HPLC analysis using an Agilent HPLC 1100 Series and a Prontosil 120-5-C18 ace-EPS 5.0 μm column (250 mm × 4.6 mm; Bischoff). The mobile phase was water/acetonitrile/formic acid (94.9 : 5 : 0.1, v/v; A) and water/acetonitrile/formic acid (5 : 94.9 : 0.1, v/v; B) in a gradient mode (from 10 to 100 % B within 32 min, then 100 % B for 8 min). The flow rate was 1.0 ml min−1 and compounds were detected with a diode array detector at 300 nm. Retention times for daidzein and equol were 19.5 and 23.5 min, respectively. Calibration used two independent dilution series of daidzein that included the following concentrations: 0.1, 1, 10, 25, 50, 100 and 200 μM. Growing cells of strain B7T were not capable of converting daidzein under the experimental conditions used (Supplementary Fig. S2, available in IJSEM Online).
On the basis of the phylogenetic, chemotaxonomic and phenotypic analyses presented above, strain B7T can be distinguished from the type strain of E. mucosicola and thus represents a novel species, for which the name Enterorhabdus caecimuris sp. nov. is proposed.
Emended description of the genus Enterorhabdus Clavel et al. 2009
The description is as given previously (Clavel et al., 2009) with the following modifications. Members of the genus are aerotolerant anaerobes. No growth occurs in the presence of 0.5 % (w/v) bile salts. Do not possess glycosidases. Cysteine is not required for growth. The diamino acid in the peptidoglycan is meso- or ll-diaminopimelic acid. The main cellular fatty acid is C16 : 0 (approx. 20 % of total fatty acids). Major polar lipids are diphosphatidylglycerol and two glycolipids. Whole-cell sugars include galactose and ribose. Respiratory menaquinones are mainly monomethylated (≥60 % total lipoquinones). The G+C content is 64.2–64.5 mol%.
Description of Enterorhabdus caecimuris sp. nov.
Enterorhabdus caecimuris (ca.e.ci.mu′ris. L. n. caecum caecum; L. n. mus muris mouse; N.L. gen. n. caecimuris of the caecum of a mouse).
Gram-positive, non-motile, non-spore-forming rods (0.5×2.0 μm) growing as single cells under strictly anoxic conditions. Colonies are small (pinpoint), circular, entire and non-haemolytic after 48 h at 37 °C on Columbia blood agar. Grows in the presence of 2 % (w/v) NaCl and at 27–40 °C. Major fatty acids are C14 : 0, C16 : 0 and C18 : 1c9. Whole-cell sugars are galactose, glucose and ribose. Quinones are mono- and dimethylated. Produces glutamate decarboxylase and arginine dihydrolase but not aminopeptidase. The type strain is resistant to cefotaxime and does not convert daidzein to equol. The DNA G+C content of the type strain is 64.5 mol%.
The type strain is B7T (=DSM 21839T =CCUG 56815T), isolated from the caecum of a C3H/HeJBir mouse.
Supplementary Material
Acknowledgments
We are grateful to Professor J. P. Euzéby (Ecole Nationale Vétérinaire, Toulouse, France) for his help with Latin etymology, to Benjamin Tiemann for technical assistance and to members of the Culture Collection University of Göteborg (CCUG), especially Dr Enevold Falsen. This research was supported in part by National Institutes of Health grants DK071176 and DK64400.
Abbreviations
FT-IRS, Fourier-transform infrared spectroscopy
Footnotes
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA and gyrase B gene sequences of strain B7T are DQ789120 and GQ409830, respectively.
Supplementary figures showing the polar lipid compositions and daidzein conversion abilities of strain B7T and the type strain of Enterorhabdus mucosicola are available with the online version of this paper.
References
- Cashion, P., Holder-Franklin, M. A., McCully, J. & Franklin, M. (1977). A rapid method for the base ratio determination of bacterial DNA. Anal Biochem 81, 461–466. [DOI] [PubMed] [Google Scholar]
- Clavel, T., Charrier, C., Braune, A., Wenning, M., Blaut, M. & Haller, D. (2009). Isolation of bacteria from the ileal mucosa of TNFdeltaARE mice and description of Enterorhabdus mucosicola gen. nov., sp. nov. Int J Syst Evol Microbiol 59, 1805–1812. [DOI] [PubMed] [Google Scholar]
- De Ley, J., Cattoir, H. & Reynaerts, A. (1970). The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12, 133–142. [DOI] [PubMed] [Google Scholar]
- Duck, L. W., Walter, M. R., Novak, J., Kelly, D., Tomasi, M., Cong, Y. & Elson, C. O. (2007). Isolation of flagellated bacteria implicated in Crohn's disease. Inflamm Bowel Dis 13, 1191–1201. [DOI] [PubMed] [Google Scholar]
- Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41, 95–98. [Google Scholar]
- Huß, V. A. R., Festl, H. & Schleifer, K. H. (1983). Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst Appl Microbiol 4, 184–192. [DOI] [PubMed] [Google Scholar]
- Kirschner, C., Maquelin, K., Pina, P., Ngo Thi, N. A., Choo-Smith, L. P., Sockalingum, G. D., Sandt, C., Ami, D., Orsini, F. & other authors (2001). Classification and identification of enterococci: a comparative phenotypic, genotypic, and vibrational spectroscopic study. J Clin Microbiol 39, 1763–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley, R. E., Backhed, F., Turnbaugh, P., Lozupone, C. A., Knight, R. D. & Gordon, J. I. (2005). Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A 102, 11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley, R. E., Turnbaugh, P. J., Klein, S. & Gordon, J. I. (2006). Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023. [DOI] [PubMed] [Google Scholar]
- Maruo, T., Sakamoto, M., Ito, C., Toda, T. & Benno, Y. (2008). Adlercreutzia equolifaciens gen. nov., sp. nov., an equol-producing bacterium isolated from human faeces, and emended description of the genus Eggerthella. Int J Syst Evol Microbiol 58, 1221–1227. [DOI] [PubMed] [Google Scholar]
- Matthies, A., Clavel, T., Gutschow, M., Engst, W., Haller, D., Blaut, M. & Braune, A. (2008). Conversion of daidzein and genistein by an anaerobic bacterium newly isolated from the mouse intestine. Appl Environ Microbiol 74, 4847–4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesbah, M., Premachandran, U. & Whitman, W. (1989). Precise measurement of the G+C content of deoxyribonucleic acid by high performance liquid chromatography. Int J Syst Bacteriol 39, 159–167. [Google Scholar]
- Minamida, K., Ota, K., Nishimukai, M., Tanaka, M., Abe, A., Sone, T., Tomita, F., Hara, H. & Asano, K. (2008). Asaccharobacter celatus gen. nov., sp. nov., isolated from rat caecum. Int J Syst Evol Microbiol 58, 1238–1240. [DOI] [PubMed] [Google Scholar]
- Rhuland, L. E., Work, E., Denman, R. F. & Hoare, D. S. (1955). The behavior of the isomers of α,ε-diaminopimelic acid on paper chromatograms. J Am Chem Soc 77, 4844–4846. [Google Scholar]
- Rossello-Mora, R. & Amann, R. (2001). The species concept for prokaryotes. FEMS Microbiol Rev 25, 39–67. [DOI] [PubMed] [Google Scholar]
- Santos, S. R. & Ochman, H. (2004). Identification and phylogenetic sorting of bacterial lineages with universally conserved genes and proteins. Environ Microbiol 6, 754–759. [DOI] [PubMed] [Google Scholar]
- SasserM. (1990). Identification of bacteria by gas chromatography of cellular fatty acids, MIDI Technical Note 101. Newark, DE: MIDI Inc.
- Stackebrandt, E. & Goebel, B. M. (1994). Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44, 846–849. [Google Scholar]
- Staneck, J. L. & Roberts, G. D. (1974). Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol 28, 226–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg, J. P., Elson, C. O., Bedigian, H. & Birkenmeier, E. H. (1994). Spontaneous, heritable colitis in a new substrain of C3H/HeJ mice. Gastroenterology 107, 1726–1735. [DOI] [PubMed] [Google Scholar]
- Tamaoka, J. & Komagata, K. (1984). Determination of DNA base composition by reversed-phase high-performance liquid chromatography. FEMS Microbiol Lett 25, 125–128. [Google Scholar]
- Visuvanathan, S., Moss, M. T., Standord, J. L., Hermon-Taylor, J. & McFadden, J. J. (1989). Simple enzymatic method for isolation of DNA from diverse bacteria. J Microbiol Methods 10, 59–64. [Google Scholar]
- Wenning, M., Scherer, S. & Naumann, D. (2008). Infrared spectroscopy in the identification of microorganisms. In Vibrational Spectroscopy for Medical Diagnosis, pp. 71–96. Edited by M. Diem, P. R. Griffith & J. M. Chalmers. Chichester, UK: Wiley.
- Whiton, R. S., Lau, P., Morgan, S. L., Gilbart, J. & Fox, A. (1985). Modifications in the alditol acetate method for analysis of muramic acid and other neutral and amino sugars by capillary gas chromatography-mass spectrometry with selected ion monitoring. J Chromatogr 347, 109–120. [DOI] [PubMed] [Google Scholar]
- Würdemann, D., Tindall, B. J., Pukall, R., Lünsdorf, H., Strömpl, C., Namuth, T., Nahrstedt, H., Wos-Oxley, M., Ott, S., Schreiber, S., Timmis, K. N. & Oxley, A. P. A. (2009). Gordonibacter pamelaeae gen. nov., sp. nov., a new member of the Coriobacteriaceae isolated from a patient with Crohn's disease and reclassification of Eggerthella hongkongensis Lau et al. 2006 as Paraeggerthella hongkongensis gen. nov., comb. nov. Int J Syst Evol Microbiol 59, 1405–1415. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.