Abstract
GB virus C (GBV-C) is a common, non-pathogenic human virus that infects lymphocytes. Persistent GBV-C infection of humans with coexistent human immunodeficiency virus (HIV) infection is associated with prolonged survival, and GBV-C replication inhibits HIV replication in vitro. A GBV-C virus variant was identified in chimpanzees in 1998 and was named GBV-Ctrog or GBV-Ccpz. The prevalence and natural history of GBV-C in chimpanzees remains uncharacterized. We examined the sera from 235 captive chimpanzees for the presence of GBV-C viraemia, viral persistence and clearance, E2 antibody kinetics and RNA sequence diversity. Sequences from six isolates shared more sequence identity with GBV-Ccpz than with human GBV-C. The prevalence of GBV-Ccpz viraemia and E2 antibody in chimpanzees (2.5 and 11 %, respectively) was similar to human GBV-C prevalence in healthy human blood donors (1.8 and 9 %, respectively). Persistent GBV-Ccpz infection occurred in two of the six viraemic animals and was documented for 19 years in one animal. Host subspecies troglodyte GBV-C isolates and published verus GBV-C isolates shared a high degree of sequence identity, suggesting that GBV-C in chimpanzees should be identified with a chimpanzee designation (GBV-Ccpz). The prevalence and natural history of chimpanzee GBV-C variant (GBV-Ccpz) appears to be similar to human GBV-C infection. The chimpanzee could serve as an animal model to study HIV–GBV-C co-infection.
INTRODUCTION
Following the discovery of hepatitis C virus (HCV) in 1989, virus discovery groups searched for novel aetiological agents responsible for non-A, non-B and non-C hepatitis. In the process, human and primate viruses related to HCV were identified. Abbott Laboratories identified two viruses in tamarins which they named GB virus A (GBV-A) and GB virus B (GBV-B) (Schaluder et al., 1995). The tamarins in which these viruses were identified had been inoculated with the 12th passage of tamarin plasma. The initial tamarin had been inoculated with serum from a surgeon with non-A, non-B hepatitis whose initials were G. B. (Schaluder et al., 1995). GBV-A and GBV-B were not identified in any human sera (Schaluder et al., 1995); however, using degenerate oligonucleotides to amplify related viral sequences, these investigators discovered a human virus which they named GB virus C (GBV-C) (Simons et al., 1995). Concurrently, Genelabs Inc. discovered a virus in a patient with HCV infection that they called hepatitis G virus (HGV) (Linnen et al., 1996). Sequence comparisons revealed that HGV and GBV-C were different isolates of the same species (Linnen et al., 1996). Based on phylogenetic relationships, the GB viruses and HCV are classified as members of the family Flaviviridae. GBV-A is closely related to GBV-C, and neither virus is associated with hepatitis or any other disease. GBV-B is more closely related to HCV and causes hepatitis in tamarins and owl monkeys (Schaluder et al., 1995).
GBV-C is a lymphotropic virus associated with improved survival in HIV-infected individuals (Stapleton et al., 2004; Zhang et al., 2006). The prevalence of GBV-C viraemia ranges from 1 to 5 % in healthy human blood donors and is significantly higher (up to 42 %) in individuals with other blood-borne or sexually transmitted infections (Barnes et al., 2007; Mohr & Stapleton, 2009; Rey et al., 2000; Stapleton, 2003; Thomas et al., 1998; Williams et al., 2004). GBV-C may cause persistent infection, and approximately 80 % of HIV-infected individuals maintain GBV-C viraemia for at least 5 years (Williams et al., 2004). However, the majority of immune-competent individuals appear to clear GBV-C viraemia within 2 years (Hitzler & Runkel, 2004; Theodore & Lemon, 1997; Thomas et al., 1998). Unlike HCV, antibodies to GBV-C are not readily detected during viraemia (Heuft et al., 1998); however, antibodies to the GBV-C envelope glycoprotein E2 are detected in individuals following clearance of viraemia. Concurrent detection of GBV-C E2 antibodies and viraemia is uncommon, with fewer than 7 % of individuals having concurrent anti-E2 antibody and GBV-C viraemia (Lefrère et al., 1997; Sauleda et al., 1999). Anti-E2 antibodies appear to partially protect against reinfection (Hassoba et al., 1998; Thomas et al., 1998; Tillmann et al., 1998). Based on the prevalence of E2 antibody to GBV-C RNA in healthy blood donors, it appears that approximately 75–80 % of GBV infections are cleared (Heuft et al., 1998).
Abbott Laboratories identified a variant of GBV-C (GBV-Ctrog) in an HCV-infected chimpanzee with resolving hepatitis (Birkenmeyer et al., 1998), and reported a near-complete genome sequence (GenBank accession no. AF070476) (Birkenmeyer et al., 1998). Adams et al. (1998) also identified GBV-C RNA in three of 39 non-captive chimpanzees (subspecies troglodytes and verus) that they called GBV-Ccpz. For the remainder of the paper, the chimpanzee variant of GBV-C will be noted with the designation GBV-Ccpz rather than GBV-Ctrog. GBV-Ccpz infection was not found in human or macaque monkey blood samples (Birkenmeyer et al., 1998). The GBV-Ccpz polyprotein shares 83.6 % aa identity with GBV-C, while human GBV-C isolates are >95 % identical (Mohr & Stapleton, 2009; Muerhoff et al., 2005; Pavesi, 2001). Based on limited phylogenetic analysis of sequences from the 5′ NTR region, helicase and RNA-dependent RNA polymerase (RdRp), all of the GBV-Ccpz sequences are monophyletic within a group of GBV-C viruses from humans and chimpanzees (Adams et al., 1998). Thus, GBV-Ccpz is considered a chimpanzee variant of GBV-C rather than a separate genotype. Sequence analyses of all available chimpanzee GBV-C sequences from Abbott Laboratories (named GBV-Ctrog) and Adams et al. (named GBV-Ccpz) demonstrated that these viruses were different isolates of the GBV-Ccpz variant (Adams et al., 1998; Birkenmeyer et al., 1998).
Although Adams et al. detected GBV-Ccpz viraemia in three of 39 non-captive chimpanzees and generated partial sequences for one chimpanzee with samples 24 months apart, the prevalence and natural history of GBV-Ccpz has not been otherwise examined (Adams et al., 1998). At the time GBV-Ccpz was identified, serological reagents to detect GBV-C E2 antibodies were not available, so there are no data published on the presence of E2 antibodies in chimpanzees. In this study, we examine the prevalence and natural history of GBV-Ccpz in a large cohort of captive chimpanzees.
RESULTS
GBV-Ccpz prevalence and natural history in captive chimpanzees
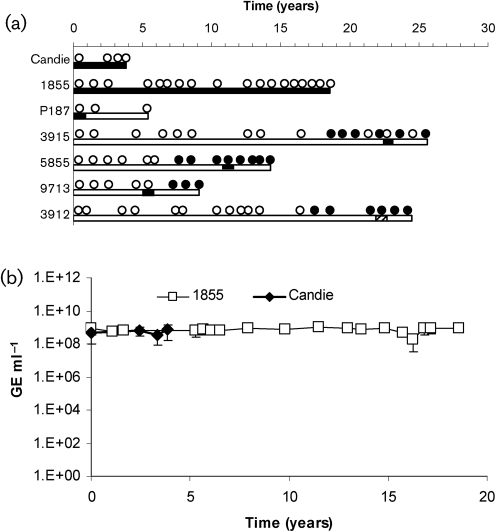
Serum samples from 235 captive chimpanzees were tested by nested RT-PCR using primers designed from a human GBV-C 5′ NTR sequence (GenBank accession no. AF121950) or by real-time RT-PCR using primers and probe designed to amplify GBV-Ccpz. Seven of the 235 (3.0 %) samples contained GBV-C RNA. One of these samples came from the South west Foundation for Biomedical Research (SFBR) and the remaining six samples came from the University of Texas MD Anderson Cancer Center. Sequence analysis was successful for five of the samples (Candie, 1855, P187, 3915 and 3912) and alignments of four of the sequences demonstrated that the sequences aligned more closely with GBV-Ccpz than with human GBV-C and one sequence aligned most closely with human GBV-C (Fig. 1a). Sequence analysis was not successful for two of the seven animals, although GBV-Ccpz viraemia is presumed because the amplification was successful only when GBV-Ccpz-specific primers were utilized. Two of the GBV-Ccpz-positive samples tested positive for all available samples, demonstrating persistent infection of at least 4 or 19 years, respectively (chimpanzees Candie and 1855) (Fig. 1a). The remaining four animals had transient viraemia with only one sample containing GBV-Ccpz RNA (Fig. 1a).
Fig. 1.
Evolution of GBV-C viraemia and E2 antibody in chimpanzees with GBV-C infection. (a) Results of the serum of seven animals tested for GBV-C E2 antibody (•, positive result; ○, negative result) and GBV-Ccpz RNA (filled bars, positive result for GBV-Ccpz RNA; dashed bars, positive result for human GBV-C RNA; open bars, negative result for either RNA). (b) GBV-Ccpz viraemia titres [genome equivalents (GE) ml−1] were measured by real-time PCR at multiple time points during persistent infection for chimpanzees Candie (◊) and 1855 (□). The first available samples are marked at t=0.
E2 antibody was detected in 26 of the 235 chimpanzee serum samples (11.1 %). The two persistently infected chimpanzees did not have E2 antibody detected in any of their samples, and one chimpanzee with transient viraemia did not develop E2 antibodies (Fig. 1a). In contrast, E2 antibodies were detected in the other three GBV-Ccpz transiently infected chimpanzees. One of these animals had detectable E2 antibody levels after GBV-Ccpz viraemia, consistent with seroconversion, while the other two transiently viraemic animals had E2 antibody detected before and after GBV-Ccpz viraemia. The chimpanzee with transient human GBV-C viraemia was also positive for E2 antibody on multiple sample dates surrounding the period of viraemia. None of the chimpanzees received human blood products (Table 1).
Table 1.
Chimpanzee demographic information, blood-product exposure history and virus exposure history
M, Male; F, female; DOB, date of birth; RBC, red blood cells.
| Chimpanzee ID | GBV-C RNA+ sample date (day/month/year) | Sex | DOB (day/month/year) | HIV | HCV | Blood-product/virus exposure history | ||
|---|---|---|---|---|---|---|---|---|
| Exposure | Status | Exposure | Status | |||||
| P187 | 24/3/1999 | M | 24/11/1984 | No | Negative | Yes | Negative | Sheep RBC in 7/10/1998; chimpanzee plasma in 1/12/2000 |
| Candie | 1/2/1988–17/12/1991 | F | 27/4/1982 | No | Negative | No | Negative | None |
| 1855 | 19/9/1991–31/3/2009 | F | 1/1/1965 | No | Negative | No | Negative | Slow virus; Aleutian disease |
| 3915 | 9/5/2006 | F | 25/4/1978 | No | Negative | No | Negative | Hepatitis A virus; hepatitis B virus; vaccinia virus |
| 5855 | 22/5/2006 | M | 24/7/1992 | No | Negative | Yes | Negative | Respiratory syncytial virus; hepatitis B virus |
| 9713 | 27/1/2005 | F | 24/8/1999 | No | Negative | No | Negative | None |
| 3912 | 11/9/2006 | M | 18/4/1978 | No | Negative | No | Negative | Hepatitis E virus; Respiratory syncytial virus |
Among the chimpanzees with persistent viraemia (Candie and 1855), the serum viral load remained constant with a mean of 5.3×108 genome equivalents (GE) ml−1 for chimpanzee Candie and 7.3×108 GE ml−1 for chimpanzee 1855 over 4 and 19 years, respectively (Fig. 1b).
Sequences from the 5′ NTR and non-structural protein (NS)5A/B coding region were determined at early and late infection time points in the animals with persistent infection, chimpanzees 1855 and Candie (Fig. 2). No nucleotide changes were observed in a 329 nt sequence from the 5′ NTR of chimpanzees 1855 and Candie (data not shown; GenBank accession nos HM626487, HM626488, HM626489 and HM626490). The rate of substitution in a 394 nt segment in the NS5A/B coding region was similarly low, with Candie showing only 1 nt substitution over a 4-year period and 1855 showing 6 nt substitutions over a 16-year period (Fig. 2). The amino acid sequences of this NS5A/B region from these longitudinal samples were identical in both animals.
Fig. 2.
Nucleotide alignment of GBV-Ccpz partial NS5A/B sequences obtained at early and late time points during persistent infections of chimpanzees Candie (early time point; HM638235) and 1855 (early time point; HM638234). Consensus sequences of six quasispecies (as described in Fig. 3) from the late infection time points were compared. Arrowhead denotes the putative start side of NS5B and dots represent identical bases.
Sequence diversity among GBV-Ccpz isolates
To study GBV-Ccpz NS5A/B sequence heterogeneity, six clones each from chimpanzees Candie and 1855 were compared with the consensus sequence (the sequence which occurs with the highest frequency for each nucleotide position; Ruiz et al., 2010). Chimpanzee Candie had only one of six clones identical to the consensus sequence after 4 years of infection, resulting in a heterogeneity index of 0.83 (the proportion of GBV-Ccpz clones not bearing the predominant sequence) (Fig. 3a). Chimpanzee Candie demonstrated a mean of 2.8 substitutions per clone in the 394 nt sequence examined, with transitions (A↔G or C↔T) accounting for 64.7 % of the total number of substitutions. At 16 years post-infection (p.i.), chimpanzee 1855 did not have a predominant nucleotide sequence (Fig. 3b), which accounted for a heterogeneity index of 1.0, consistent with the prediction of an error-prone RdRp and the generation of quasispecies in serum. Chimpanzee 1855 demonstrated a mean of 4.2 substitutions per clone, and transitions accounted for 84 % of these substitutions (Fig. 3b). Comparison of non-synonymous to synonymous substitutions (dN/dS ratio) in the six RdRp sequences demonstrated a ratio of <0.25 for chimpanzees Candie and 1855, indicating that there was not positive selection. Sequence diversity was not detected in another chimpanzee (3915) with transient GBV-Ccpz infection, with all five clones having an identical sequence (heterogeneity index of 0; data not shown; GenBank accession no. HM626492).
Fig. 3.
Nucleotide alignment of GBV-Ccpz partial NS5A/B sequences recovered from individual clones (1–6) isolated from serum from two chimpanzees: (a) Candie (HM626501–HM626506), 4 years p.i. and (b) 1855 (HM626495–HM626500), 16 years p.i. Arrowhead denotes the putative start site of NS5B and dots represent identical bases.
Phylogenetic relationships of GBV-Ccpz isolates
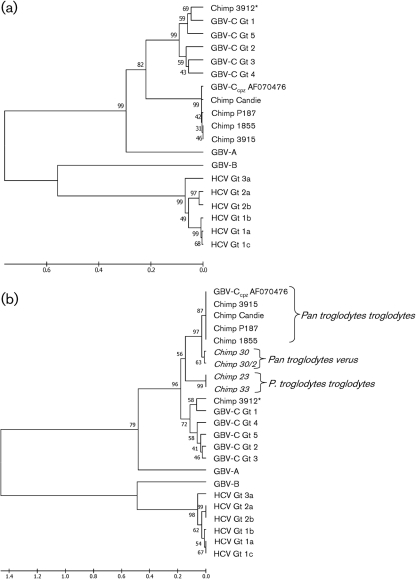
Human GBV-C isolates can be grouped into five, or possibly six, genotypes (Muerhoff et al., 2006). GBV-C, HCV, GBV-A and GBV-B 5′ NTR sequences were compared with the published GBV-Ccpz 5′ NTR sequence (GenBank accession no. AF070476), and the GBV-Ccpz and human GBV-C 5′ NTR sequences identified in this captive chimpanzee population. As predicted, the newly generated GBV-Ccpz sequences and AF070476 form a monophyletic group separate from the human GBV-C sequences (Fig. 4a).
Fig. 4.
Phylogenetic relationships of 5′ NTR of GB viruses and hepaciviruses. (a) 5′ NTR nucleotide sequences from GBV-Ccpz, GBV-C, HCV, GBV-A and GBV-B, and the sequences from chimpanzees P187 (HM638236), 1855 (HM626488), Candie (HM626490), 3912 (HM769722) and 3915 (HM626491) were aligned with clustal w. There are 308 nt in the final dataset, although only 137 nt were available for sequence alignment with 3912 (marked with an asterisk). (b) GBV-Ccpz 5′ NTR sequences from non-captive chimpanzees (noted in italics; see text) were included in the comparison. There are 98 nt in the final dataset except for 3912, for which there were 45 nt available. The evolutionary distances were computed using the maximum composite likelihood method. Bootstrap values are shown for each branch point. Scales indicate the number of base substitutions per site.
A GBV-C isolate with a 5′ NTR sequence that aligned more closely with human GBV-C sequences from genotype 1 was identified in one chimpanzee (number 3912) (Fig. 4). This GBV-C isolate was genotype 1, which correlates with African human isolates (Muerhoff et al., 2006). This animal did not receive human blood products, and the mode of transmission is not known. Since chimpanzees can support experimental human GBV-C infection (Bukh et al., 1998), and other animals in the colony received human blood products including blood from humans with HIV and HCV infection, it is possible that the animal acquired human GBV-C via intra-colony transmission.
Adams et al. (1998) published partial 5′ NTR sequences from three non-captive chimpanzees (subspecies troglodytes and verus), including one animal with two samples obtained 24 months apart. The 5′ NTR sequences of these isolates share less sequence identity with the published GBV-Ccpz sequence (AF070476) and the GBV-Ccpz sequences that we characterized (Fig. 4b), suggesting that GBV-Ccpz sequences from non-captive chimpanzees differ from captive chimpanzees. 5′ NTR sequences obtained from the chimpanzee of the subspecies verus (chimpanzee 30), are more similar to the GBV-Ccpz sequence AF070476 obtained from the subspecies troglodytes than are the sequences from the remaining non-captive chimpanzees (23 and 33), which were also obtained from troglodytes subspecies hosts. Thus, GBV-Ccpz infects both subspecies of chimpanzee, troglodytes and verus, and does not strictly co-speciate with either animal host. However, because the newly studied chimpanzees are captive animals, it is possible that the virus was transmitted in captivity, and our results may not accurately reflect the species diversity of GBV-Ccpz infection found in the wild.
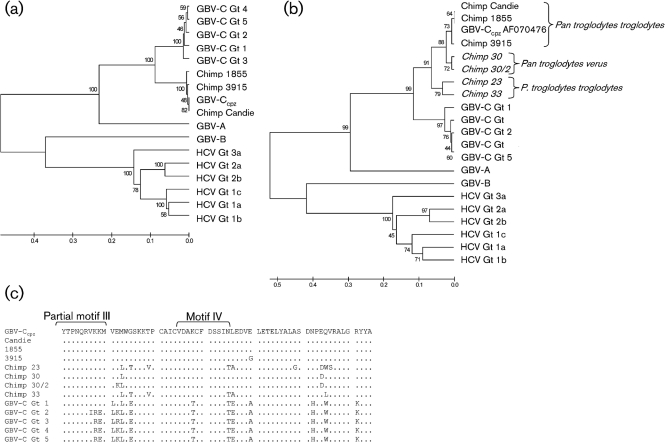
Phylogenetic relationships are best determined by comparing highly conserved functional domains including regions of the RdRp. The deduced amino acid sequences of GBV-Ccpz isolates were determined and compared to the published GBV-Ccpz sequence (AF070476), human GBV-C, HCV, GBV-A and GBV-B sequences. The GBV-Ccpz NS5B sequences we characterized shared considerable sequence identity with AF070476 and, like the 5′ NTR sequences, formed a monophyletic group separate from the human GBV-C genotypes (Fig. 5a). NS5B sequences generated from non-captive chimpanzees by Adams et al. (1998) diverged from the GBV-Ccpz sequences obtained from captive chimpanzees (Fig. 5b).
Fig. 5.
Phylogenetic relationships of RdRp of GB viruses and hepaciviruses. (a) RdRp amino acid sequences from GBV-Ccpz, GBV-C, HCV, GBV-A and GBV-B, and chimpanzees 1855 (HM626494), 3915 (HM626492) and Candie (HM626493) were aligned with clustal w. There are 231 aa in the final dataset. (b) GBV-Ccpz RdRp sequences from non-captive chimpanzees (noted in italics; see text) were included in the comparison. There are 61 aa in the final dataset. The evolutionary distances were computed using the Poisson correction method. Bootstrap values are shown for each branch point. Scales in (a) and (b) indicate the number of amino acid substitutions per site. (c) NS5B functional motifs III and IV are marked as described by Koonin (1991) from the NS5B alignment in (b).
The chimpanzee GBV-Ccpz RdRp functional motifs, as defined by Koonin (1991), were highly conserved with human GBV-C sequences. The NS5B sequences from chimpanzees 1855, 3915, Candie and the published GBV-Ccpz sequence (AF070476) were identical within the eight RdRp conserved motifs (data not shown). The partial GBV-Ccpz NS5B sequences from non-captive chimpanzees only contain sequence for RdRp motifs III and IV and differed from the GBV-Ccpz AF070476, chimpanzee 1855, 3915 and Candie sequences (Fig. 5c) (Adams et al., 1998; Koonin, 1991). The four non-captive chimpanzee sequences were identical to the AF070476 sequence in RdRp motif III, and two of the four non-captive chimpanzee sequences (23 and 33) had 1 aa substitution in the RdRp motif IV compared to AF070476 (asparagine to threonine; Fig. 5c). The substitutions were identical to the human GBV-C RdRp motif IV amino acid sequences instead of the GBV-Ccpz AF070476 sequence. In contrast, human GBV-C sequences differ from the AF070476 sequence by up to 3 aa substitutions in RdRp motif III and 2 aa substitutions in RdRp motif IV. The observation that two of the chimpanzees have 1 aa substitution in the RdRp motif IV when compared with the other chimpanzee GBV-Ccpz sequences suggests that there is sequence diversity among GBV-Ccpz sequences. More GBV-Ccpz sequences from captive and non-captive chimpanzees are necessary to determine whether there are multiple genotypes as with human GBV-C.
DISCUSSION
Human GBV-C infection may persist in human hosts for decades, although the majority of humans studied cleared infection within 2 years following infection (Alter, 1997; Hitzler & Runkel, 2004; Theodore & Lemon, 1997). Although most GBV-Ccpz infections were transient in captive chimpanzees, persistent infection was documented for up to 19 years in one animal. Serum GBV-Ccpz viral loads were high and constant in persistently infected animals (∼1×108 GE ml−1). Thus, GBV-Ccpz viral loads are similar to that observed for human GBV-C (Sauleda et al., 1999; Tillmann et al., 2001). The fact that the animals did not have a documented exposure to human blood products or tissues, and that their viral genome sequences align most closely with chimpanzee GBV-C sequences, suggests that chimpanzee infection was acquired via intra-colony transmission (Brook, 1998).
As with human GBV-C infection, seroconversion may occur with GBV-Ccpz clearance, although E2 antibody may be intermittent and was detected before and after GBV-Ccpz viraemia. The detection of E2 antibodies prior to the detection of GBV-Ccpz viraemia may reflect a GBV-Ccpz viral load below the limit of detection of the assay. Of the four chimpanzees that had both viraemia and E2 antibodies detected, three animals had concurrent detection of GBV-C E2 antibodies and viraemia. This prevalence is higher than the prevalence of concurrent viraemia and E2 antibody in humans (7 %) (Lefrère et al., 1997; Sauleda et al., 1999). E2 antibodies have not been examined in chimpanzee serum prior to this study, and more information is needed to provide a clear understanding of the relationships between E2 antibody and GBV-Ccpz viraemia. Nevertheless, the increased frequency of coexisting E2 antibody and viraemia may reflect the limited sample size or suggest that the immune mechanism for clearing human and chimpanzee GBV-C infections differs.
Like human GBV-C, GBV-Ccpz quasispecies are detected in serum (Sauleda et al., 1999; Thomas et al., 1998; Tillmann et al., 1998). GBV-Ccpz NS5A/B sequence diversity was detected in chimpanzees with persistent infection and not in a chimpanzee with transient GBV-Ccpz infection, suggesting that the generation of sequence diversity may require persistent infection. Human GBV-C quasispecies have nucleotide substitution rates of up to 8.7 % in the 5′ NTR region, 2.0 % in the E2 region and 3.3 % in the NS3 region (Ruiz et al., 2010; Zampino et al., 1999). We found lower rates of GBV-Ccpz NS5A/B nucleotide substitution in chimpanzees, with rates of 0.7 % (Candie) and 1.0 % (1855) in the NS5A/B region. None of the substitutions in NS5A/B quasispecies correlated with the mutations observed by Bukh et al. (1998) during the experimental human GBV-C infection of chimpanzees, and most GBV-Ccpz nucleotide substitutions in the NS5A/B region were silent mutations. Positive selection was not detected (dN/dS ratio <1) in the two chimpanzees with persistent infection, suggesting a lack of immunological selective pressure. Even though sequence diversity was detected, a minority of sequences predominated in human GBV-C and GBV-Ccpz during persistent infection (Radkowski et al., 1999; Ruiz et al., 2010). The mutation rate observed between early and late samples during persistent infection was only 0.2–1.5 %, and none of the mutations resulted in a change in the amino acid sequence. RNA secondary structure constraints in the NS5A and NS5B regions of the GBV-C genome may contribute to the low mutation rate over time (Davis et al., 2008; Thurner et al., 2004).
Given the worldwide distribution and presence of quasispecies of human GBV-C, there is a surprising lack of genetic diversity among human GBV-C isolates. Although few GBV-Ccpz sequences are available, the extent of sequence diversity observed between GBV-Ccpz isolates may be similar to that of human GBV-C genotypes. More GBV-Ccpz sequences need examining to determine if the GBV-Ccpz sequence diversity is significant enough to form separate genotypes as with human GBV-C.
Adams et al. (1998) found that GBV-Ccpz isolates from non-captive chimpanzees of the Pan troglodytes subspecies verus aligned as a separate group from those found in P. troglodytes subspecies troglodytes. The P. troglodytes verus chimpanzees originated from West Africa and P. troglodytes troglodytes animals originated from Cameroon and Nigeria (Adams et al., 1998). Alignment of the previously published GBV-Ccpz sequences and the sequences we characterized does not confirm that GBV-Ccpz segregates into separate verus and troglodytes subspecies groups. The GBV-Ccpz sequences we studied aligned more closely with GBV-Ccpz sequences from non-captive chimpanzees of the subspecies verus, and to a lesser extent with non-captive chimpanzees of the subspecies troglodytes, raising the possibility of interspecies transmission. Chimpanzee inter-subspecies transmission of GBV-Ccpz is feasible because human GBV-C can infect both chimpanzees (Bukh et al., 1998) and humans. Our data suggest that it may be more appropriate to call the chimpanzee variant GBV-Ccpz to signify that this virus infects both verus and troglodytes subspecies as suggested by Adams et al. (1998).
Another GB virus, GBV-A, has a species-specific pattern of sequence divergence, and the levels of sequence variation between GBV-A found in different species are similar to the relative sequence distance between human GBV-C and GBV-Ccpz. This is consistent with the hypothesis that GBV-C may have evolved with a common ancestor of humans and chimpanzees into the distinct GBV-C and GBV-Ccpz variants (Adams et al., 1998). The similarities of human GBV-C and GBV-Ccpz (serum viral load, seroconversion and persistence) and the length of time that each virus has existed in its host since humans and primates evolved separately suggests that chimpanzee GBV-Ccpz infection could serve as an animal model of GBV-C–HIV-1 interaction in vivo. Chimpanzees also support experimental infection with human GBV-C with viral loads of 106–107 GE ml−1 (Bukh et al., 1998). Thus, HIV-1 co-infection with either GBV-C or GBV-Ccpz could be used to examine HIV–GBV-C interactions in vivo. Finally, it is unclear why human GBV-C or GBV-Ccpz viraemia persists in some hosts but not others. The chimpanzee may provide a model to study host factors related to clearance and persistence of infection.
METHODS
Sample identification.
Chimpanzee (P. troglodytes troglodytes) serum samples (frozen serum samples) were obtained from repositories located at the SFBR [National Heart, Lung and Blood Institute (NHLBI) colony and South-west National Primate Research Center colony (n=81)] San Antonio, TX and from the University of Texas MD Anderson Cancer Center (n=154) Bastrop, TX. A single serum sample was tested for GBV-C RNA, and longitudinal samples were studied in animals that tested positive when available. Samples were also tested for the presence of anti-GBV-C E2 antibodies. Demographic information, infection history and blood product history for GBV-C RNA+ animals are shown in Table 1.
GBV-Ccpz RNA detection.
RNA was extracted from chimpanzee serum samples using the QIAamp Viral RNA Mini kit (Qiagen). RNA was stored at −80 °C until use. Reverse transcription was performed using a Moloney murine leukaemia virus reverse transcriptase mutant with reduced RNase H activity (SuperScript II; Invitrogen), and PCR was performed using high-fidelity Taq polymerase (Platinum Taq DNA Polymerase High Fidelity; Invitrogen). Oligonucleotide primers employed are shown in Table 2.
Table 2.
Oligonucleotide primer sequences utilized to detect GBV-C RNA
NS5A, Non-structural protein 5A; NS5B, non-structural protein 5B; +, sense primer; −, antisense primer; Pr, probe sequence.
| Region | RT-PCR product size (bp) | GBV-C primer sequence (5′−3′) | |
|---|---|---|---|
| Outer | Inner | ||
| GBV-C 5′ NTR | 92 | +GGCGACCGGCCAAAA | Pr-AGGGTTGGTAGGTCGTAAATCCCGGTCA |
| −CTTAAGACCCACCTATAGTGGCTACC | |||
| GBV-Ctrog/cpz 5′ NTR | 77 | +AATGCATGGGGCCACCC | Pr-CTGCAGCCGGGGTAGACCAA |
| −ATGCCACCCGACCTCAC | |||
| GBV-C 5′ NTR | 203 | +AAGCCCCATAAACCGACGCC | +CGGCCAAAAGGTGGTGGATG |
| −TGAAGGGCGACGTGGACCGT | −GTAACGGGCTCGGTTTAACG | ||
| GBV-Ctrog/cpz 5′ NTR | 364 | +TTGGCAGGTCGTAAATCC | +GCCATTCTGGTAGCACCT |
| −GCGCAACAGTTTGTGAGG | −GACCTCACCCGAAGGATT | ||
| GBV-Ctrog/cpz NS5A/B | 388 | +GCAGCCATGGGCTGGGGATCTAAG | +GAAACACCTGAAGGGAAAATGGC |
| −TCTGATGCCAAAGCATAGAGCTCAGTCTC | −TCTGATGCCAAAGCATAGAGCTCAGTCTC | ||
| GBV-Ctrog/cpz NS5B | 781 | +GAAACACCTGAAGGGAAAATGGC | +GGAGGTCTTCTACCGAGATCGGAA |
| −GGTGCCAAGGGTAGAGCAAACAA | −GGTGCCAAGGGTAGAGCAAACAA | ||
PCR products were purified using the QIAquick PCR purification kit (Qiagen), ligated with pCR2.1 (TA Cloning kit, Invitrogen), and INVFα or DH5α competent cells (Invitrogen) were transformed. Six colonies were randomly selected to study sequence diversity. Plasmid DNA was purified (WizardPlus SV Miniprep DNA Purification System; Promega) and sequenced (ABI sequencer; University of Iowa DNA Facility). Nucleotide sequences were entered into GenBank with accession numbers HM626487–HM626506, HM638234–HM638236 and HM769722.
Sequence analysis was performed using DNAman (Linnen, Biosoft), and phylogenetic and molecular evolutionary analyses were conducted using mega version 4 (Tamura et al., 2007). Sequences were aligned with the clustal w method, evolutionary histories were inferred using the unweighted pair-group method with arithmetic mean (UPGMA) method (Sneath & Sokal, 1973) and bootstrap consensus trees were inferred from 2000 replicates (Felsenstein, 1985). The evolutionary distances were computed using the Maximum Composite Likelihood method (Tamura et al., 2004) or the Poisson correction method (Zuckerkandl & Pauling, 1965). dN and dS values were estimated using the Nei-Gojobori method in mega4 (Nei & Gojobori, 1986; Tamura et al., 2007). GBV-C isolates representing the different genotypes were: AB003291, genotype 1; AF121950, genotype 2; U94695, genotype 3; AB003292, genotype 4; and AY949771, genotype 5. Representative isolates of three HCV genotypes were: AF0011753, genotype 1a; AF333324, genotype 1b; D14853, genotype 1c; D00944, genotype 2a; D10988, genotype 2b; and AF046866, genotype 3a. GBV-A (U94421) and GBV-B (AJ277947) sequences were also analysed.
For real-time PCR, RNA was amplified using 5′-NTR primers and a 6-carboxyfluorescein/6-carboxytetramethylrhodamine-labelled probe as described previously (Souza et al., 2006) (Table 2) using the SuperScript II Platinum One-step Quantitative RT-PCR (Invitrogen) as recommended by the manufacturer. A standard curve was generated using an 842 nt GBV-C 5′ NTR (GenBank accession no. AF121950) synthetic RNA and was confirmed for GBV-C amplification by terminal dilution. This standard curve was also used for GBV-Ccpz. Results were analysed with 7500 System SDS Software.
E2 antibody detection.
Serum samples were tested for E2 antibodies with either the μPlate anti-HGenv test (kindly provided by Dr Georg Hess, Roche Diagnostics, Mannheim, Germany), or by using an in-house assay when the commercial assay was no longer available. The sensitivity and specificity of the μPlate anti-HGenv test and in-house assay correlated (overall regression=0.76). For the in-house assay, Nunc Immobilizer plates were coated with recombinant GBV-C E2 expressed in CHO cells as described previously (McLinden et al., 2006), blocked with PBS containing 0.02 % Triton X, 0.02 % azide, 1 % BSA and 2.5 % FCS. Serum diluted 1 : 50 was added to the wells for 1 h at 37 °C. Wells were washed and bound antibody was detected using alkaline-phosphatase-labelled anti-human Fc antibodies (Sigma) followed by incubation with p-nitrophenylphosphate (Sigma) diluted in diethanolamine buffer for 1 h at 37 °C. The absorbance was measured at 405 nm after 30 min.
Acknowledgments
We thank Dr Inara Souza and Donna Klinzman from the University of Iowa for technical assistance. We thank Dr Kristin Barnhart and Rebekah Jones from the University of Texas MD Anderson Cancer Center, Department of Veterinary Sciences, for providing chimpanzee serum samples. We thank La Shayla Morrow at SFBR for providing assistance with the samples. This work was supported in part by a Merit Review grant from the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development (J. T. S.), a grant from the National Institutes of Health (RO1 AI-58740, J. T. S.) and E. L. M. received support from the University of Iowa MSTP program and Virology NIH Training Grant (T-32). The NHLBI colony was supported by a contract from NHLBI and the South-west National Primate Research Center colony is supported by a P51 centre grant from NCRR, NIH.
Footnotes
The GenBank/EMBL/DDBJ accession numbers for the GBV-C sequences determined in this study are HM626487–HM626506, HM638234–HM638236 and HM769722.
References
- Adams, N. J., Prescott, L. E., Jarvis, L. M., Lewis, J. C. M., McClure, M. O., Smith, D. B. & Simmonds, P. (1998). Detection in chimpanzees of a novel flavivirus related to GB virus-C/hepatitis G virus. J Gen Virol 79, 1871–1877. [DOI] [PubMed] [Google Scholar]
- Alter, H. J. (1997). G-pers creepers, where'd you get those papers? A reassessment of the literature on the hepatitis G virus. Transfusion 37, 569–572. [DOI] [PubMed] [Google Scholar]
- Barnes, A., Allen, J. B., Klinzman, D., Zhang, W., Chaloner, K. & Stapleton, J. T. (2007). GBV-C persistence does not require CD4+ T cell preservation, and GBV-C viral load (VL) is weakly inversely related to HIV VL. In 4th IAS Conference on HIV, Sydney.
- Birkenmeyer, L. G., Desai, S. M., Muerhoff, A. S., Leary, T. P., Simons, J. N., Montes, C. C. & Mushahwar, I. K. (1998). Isolation of a GB virus-related genome from a chimpanzee. J Med Virol 56, 44–51. [DOI] [PubMed] [Google Scholar]
- Brook, M. G. (1998). Sexual transmission and prevention of the hepatitis viruses A–E and G. Sex Transm Infect 74, 395–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukh, J., Kim, J. P., Govindarajan, S., Apgar, C. L., Foung, S. K., Wages, J., Jr, Yun, A. J., Shapiro, M. & Purcell, R. H. (1998). Experimental infection of chimpanzees with hepatitis G virus and genetic analysis of the virus. J Infect Dis 177, 855–862. [DOI] [PubMed] [Google Scholar]
- Davis, M., Sagan, S. M., Pezacki, J. P., Evans, D. J. & Simmonds, P. (2008). Bioinformatic and physical characterizations of genome-scale ordered RNA structure in mammalian RNA viruses. J Virol 82, 11824–11836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. [DOI] [PubMed] [Google Scholar]
- Hassoba, H. M., Pessoa, M. G., Terrault, N. A., Lewis, N. J., Hayden, M., Hunt, J. C., Qiu, X., Lou, S. C. & Wright, T. L. (1998). Antienvelope antibodies are protective against GBV-C reinfection: evidence from the liver transplant model. J Med Virol 56, 253–258. [PubMed] [Google Scholar]
- Heuft, H. G., Berg, T., Schreier, E., Kunkel, U., Tacke, M., Schwella, N., Hopf, U., Salama, A. & Huhn, D. (1998). Epidemiological and clinical aspects of hepatitis G virus infection in blood donors and immunocompromised recipients of HGV-contaminated blood. Vox Sang 74, 161–167. [PubMed] [Google Scholar]
- Hitzler, W. E. & Runkel, S. (2004). Prevalence, persistence and liver enzyme levels of HGV RNA-positive blood donors determined by large-scale screening and transmission by blood components. Clin Lab 50, 25–31. [PubMed] [Google Scholar]
- Koonin, E. V. (1991). The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol 72, 2197–2206. [DOI] [PubMed] [Google Scholar]
- Lefrère, J.-J., Loiseau, P., Maury, J., Lasserre, J., Mariotti, M., Ravera, N., Lerable, J., Lefevre, G., Morand-Jouberg, L. & other authors (1997). Natural history of GBV-C/hepatitis G virus infection through the follow-up of GBV-C/hepatitis G virus-infected blood donors and recipients studied by RNA polymerase chain reaction and anti-E2 serology. Blood 90, 3776–3780. [PubMed] [Google Scholar]
- Linnen, J., Wages, J., Zhang-Keck, Z.-Y., Fry, K. E., Krawczynski, K. Z., Alter, H., Koonin, E., Gallagher, M., Alter, M. & other authors (1996). Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science 271, 505–508. [DOI] [PubMed] [Google Scholar]
- McLinden, J. H., Kaufman, T. M., Xiang, J., Chang, Q., Klinzman, D., Engel, A. M., Hess, G., Schmidt, U., Houghton, M. & other authors (2006). Characterization of an immunodominant antigenic site on GB virus C glycoprotein E2 that is involved in cell binding. J Virol 80, 12131–12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr, E. L. & Stapleton, J. T. (2009). GB virus type C interactions with HIV: the role of envelope glycoproteins. J Viral Hepat 16, 757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muerhoff, A. S., Leary, T. P., Sathar, M. A., Dawson, G. J. & Desai, S. M. (2005). African origin of GB virus C determined by phylogenetic analysis of a complete genotype 5 genome from South Africa. J Gen Virol 86, 1729–1735. [DOI] [PubMed] [Google Scholar]
- Muerhoff, A. S., Dawson, G. J. & Desai, S. M. (2006). A previously unrecognized sixth genotype of GB virus C revealed by analysis of 5′-untranslated region sequences. J Med Virol 78, 105–111. [DOI] [PubMed] [Google Scholar]
- Nei, M. & Gojobori, T. (1986). Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3, 418–426. [DOI] [PubMed] [Google Scholar]
- Pavesi, A. (2001). Origin and evolution of GBV-C/hepatits G virus and relationships with ancient human migrations. J Mol Evol 53, 104–113. [DOI] [PubMed] [Google Scholar]
- Radkowski, M., Wang, L. F., Cianciara, J., Rakela, J. & Laskus, T. (1999). Analysis of hepatitis G virus/GB virus C quasispecies and replication sites in human subjects. Biochem Biophys Res Commun 258, 296–299. [DOI] [PubMed] [Google Scholar]
- Rey, D., Vidinic-Moularde, J., Meyer, P., Schmitt, C., Fritsch, S., Lang, J. M. & Stoll-Keller, F. (2000). High prevalence of GB virus C/hepatitis G virus RNA and antibodies in patients infected with human immunodeficiency virus type 1. Eur J Clin Microbiol Infect Dis 19, 721–724. [DOI] [PubMed] [Google Scholar]
- Ruiz, V., Giordano, M., Rivero, C. W., Minassian, M. L., Cuestas, M. L., Trinks, J., Mathet, V. L. & Oubina, J. R. (2010). GB virus C quasispecies detected in plasma and lymphocyte subsets in a natural human infection. J Gen Virol 91, 1687–1692. [DOI] [PubMed] [Google Scholar]
- Sauleda, S., Esteban, J. I., Hernandez, J. M., Reesink, H., Castella, D., Quer, J., Hess, G., Esteban, R. & Guardia, J. (1999). Evaluation of RNA and E2 antibodies in prospectively followed recipients of hepatitis G virus-infected blood. Transfusion 39, 633–638. [DOI] [PubMed] [Google Scholar]
- Schaluder, G. G., Dawson, G. J., Simons, J. N., Pilot-Matias, T. J., Gutierrez, R. A., Heynen, C. A., Knigge, M. F., Kurpiewski, G. S., Buijk, S. L. & other authors (1995). Molecular and serologic analysis in the transmission of the GB hepatitis agents. J Med Virol 46, 81–90. [DOI] [PubMed] [Google Scholar]
- Simons, J. N., Leary, T. P., Dawson, G. J., Pilot-Matias, T. J., Muerhoff, A. S., Schlauder, G. G., Desai, S. M. & Mushahwar, I. K. (1995). Isolation of novel virus-like sequences associated with human hepatitis. Nat Med 1, 564–569. [DOI] [PubMed] [Google Scholar]
- Sneath, P. H. A. & Sokal, R. R. (1962). Numerical Taxonomy Nat 193, 855–860. [DOI] [PubMed]
- Souza, I. E., Allen, J. B., Xiang, J., Klinzman, D., Diaz, R., Zhang, S., Chaloner, K., Zdunek, D., Hess, G. & other authors (2006). Optimal testing for GB virus C viremia: effect of primer selection on estimates of GBV-C prevalence and response to antiretroviral therapy. J Clin Microbiol 44, 3105–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton, J. T. (2003). GB virus type C/hepatitis G virus. Semin Liver Dis 23, 137–148. [DOI] [PubMed] [Google Scholar]
- Stapleton, J. T., Williams, C. F. & Xiang, J. (2004). GB virus C: a beneficial infection? J Clin Microbiol 42, 3915–3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., Nei, M. & Kumar, S. (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A 101, 11030–11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., Dudley, J., Nei, M. & Kumar, S. (2007). mega4: Molecular Evolutionary Genetics Analysis (mega) software version 4.0. Mol Biol Evol 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Theodore, D. & Lemon, S. M. (1997). GB virus C, hepatitis G virus, or human orphan flavivirus? Hepatology 25, 1285–1286. [DOI] [PubMed] [Google Scholar]
- Thomas, D. L., Vlahov, D., Alter, H. J., Marshall, R., Astemborski, J. & Nelson, K. E. (1998). Association of antibody to GB virus C (hepatitis G virus) with viral clearance and protection from reinfection. J Infect Dis 177, 539–542. [DOI] [PubMed] [Google Scholar]
- Thurner, C., Witwer, C., Hofacker, I. L. & Stadler, P. F. (2004). Conserved RNA secondary structures in Flaviviridae genomes. J Gen Virol 85, 1113–1124. [DOI] [PubMed] [Google Scholar]
- Tillmann, H. L., Heringlake, S., Trauwein, C., Meissner, D., Nashan, B., Schlitt, H. J., Kratochvil, J., Hunt, J., Qiu, X. & other authors (1998). Antibodies against the GB virus C envelope 2 protein before liver transplantation protect against GB virus C de novo infection. Hepatology 28, 379–384. [DOI] [PubMed] [Google Scholar]
- Tillmann, H. L., Heiken, H., Knapik-Botor, A., Heringlake, S., Ockenga, J., Wilber, J. C., Goergen, B., Detmer, J., Manns, M. P. & other authors (2001). Infection with GB virus C and reduced mortality among HIV-infected patients. N Engl J Med 345, 715–724. [DOI] [PubMed] [Google Scholar]
- Williams, C. F., Klinzman, D., Yamashita, T. E., Xiang, J., Polgreen, P. M., Rinaldo, C., Liu, C., Phair, J., Margolick, J. B. & other authors (2004). Persistent GB virus C infection and survival in HIV-infected men. N Engl J Med 350, 981–990. [DOI] [PubMed] [Google Scholar]
- Zampino, R., Pickering, J., Iqbal, M., Gaud, U., Thomas, H. C. & Karayiannis, P. (1999). Hepatitis G virus/GBV-C persistence: absence of hypervariable E2 region and genetic analysis of viral quasispecies in serum and lymphocytes. J Viral Hepat 6, 209–218. [DOI] [PubMed] [Google Scholar]
- Zhang, W., Chaloner, K., Tillmann, H. L., Williams, C. F. & Stapleton, J. T. (2006). Effect of early and late GBV-C viremia on survival of HIV infected individuals: a meta-analysis. HIV Med 7, 173–180. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl, E. & Pauling, L. (1965). Evolutionary divergence and convergence in proteins. In Evolving Genes and Proteins, pp. 97–166. Edited by Bryson, V. & Vogel, H. J.. New York. : Academic Press.