Abstract
Interaction between human cytomegalovirus uracil DNA glycosylase (UL114) and the viral DNA polymerase accessory subunit (UL44) has been reported; however, no such association was found in proteomic studies of UL44-interacting proteins. Utilizing virus expressing FLAG-tagged UL114, nuclease-resistant association of UL44 and the DNA polymerase catalytic subunit UL54 with UL114 was observed by co-immunoprecipitation. Contrary to a previous report, we observed that UL114 was much less abundant than UL44. Interaction of UL114 with UL54, independent of the UL54 carboxyl terminus, but not with UL44 was detected in vitro. Our data are consistent with a direct UL114–UL54 interaction, and suggest that UL114 and UL54 act in concert during base excision repair of the viral genome.
The presence of uracil in DNA can cause mutations. Many organisms, including some viruses, use the base excision repair (BER) pathway to remove uracil from their genomes. Human cytomegalovirus (HCMV) encodes a protein, UL114, that has uracil DNA glycosylase (UNG) activity (Ranneberg-Nilsen et al., 2008). UNG catalyses the first step in BER, hydrolysis of the N-glycosyl bond between uracil and the deoxyribose moiety in the DNA backbone. An apyrimidinic/apurinic (AP) endonuclease then generates a 3′-OH terminus at the site of hydrolysis and the lesion is repaired by a DNA polymerase and a DNA ligase. The enzymes that perform these steps in BER of the HCMV genome are unknown.
Association of UL114 and the HCMV DNA polymerase subunit UL44, detected by immunoprecipitation (IP) in infected cell lysate, and DNA-dependent interaction of purified UL144 and UL44 proteins have been reported (Prichard et al., 2005; Ranneberg-Nilsen et al., 2008). UL44 stimulates long-chain DNA synthesis, probably by holding the viral DNA polymerase catalytic subunit, UL54, on DNA (Ertl & Powell, 1992). UL44 has structural similarity to eukaryotic DNA polymerase processivity factor proliferating cell nuclear antigen (PCNA) (Appleton et al., 2004, 2006), which interacts with numerous proteins involved in DNA repair, including cellular UNG (Maga & Hubscher, 2003; Moldovan et al., 2007). There are reasons, therefore, to consider a UL114–UL44 interaction as a plausible mechanism by which UL114 could be recruited to DNA to catalyse BER.
We have previously conducted proteomic analysis under a variety of conditions to identify proteins associated with UL44 in infected cell lysate (Strang et al., 2009, 2010). In no experiment did we detect the association of UL114 with UL44. We sought to clarify why our results would differ from those reported previously.
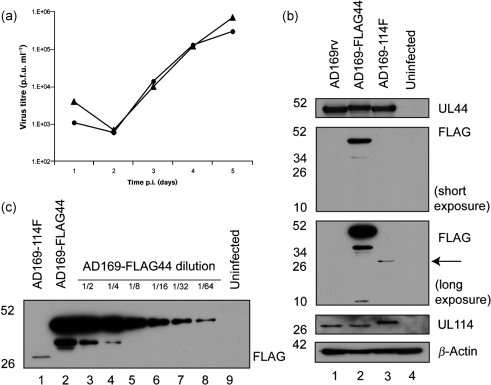
To this end, recombinant HCMV expressing a FLAG-tagged version of UL114 (UL114–FLAG) was generated. Using Red two-step recombination (Tischer et al., 2006), a single FLAG epitope (DYKDDDDK) was inserted immediately before the termination codon of the UL114 coding sequence in the bacterial artificial chromosome (BAC) AD169-BAC (Hobom et al., 2000). Viruses from this BAC (AD169-114F) and the parental BAC (AD169rv) were reconstituted in human foreskin fibroblast (HFF) cells (ATCC). The replication kinetics of AD169rv and AD169-114F infection were analysed (Fig. 1a). Incorporation of the FLAG epitope into UL114 exerted little or no effect on virus replication, as replication of AD169-114F in HFF cells was comparable to that of AD169rv, and did not display the delayed replication kinetics typical of virus that does not express UL114 (Prichard et al., 1996). HFF cells were infected with AD169rv, AD169-114F or a recombinant virus, AD169-FLAG44 (Strang et al., 2010) that expresses FLAG-tagged UL44 (FLAG–UL44). Similar levels of UL44 were observed in cells infected with each virus (Fig. 1b). Only upon long exposure of the blot could UL114–FLAG be detected in cells infected with AD169-114F with an antibody recognizing the FLAG tag. Comparing UL114–FLAG levels in AD169-UL114F-infected cell lysate with FLAG–UL44 levels in dilutions of AD169-FLAG44-infected cell lysate (Fig. 1c), UL114 was present at <1/64 the level of UL44, which contrasts with a previous report that UL114 is more abundant than UL44 (Ranneberg-Nilsen et al., 2008). This discrepancy was not due to an effect of the FLAG epitope on UL114 accumulation, as similar amounts of UL114 were detected in cells infected with all three viruses (Fig. 1b), using anti-UL114 antisera graciously provided by Toril Ranneberg-Nilsen (Rikshospitalet Medical Centre, Oslo, Norway). It is unknown why there are differences between our findings and previous conclusions (Ranneberg-Nilsen et al., 2008). We have found that the colorimetric assay used in the previous study (Ranneberg-Nilsen et al., 2008) can be rather inaccurate, and speculate that this inaccuracy may contribute to the discrepancy. A second possibility, which we think less likely, is that refolding of UL114 during Western blotting could restrict accessibility of the epitope tag.
Fig. 1.
Characterization of AD169-114F virus. (a) Replication of AD169rv (▴) and AD169-114F (▪) viruses. HFF cells were infected at an m.o.i. of 1. Virus supernatants were harvested at the indicated time points. Virus titre is represented as p.f.u. ml−1 on HFF cells. Identical results were observed in an independent experiment. (b) Western blotting of AD169rv-, AD169-FLAG44- and AD169-114F-infected cells. HFF cells were infected at an m.o.i. of 1. Cell lysates were prepared 72 h post-infection (p.i.). Blots were probed with antibodies recognizing UL44 (Virusys), β-actin (Sigma), UL114 (Ranneberg-Nilsen et al., 2008) (a kind gift from Dr Toril Ranneberg-Nilsen, Rikshospitalet Medical Centre, Oslo, Norway) and FLAG (Sigma), as indicated on the right. Short and long exposures of the anti-FLAG blot (5 and 30 s exposure, respectively) are shown. The position of UL114–FLAG is indicated by an arrow on the long-exposure blot. (c) Western blotting of uninfected (lane 9) and AD169-114F- and AD169-FLAG44-infected (lanes 1 and 2) cell lysate and a 2-fold dilution series of AD169-FLAG44-infected cell lysate (lanes 3–8) probed with anti-FLAG antibody. All samples used are those seen in Fig. 1(b). The positions of molecular mass markers (kDa) are indicated on the left.
IP was performed on lysate from HFF cells infected with AD169rv or AD169-114F using anti-FLAG antibody. This experiment was carried out as described elsewhere (Strang et al., 2010) except that, in this instance, beads bearing anti-FLAG antibody were incubated with infected cell lysate three times in order to gather enough UL114–FLAG so that this protein could be detected robustly in immunoprecipitated protein by Western blotting. This is probably due to the low levels of UL114–FLAG in the infected cell (Fig. 1). Using Western blotting, UL114–FLAG and UL44 could be detected in protein immunoprecipitated from AD169-114F-infected cell lysate (Fig. 2a, lane 2), but not in protein immunoprecipitated from AD169rv-infected cell lysate (lane 1), indicating an association of the two proteins in the lysate. This result differs from our previous observations when immunoprecipitating UL44 followed by mass spectrometry analysis (Strang et al., 2009, 2010), probably due to the low levels of UL114 in the infected cell (Fig. 1). Protein immunoprecipitated with UL114–FLAG was also examined by Western blotting using a mAb recognizing UL54. UL54 was found in protein immunoprecipitated with UL114–FLAG (Fig. 2a, lane 2). Note that both UL54 and UL114–FLAG were present in infected cell lysate below the level of detection by Western blotting (Fig. 2a, lanes 4 and 5).
Fig. 2.
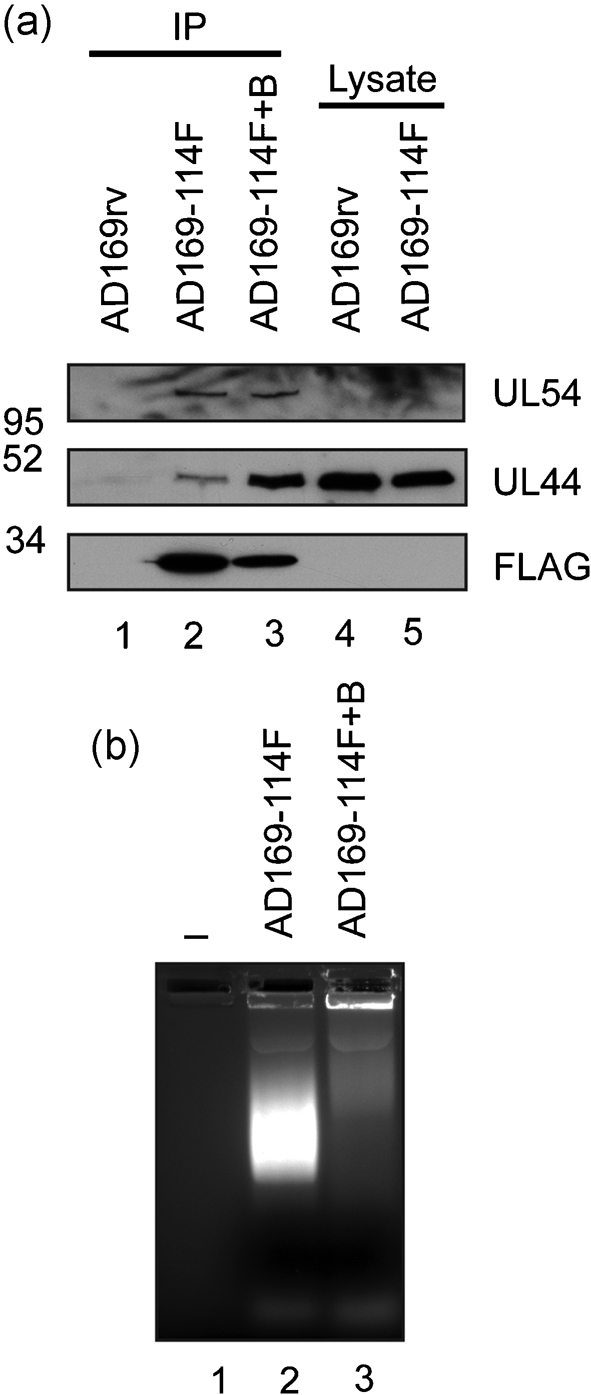
Detection of protein immunoprecipitated from AD169-114F-infected cell lysate by Western blotting. (a) IP using anti-FLAG antibodies. Lysates of HFF cells infected (m.o.i. of 3) with AD169rv or AD169-114F were prepared 72 h p.i. Protein was immunoprecipitated using an anti-FLAG antibody in the presence (lane 3) or absence (lanes 1 and 2) of the nuclease Benzonase (400 U) essentially as described elsewhere (Strang et al., 2010), except that beads bearing anti-FLAG antibody were incubated with lysate three times before washing. Protein immunoprecipitated from AD169rv (lane 1) or AD169-114F (lanes 2 and 3) or the cell lysate used for IP (lanes 4 and 5) was examined by Western blotting using antibodies recognizing FLAG (Sigma) (bottom panel), UL44 (Virusys) (middle panel) or UL54 (a kind gift from Dr Nigel Stow, MRC Virology Unit, University of Glasgow, UK) (top panel). The positions of molecular mass markers (kDa) are indicated on the left. (b) Analysis of Benzonase activity. Samples used in the IP in (a) were run out onto an ethidium bromide-stained 0.8 % agarose gel. Lanes: 1, no sample; 2, IP in the absence of Benzonase (Fig. 2a, lane 2); 3, IP in the presence of Benzonase (Fig. 2a, lane 3).
As DNA-binding proteins can associate during IP due to their adjacent binding on DNA rather than due to protein–protein interactions (Lai & Herr, 1992; Taylor & Knipe, 2004), the ability of proteins to associate with UL114–FLAG was also tested in the presence of the nuclease Benzonase (Novagen). Similar levels of UL54 and an increased level of UL44 could be observed when protein was immunoprecipitated from infected cell lysate in the presence of Benzonase (Fig. 2a, lane 3). The cell lysates analysed in Fig. 2(a) were examined on an ethidium bromide-stained agarose gel (Fig. 2b). In the absence of Benzonase, robust staining of nucleic acid could be seen (lane 2), whereas in the presence of Benzonase (lane 3), little staining of DNA was observed, indicating that Benzonase had efficiently degraded the nucleic acid in the cell lysate. The association of UL44 and UL54 with UL114–FLAG during IP was, therefore, unlikely to be due to their adjacent binding on DNA. It is possible that increased levels of UL44 were observed in immunoprecipitated protein because the action of Benzonase liberates UL44 from DNA, allowing it to interact with immunoprecipitated proteins. Similar results have been observed previously, where the presence of Benzonase resulted in greater amounts of the HCMV replication factor UL84 associating with UL44 during IP (Strang et al., 2009).
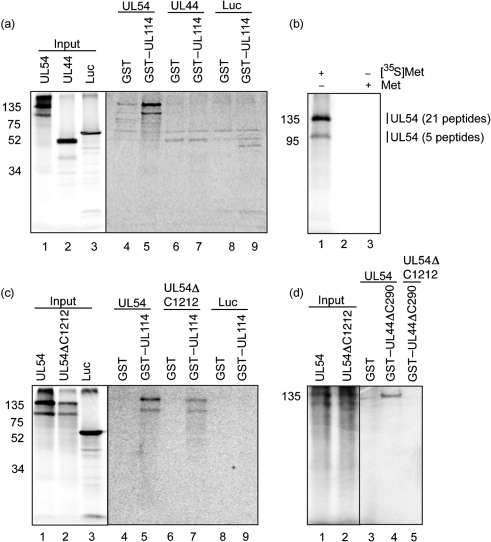
To investigate whether UL114 can bind UL44 or UL54 in the absence of any other viral proteins, the ability of radiolabelled UL44 or UL54 generated by in vitro transcription/translation to bind bacterially purified glutathione S-transferase (GST) or a fusion protein of UL114 protein fused to GST (GST–UL114) was tested as described elsewhere (Strang et al., 2009) (Fig. 3). All reagents have been described previously (Strang et al., 2009), except GST–UL114, which was generated from plasmid pGEX-6X-1 (GE Healthcare) into which UL114 had been cloned from BAC AD169-BAC (Hobom et al., 2000). To control for non-specific binding of proteins to GST or GST–UL114, the ability of radiolabelled luciferase to bind these proteins was also tested. Both UL44 and luciferase bound GST and GST–UL114 weakly at similar levels (lanes 6–9), indicating no specific interaction of UL44 with GST–UL114. However, considerably greater levels of UL54 (140 kDa) bound to GST–UL114 (lane 5) and much more than bound to GST (lane 4).
Fig. 3.
Interaction of UL54 with a GST–UL114 fusion protein. GST-pulldown assays were performed as described previously (Strang et al., 2009). (a, c, d) GST, GST–UL114 or GST–UL44ΔC290 fusion proteins were incubated with UL54, UL54ΔC1212, UL44 or luciferase [generated by using an in vitro transcription/translation reaction (Promega) in the presence of [35S]methionine (Amersham Pharmacia Biotech)] and passed over a glutathione column. The input radiolabelled proteins and proteins eluted with glutathione from each reaction are shown as indicated. The radiolabelled and GST proteins used in each reaction are noted. (b) UL54 was produced by in vitro transcription/translation in the presence of methionine (lane 3) or [35S]methionine (lane 1). Lane 2, no sample. Sections cut from the gel for LC/MS/MS analysis are indicated by vertical lines to the right of the figure, as are the number of UL54 peptides detected. In all panels, the positions of molecular mass markers (kDa) are indicated on the left.
A protein of approximately 100 kDa, which is observed routinely during in vitro transcription/translation of UL54 and is thought to be a truncated form of UL54 produced by premature termination of UL54 translation (Cihlar et al., 1997; Loregian et al., 2004), was also able to interact specifically with GST–UL114 (lanes 1 and 5). To investigate this directly, UL54 was produced by in vitro transcription/translation in the presence of either methionine or [35S]methionine (Fig. 3b). After proteins from the in vitro transcription/translation reactions had been separated by SDS-PAGE, non-radiolabelled proteins from sections of the SDS-PAGE gel were subjected to tryptic digest and tandem liquid chromatography–mass spectrometry (LC/MS/MS) to determine their identity. Peptides spanning aa 48–1064 of UL54 (1242 aa) and aa 40–581 of UL54 could be detected in 140 and 100 kDa gel sections, respectively. Given the size of the 100 kDa protein and the detection of peptides within 40 residues of the amino terminus, it is likely that this protein lacks the carboxyl terminus of UL54, as suggested elsewhere (Cihlar et al., 1997). This in turn suggests that the UL54 carboxyl terminus is dispensable for UL54–UL114 association.
To investigate whether the UL54 carboxyl terminus is required for interaction with UL114, we tested the ability of UL54ΔC1212, a UL54 mutant lacking the carboxyl-terminal 30 residues and thus incapable of binding UL44 (Loregian et al., 2004), to bind GST and GST–UL114 (Fig. 3c). As before (Fig. 3a), binding of wild-type UL54 to GST–UL114 but not GST was observed (Fig. 3c, lanes 4 and 5), whilst luciferase did not interact with either GST or GST–UL114 at any detectable level (lanes 8 and 9). Binding of UL54ΔC1212 to GST–UL114 but not GST was observed (lanes 6 and 7) (note that input levels of this protein were lower than those of UL54). The UL54 carboxyl terminus is required for UL44 binding (Loregian et al., 2004). As expected, UL54 (140 kDa species), but neither the 100 kDa species nor UL54ΔC1212, was able to bind UL44 (Fig. 3d). The UL54 carboxyl terminus is therefore not required for UL54–UL114 interaction, indicating that the mechanism of UL54–UL114 interaction differs from that of UL54–UL44 interaction. This raises that possibility that UL54 can interact with UL114 and UL44 simultaneously. This is supported by the observation that both UL44 and UL54 can be found in protein co-immunoprecipitating with UL114–FLAG (Fig. 2a).
Our experiments demonstrate association of UL114, UL44 and UL54 in infected cell lysate, but only association of UL54 with UL114 in vitro. It seems likely, therefore, that UL114–FLAG interacts with UL54 in the infected cell. The association of UL44 with UL114 in infected cell lysate (Prichard et al., 2005; Ranneberg-Nilsen et al., 2008) may, therefore, be mediated by UL54. As we have discussed elsewhere (Strang et al., 2009), the observation that purified UL44 and UL114 proteins interact in a DNA-dependent fashion in vitro (Ranneberg-Nilsen et al., 2008) may be explained by non-specific interactions between UL114 and aggregates of full-length UL44 protein prepared from Escherichia coli under conditions of low ionic strength. Moreover, it has been reported that no UL114–UL44 interaction was detected in yeast two-hybrid assays, in pull-down assays using purified His-tagged UL44 and UL114 or in co-IP of in vitro-transcribed/translated UL44 and UL114 (Ranneberg-Nilsen et al., 2008).
BER of the HCMV genome is poorly understood, but insights into this process may be gleaned from studies of a related herpesvirus, herpes simplex virus 1 (HSV-1). Interaction of the HSV-1 DNA polymerase catalytic subunit UL30 with the HSV-1 UNG UL2 has been reported (Bogani & Boehmer, 2008; Bogani et al., 2010). Also, UL30 possesses AP endonuclease and 5′-deoxyribosephosphate lyase activity (Bogani & Boehmer, 2008) and BER can be reconstituted in vitro, dependent upon the presence of UL30 and UL2 (Bogani et al., 2009). As the HSV-1 and HCMV DNA polymerase and UNG proteins are related, it is possible that, during HCMV replication, UL54 acts in concert with UL114 during BER of the HCMV genome.
Acknowledgments
We gratefully acknowledge advice and encouragement received from members of the Coen laboratory, and Nigel Stow (MRC Virology Unit, University of Glasgow, UK) and Toril Ranneberg-Nilsen (Rikshospitalet Medical Centre, Oslo, Norway) for generously providing reagents. This work was supported by NIH grants AI19838 and AI26077 to D. M. C. and an award from the William Randolph Hearst Fund to B. L. S.
References
- Appleton, B. A., Loregian, A., Filman, D. J., Coen, D. M. & Hogle, J. M. (2004). The cytomegalovirus DNA polymerase subunit UL44 forms a C clamp-shaped dimer. Mol Cell 15, 233–244. [DOI] [PubMed] [Google Scholar]
- Appleton, B. A., Brooks, J., Loregian, A., Filman, D. J., Coen, D. M. & Hogle, J. M. (2006). Crystal structure of the cytomegalovirus DNA polymerase subunit UL44 in complex with the C terminus from the catalytic subunit. Differences in structure and function relative to unliganded UL44. J Biol Chem 281, 5224–5232. [DOI] [PubMed] [Google Scholar]
- Bogani, F. & Boehmer, P. E. (2008). The replicative DNA polymerase of herpes simplex virus 1 exhibits apurinic/apyrimidinic and 5′-deoxyribose phosphate lyase activities. Proc Natl Acad Sci U S A 105, 11709–11714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogani, F., Chua, C. N. & Boehmer, P. E. (2009). Reconstitution of uracil DNA glycosylase-initiated base excision repair in herpes simplex virus-1. J Biol Chem 284, 16784–16790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogani, F., Corredeira, I., Fernandez, V., Sattler, U., Rutvisuttinunt, W., Defais, M. & Boehmer, P. E. (2010). Association between the herpes simplex virus-1 DNA polymerase and uracil DNA glycolyase. J Biol Chem [DOI] [PMC free article] [PubMed]
- Cihlar, T., Fuller, M. D. & Cherrington, J. M. (1997). Expression of the catalytic subunit (UL54) and the accessory protein (UL44) of human cytomegalovirus DNA polymerase in a coupled in vitro transcription/translation system. Protein Expr Purif 11, 209–218. [DOI] [PubMed] [Google Scholar]
- Ertl, P. F. & Powell, K. L. (1992). Physical and functional interaction of human cytomegalovirus DNA polymerase and its accessory protein (ICP36) expressed in insect cells. J Virol 66, 4126–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobom, U., Brune, W., Messerle, M., Hahn, G. & Koszinowski, U. H. (2000). Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J Virol 74, 7720–7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, J. S. & Herr, W. (1992). Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci U S A 89, 6958–6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loregian, A., Appleton, B. A., Hogle, J. M. & Coen, D. M. (2004). Residues of human cytomegalovirus DNA polymerase catalytic subunit UL54 that are necessary and sufficient for interaction with the accessory protein UL44. J Virol 78, 158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maga, G. & Hubscher, U. (2003). Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J Cell Sci 116, 3051–3060. [DOI] [PubMed] [Google Scholar]
- Moldovan, G. L., Pfander, B. & Jentsch, S. (2007). PCNA, the maestro of the replication fork. Cell 129, 665–679. [DOI] [PubMed] [Google Scholar]
- Prichard, M. N., Duke, G. M. & Mocarski, E. S. (1996). Human cytomegalovirus uracil DNA glycosylase is required for the normal temporal regulation of both DNA synthesis and viral replication. J Virol 70, 3018–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prichard, M. N., Lawlor, H., Duke, G. M., Mo, C., Wang, Z., Dixon, M., Kemble, G. & Kern, E. R. (2005). Human cytomegalovirus uracil DNA glycosylase associates with ppUL44 and accelerates the accumulation of viral DNA. Virol J 2, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranneberg-Nilsen, T., Dale, H. A., Luna, L., Slettebakk, R., Sundheim, O., Rollag, H. & Bjoras, M. (2008). Characterization of human cytomegalovirus uracil DNA glycosylase (UL114) and its interaction with polymerase processivity factor (UL44). J Mol Biol 381, 276–288. [DOI] [PubMed] [Google Scholar]
- Strang, B. L., Sinigalia, E., Silva, L. A., Coen, D. M. & Loregian, A. (2009). Analysis of the association of the human cytomegalovirus DNA polymerase subunit UL44 with the viral DNA replication factor UL84. J Virol 83, 7581–7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strang, B. L., Boulant, S. & Coen, D. M. (2010). Nucleolin can associate with the human cytomegalovirus DNA polymerase accessory subunit UL44 and is necessary for viral replication. J Virol 84, 1771–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, T. J. & Knipe, D. M. (2004). Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J Virol 78, 5856–5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischer, B. K., von Einem, J., Kaufer, B. & Osterrieder, N. (2006). Two-step Red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40, 191–197. [DOI] [PubMed] [Google Scholar]