Abstract
Human cytomegalovirus (HCMV) UL141 induces protection against natural killer cell-mediated cytolysis by downregulating cell surface expression of CD155 (nectin-like molecule 5; poliovirus receptor), a ligand for the activating receptor DNAM-1 (CD226). However, DNAM-1 is also recognized to bind a second ligand, CD112 (nectin-2). We now show that HCMV targets CD112 for proteasome-mediated degradation by 48 h post-infection, thus removing both activating ligands for DNAM-1 from the cell surface during productive infection. Significantly, cell surface expression of both CD112 and CD155 was restored when UL141 was deleted from the HCMV genome. While gpUL141 alone is sufficient to mediate retention of CD155 in the endoplasmic reticulum, UL141 requires assistance from additional HCMV-encoded functions to suppress expression of CD112.
Human cytomegalovirus (HCMV), the prototype species of the subfamily Betaherpesvirinae, has a high prevalence in populations worldwide. Although HCMV is recognized to be an important human pathogen, particularly in immunocompromised individuals or following congenital infection, the vast majority of primary infections are subclinical and accompanied by asymptomatic lifelong carriage. HCMV encodes highly effective systems to provide for latency, persistent reactivation and transmission; as part of this process the virus acquired an impressive array of genes that act both to evade and redirect the host immune response (Wilkinson et al., 2008). The fact that individuals with genetic defects in their natural killer (NK) cell response are particularly susceptible to severe HCMV disease (Biron et al., 1989; Gazit et al., 2004) provided a rationale to focus attention on this arm of the immune response.
NK cells are composed of heterogeneous populations expressing a ‘mosaic’ of different activating and inhibitory receptors, the function of each cell being regulated by integration of signals received from ligands presented on potential target cells (Lanier, 2008). Inhibitory signals received mainly from autologous MHC class-I molecules normally dominate, to maintain NK cells in a resting state. However, HCMV not only efficiently downregulates MHC-I (Ahn et al., 1997; Furman et al., 2002; Jones et al., 1996; Trgovcich et al., 2006; Wiertz et al., 1996a, b), but also stimulates the expression of recognized NK cell activating ligands, e.g. MHC-I-related chains (MIC) A and B, UL16-binding proteins (ULBP) 1–3, retinoic acid early transcripts (RAET)1E/ULBP4, RAET1G/ULBP5, RAET1L/ULBP6 and CD155 (Bacon et al., 2004; Bahram et al., 1994; Bauer et al., 1999; Chalupny et al., 2003; Cosman et al., 2001; Eagle et al., 2009; Groh et al., 2001; Tomasec et al., 2005). Despite this, HCMV-infected cells actually prove to be highly resistant to NK cells in functional assays (Cerboni et al., 2000; Tomasec et al., 2005). This resilience can be attributed to a substantial proportion of HCMV genome being directed towards evading the NK cell response.
Although HCMV downregulates endogenous MHC-I, the virus also encodes its own MHC-I homologue (gpUL18) that binds the inhibitory receptor LIR-1 (ILT-2) with high affinity (Beck & Barrell, 1988; Chapman et al., 1999; Prod'homme et al., 2007) and a peptide in the UL40 leader sequence that acts to promote cell surface expression of the non-classical MHC-I molecule HLA-E, the ligand for the inhibitory receptor CD94/NKG2A (Tomasec et al., 2000; Ulbrecht et al., 2000; Wang et al., 2002). The activating receptor NKG2D is remarkable in recognizing eight ligands. To combat their activation UL16 retains MICB, ULBP1 and ULBP2 in the endoplasmic reticulum (ER); miR-UL112 targets the MICB transcript, while UL142 downregulates MICA (Chalupny et al., 2006; Cosman et al., 2001; Stern-Ginossar et al., 2007; Wills et al., 2005). The NK cell activating receptor DNAM-1 (CD226) recognizes both CD155 and CD112 (Bottino et al., 2003; Fuchs et al., 2004). We previously demonstrated that UL141 elicits efficient protection against NK cell-mediated cytolysis by sequestering CD155 in the ER yet, in isolation, had no effect on CD112 (Tomasec et al., 2005).
CD155 is the poliovirus receptor (PVR) or nectin-like molecule-5 (necl-5), while CD112 is also referred to as nectin-2, herpesvirus entry mediator B (HVEB) or poliovirus receptor-related protein 2 (PRR2). CD112 and CD155 are both structurally and functionally related. Nectins and necls are immunoglobulin-like molecules involved in cell adhesion, movement, proliferation, differentiation, polarization, virus entry and immune recognition (Takai et al., 2008). In view of its important role as an activating ligand for DNAM-1, we sought to analyse CD112 expression in the context of HCMV infection. Initial flow cytometry studies revealed that CD112 was downregulated by the low passage HCMV strain Merlin, but not high passage strain AD169 (not shown). Strain AD169 has a 15 kb deletion encompassing UL132–UL150 that includes the NK cell evasion genes UL141 and UL142. Merlin was derived from a bacterial artificial chromosome (BAC) containing the entire strain Merlin genome (R. J. Stanton, unpublished data). MerlinΔUL141 was generated using technologies developed previously to facilitate manipulation of the adenovirus genome (Stanton et al., 2008). Briefly, a selectable cassette comprising ampicillin resistance, lacZ and SacB was PCR amplified and recombineered into the Merlin BAC in place of nt 184597–185412 (relative to published Merlin sequence GenBank accession no. NC_006273) using primers SacBF-UL141 (5′-caggtagcataggaaacatacggtgaaaatactccaaaatcccaaaaatgccgcgattccccgagtggcccagggagacctgtgacggaagatcacttcg-3′, homology to pAL1111 underlined) and SacBR-UL141 (5′-ccgacgtttgagcggccgacacacggagcaggaacaggcgggcagcgtctctgcgaaaaagggaagaaaagaatcatcctgaggttcttatggctcttg-3′, homology to pAL1111 underlined). In a second recombineering step, the selectable cassette was removed using oligo delUL141 (5′-atactccaaaatcccaaaaatgccgcgattccccgagtggcccagggagagatgattcttttcttccctttttcgcagagacgctgcccgcctgttcctg-3′), leaving behind a seamless deletion of the first 816 bp of the UL141 ORF.
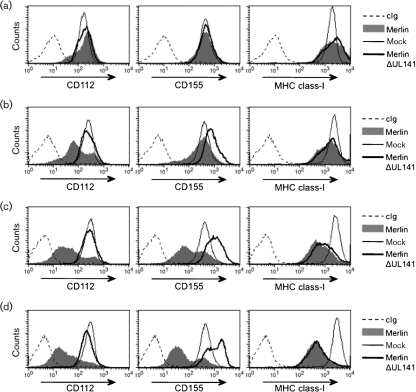
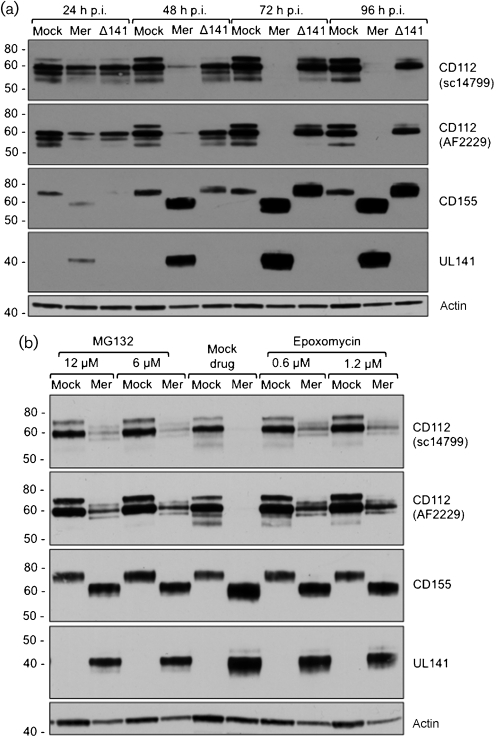
In human fetal foreskin fibroblasts (HFFF) infected with Merlin, cell surface levels of CD155, CD112 and MHC-I were progressively downregulated over the course of infection (Fig. 1), with the change in CD112 being more pronounced at 48 h post-infection (p.i.) (Fig. 1b). In accord with previous observations (Tomasec et al., 2005), cells infected with MerlinΔUL141 had elevated cell surface levels of CD155, while CD112 levels were comparable with the mock-infected HFFF (Fig. 1). Deletion of UL141 therefore ablated downregulation of both CD155 and CD112. This restoration of CD112 expression was unexpected, since UL141 had no overt effect on CD112 when expressed in isolation (Tomasec et al., 2005). Interestingly, a small reproducible decrease in CD112 persisted when MerlinΔUL141-infected and mock-infected cells were compared at 96 h p.i. (Fig. 1d). Replicate samples from the flow cytometry study were analysed by immunoblot, in order to further assess the fate of the CD112 protein within the cell. Briefly, cells were extracted with Triton X-114 (Bordier, 1981), proteins were separated on NuPAGE gels (Invitrogen) and blots were analysed with two independent polyclonal anti-CD112 antibodies. In Merlin-infected cells, the loss of CD155 from the cell surface (Fig. 1) correlated with the emergence of elevated levels of an immature (endoglycosidase H-sensitive) form of CD155 complexed with gpUL141 in the ER (Cochrane, 2009; Tomasec et al., 2005) (Fig. 2a). In contrast to CD155, the CD112 signal gradually decreased in Merlin-infected cells and was not detected by 72 h p.i. (Fig. 2a).
Fig. 1.
HFFF were infected (m.o.i.=25) for (a) 24 h (b) 48 h (c) 72 h or (d) 96 h with HCMV strain Merlin, MerlinΔUL141 or mock-infected and cell surface expression of CD112 (Santa Cruz, sc-65333) was analysed by flow cytometry. For reference, expression levels of CD155 (Abcam, ab-3142) and MHC class-I (W632; ATCC) were also monitored, alongside control Ig (cIg).
Fig. 2.
HFFF were infected (m.o.i.=25) for 24, 48, 72 or 96 h p.i. with HCMV strain Merlin (Mer), MerlinΔUL141 (Δ141) or mock-infected (Mock) and cell extracts were analysed by immunoblot using antibodies to: CD112 (R&D, AF2229; Santa Cruz, sc-14799), CD155 [5D1 (Aoki et al., 1994)], UL141 [M550 (Tomasec et al., 2005)] and actin (A-2066; Sigma). (b) HFFF were infected (m.o.i.=25) for 48 h with HCMV strain Merlin (Mer) or mock-infected, then treated for 12 h with proteasome inhibitors MG132 or Epoxomycin as indicated and analysed by immunoblot as in (a).
Quantitative real time-PCR showed CD112 mRNA levels to be marginally increased throughout the infection (not shown), consistent with CD112 expression being regulated post-transcriptionally. To determine whether CD112 was targeted for proteolytic degradation, Merlin-infected cells were incubated in the presence of proteasome inhibitors. Treatment with either MG132 or Epoxomycin (Calbiochem) was able to restore CD112 expression, indicating that HCMV targeted CD112 for proteasome-mediated degradation (Fig. 2b).
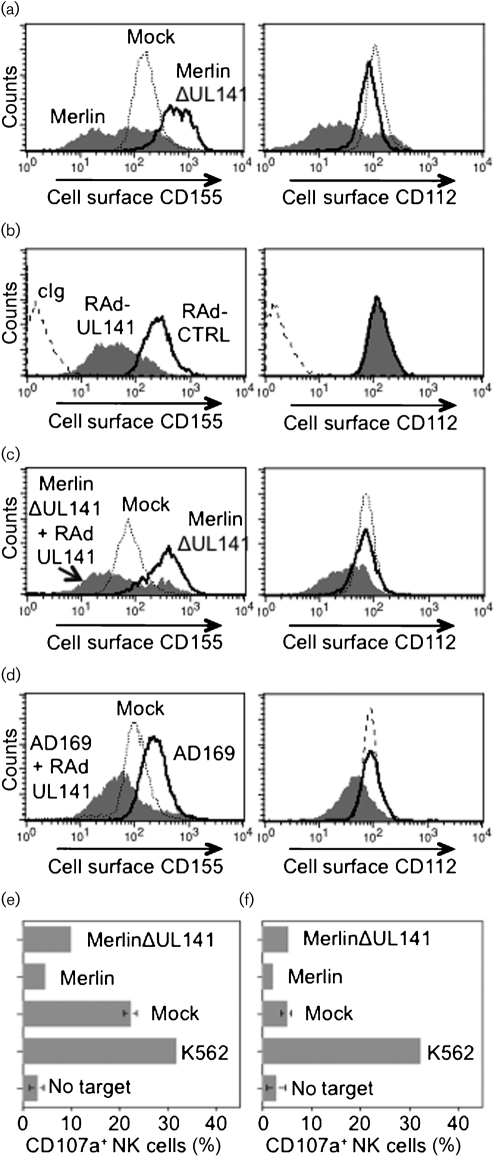
UL141 was required for efficient downregulation of both CD112 and CD155 from the cell surface in HCMV-infected cells (Figs 1 and 3a), yet had no effect on CD112 in cells infected with recombinant adenovirus vector encoding UL141 [RAdUL141 (Tomasec et al., 2005); Fig. 3b]. We reasoned that UL141 acted in partnership with an additional HCMV-encoded function(s) to downregulate CD112. Indeed, the residual level of CD112 suppression mediated by the MerlinΔUL141 (Figs 1d, 2a and 3a) could potentially be mediated by this function operating suboptimally. In cells co-infected with MerlinΔUL141 and RAdUL141, the HCMV deletion mutant was complemented; downregulation of both CD112 and CD155 was restored (Fig. 3c). Similarly, co-infection of strain AD169 with RAdUL141 also resulted in the downregulation of both CD112 and CD155 (Fig. 3d). These data are consistent with UL141 co-operating with additional HCMV-expressed function(s) to efficiently downregulate CD112, and that function also being intact within AD169 strain (thus excluding UL133–150). Through downregulation of CD112, HCMV eliminates from the cell surface an activating ligand for DNAM-1, which presumably contributes to the enhanced killing of HCMV-infected cells observed when UL141 is deleted from the virus (Fig. 3e, f), but not to the protection elicited when UL141 is expressed in isolation (Tomasec et al., 2005). HCMV thus targets both ligands for the NK cell activating receptor DNAM-1. GpUL141 alone is sufficient to sequester CD155 in the ER, while this study predicts that gpUL141 acts in concert with an additional viral function to induce proteasome-mediated degradation of CD112. This additional viral function could either directly co-operate with UL141, or act upon a cellular intermediate.
Fig. 3.
HFFF were infected for 72 h (m.o.i.=25) with HCMV strain Merlin or MerlinΔUL141, as indicated, and analysed for cell surface expression of CD155 and CD112 by flow cytometry. (b) HFFF were infected for 72 h (m.o.i.=200) with replication-deficient adenovirus vectors encoding HCMV UL141 (RAd-UL141) or equivalent empty RAd (RAd-CTRL) (Tomasec et al., 2005), as indicated, and analysed for cell surface expression of CD155 and CD112 by flow cytometry. (c) HFFF were co-infected for 72 h with MerlinΔUL141+RAd-CTRL or MerlinΔUL141+RAd-UL141, as indicated, and analysed for cell surface expression of CD155 and CD112 by flow cytometry. (d) HFFF were co-infected for 72 h with HCMV strain AD169+RAd-CTRL (AD169) or AD169+RAd-UL141, as indicated, and analysed for cell surface expression of CD155 and CD112 by flow cytometry. Control Ig histograms (cIg) were not included in panels (a), (c) and (d) to maintain figure clarity. (e) HFFF were infected for 72 h with HCMV strain Merlin, MerlinΔUL141 or mock infected. Sensitivity to NK cells was measured using alpha interferon (IFN-α) activated PBMC in allogeneic CD107a mobilization assay (Prod'homme et al., 2007) using the following antibodies: anti-CD107a-FITC (553793; BD Biosciences), anti-CD3-PerCP (SK7; BD Biosciences), anti-CD56-APC (N901; Beckman Coulter). PBMC incubated without targets and K562 cells are shown as controls. (f) RS primary skin fibroblasts were infected for 72 h with HCMV strain Merlin, MerlinΔUL141 or mock infected. Sensitivity to NK cells was measured using IFN-α activated RS PBMC in autologous CD107a mobilization assay as described in (e).
DNAM-1 is remarkable in being expressed on all NK cells and plays a major role in regulating their function. HCMV suppression of CD112 and CD155 may have ramifications that extend beyond the regulation of NK cell function. DNAM-1 is also expressed on activated T, NKT, myeloid and mast cells, megakaryocytes, platelets and a subset of B lymphocytes thereby impacting on a wide range of immunological responses and regulating platelet activation (Bachelet et al., 2006; Bottino et al., 2003; Burns et al., 1985; Kojima et al., 2003; Pende et al., 2006; Reymond et al., 2004; Scott et al., 1989; Shibuya et al., 1996, 1999, 2003; Xu & Jin, 2010). For example, the interaction between DNAM-1 and CD112/CD155 has been associated with T-cell differentiation, proliferation, cytotoxicity and cytokine secretion (Tahara-Hanaoka et al., 2004). Furthermore, nectins and necls regulate fundamental processes in cell biology including cell adhesion, movement, proliferation, differentiation, survival, polarization and signalling (Takai et al., 2008). HCMV infection is recognized to disrupt focal adhesions and intercellular connections, while inducing cell motility and transendothelial migration (Chan et al., 2009; Stanton et al., 2007). It will be important to determine how the modulation of CD112 and CD115 influences these processes.
Acknowledgments
Flow cytometers and real-time PCR facility were provided by the Cardiff University Central Biotechnology Service. We are grateful to Siân Llewellyn-Lacey for kindly providing technical support. This work was supported by funding from the Wellcome Trust and MRC.
References
- Ahn, K., Gruhler, A., Galocha, B., Jones, T. R., Wiertz, E. J., Ploegh, H. L., Peterson, P. A., Yang, Y. & Fruh, K. (1997). The ER-luminal domain of the HCMV glycoprotein US6 inhibits peptide translocation by TAP. Immunity 6, 613–621. [DOI] [PubMed] [Google Scholar]
- Aoki, J., Koike, S., Ise, I., Sato-Yoshida, Y. & Nomoto, A. (1994). Amino acid residues on human poliovirus receptor involved in interaction with poliovirus. J Biol Chem 269, 8431–8438. [PubMed] [Google Scholar]
- Bachelet, I., Munitz, A., Mankutad, D. & Levi-Schaffer, F. (2006). Mast cell costimulation by CD226/CD112 (DNAM-1/Nectin-2): a novel interface in the allergic process. J Biol Chem 281, 27190–27196. [DOI] [PubMed] [Google Scholar]
- Bacon, L., Eagle, R. A., Meyer, M., Easom, N., Young, N. T. & Trowsdale, J. (2004). Two human ULBP/RAET1 molecules with transmembrane regions are ligands for NKG2D. J Immunol 173, 1078–1084. [DOI] [PubMed] [Google Scholar]
- Bahram, S., Bresnahan, M., Geraghty, D. E. & Spies, T. (1994). A second lineage of mammalian major histocompatibility complex class I genes. Proc Natl Acad Sci U S A 91, 6259–6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer, S., Groh, V., Wu, J., Steinle, A., Phillips, J. H., Lanier, L. L. & Spies, T. (1999). Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science 285, 727–729. [DOI] [PubMed] [Google Scholar]
- Beck, S. & Barrell, B. G. (1988). Human cytomegalovirus encodes a glycoprotein homologous to MHC class-I antigens. Nature 331, 269–272. [DOI] [PubMed] [Google Scholar]
- Biron, C. A., Byron, K. S. & Sullivan, J. L. (1989). Severe herpesvirus infections in an adolescent without natural killer cells. N Engl J Med 320, 1731–1735. [DOI] [PubMed] [Google Scholar]
- Bordier, C. (1981). Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem 256, 1604–1607. [PubMed] [Google Scholar]
- Bottino, C., Castriconi, R., Pende, D., Rivera, P., Nanni, M., Carnemolla, B., Cantoni, C., Grassi, J., Marcenaro, S. & other authors (2003). Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J Exp Med 198, 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns, G. F., Triglia, T., Werkmeister, J. A., Begley, C. G. & Boyd, A. W. (1985). TLiSA1, a human T lineage-specific activation antigen involved in the differentiation of cytotoxic T lymphocytes and anomalous killer cells from their precursors. J Exp Med 161, 1063–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerboni, C., Mousavi-Jazi, M., Linde, A., Soderstrom, K., Brytting, M., Wahren, B., Karre, K. & Carbone, E. (2000). Human cytomegalovirus strain-dependent changes in NK cell recognition of infected fibroblasts. J Immunol 164, 4775–4782. [DOI] [PubMed] [Google Scholar]
- Chalupny, N. J., Sutherland, C. L., Lawrence, W. A., Rein-Weston, A. & Cosman, D. (2003). ULBP4 is a novel ligand for human NKG2D. Biochem Biophys Res Commun 305, 129–135. [DOI] [PubMed] [Google Scholar]
- Chalupny, N. J., Rein-Weston, A., Dosch, S. & Cosman, D. (2006). Down-regulation of the NKG2D ligand MICA by the human cytomegalovirus glycoprotein UL142. Biochem Biophys Res Commun 346, 175–181. [DOI] [PubMed] [Google Scholar]
- Chan, G., Nogalski, M. T. & Yurochko, A. D. (2009). Activation of EGFR on monocytes is required for human cytomegalovirus entry and mediates cellular motility. Proc Natl Acad Sci U S A 106, 22369–22374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, T. L., Heikeman, A. P. & Bjorkman, P. J. (1999). The inhibitory receptor LIR-1 uses a common binding interaction to recognize class I MHC molecules and the viral homolog UL18. Immunity 11, 603–613. [DOI] [PubMed] [Google Scholar]
- Cochrane, D. (2009). Detailed characterisation of the human cytomegalovirus immune modulatory gene UL141. In Infection, Immunity and Biochemistry, pp. 239. Cardiff: Cardiff University.
- Cosman, D., Mullberg, J., Sutherland, C. L., Chin, W., Armitage, R., Fanslow, W., Kubin, M. & Chalupny, N. J. (2001). ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity 14, 123–133. [DOI] [PubMed] [Google Scholar]
- Eagle, R. A., Traherne, J. A., Hair, J. R., Jafferji, I. & Trowsdale, J. (2009). ULBP6/RAET1L is an additional human NKG2D ligand. Eur J Immunol 39, 3207–3216. [DOI] [PubMed] [Google Scholar]
- Fuchs, A., Cella, M., Giurisato, E., Shaw, A. S. & Colonna, M. (2004). Cutting edge: CD96 (tactile) promotes NK cell-target cell adhesion by interacting with the poliovirus receptor (CD155). J Immunol 172, 3994–3998. [DOI] [PubMed] [Google Scholar]
- Furman, M. H., Dey, N., Tortorella, D. & Ploegh, H. L. (2002). The human cytomegalovirus US10 gene product delays trafficking of major histocompatibility complex class I molecules. J Virol 76, 11753–11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazit, R., Garty, B. Z., Monselise, Y., Hoffer, V., Finkelstein, Y., Markel, G., Katz, G., Hanna, J., Achdout, H. & other authors (2004). Expression of KIR2DL1 on the entire NK cell population: a possible novel immunodeficiency syndrome. Blood 103, 1965–1966. [DOI] [PubMed] [Google Scholar]
- Groh, V., Rhinehart, R., Randolph-Habecker, J., Topp, M. S., Riddell, S. R. & Spies, T. (2001). Costimulation of CD8αβ T cells by NKG2D via engagement by MIC induced on virus-infected cells. Nat Immunol 2, 255–260. [DOI] [PubMed] [Google Scholar]
- Jones, T. R., Wiertz, E. J., Sun, L., Fish, K. N., Nelson, J. A. & Ploegh, H. L. (1996). Human cytomegalovirus US3 impairs transport and maturation of major histocompatibility complex class I heavy chains. Proc Natl Acad Sci U S A 93, 11327–11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima, H., Kanada, H., Shimizu, S., Kasama, E., Shibuya, K., Nakauchi, H., Nagasawa, T. & Shibuya, A. (2003). CD226 mediates platelet and megakaryocytic cell adhesion to vascular endothelial cells. J Biol Chem 278, 36748–36753. [DOI] [PubMed] [Google Scholar]
- Lanier, L. L. (2008). Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol 9, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pende, D., Castriconi, R., Romagnani, P., Spaggiari, G. M., Marcenaro, S., Dondero, A., Lazzeri, E., Lasagni, L., Martini, S. & other authors (2006). Expression of the DNAM-1 ligands, Nectin-2 (CD112) and poliovirus receptor (CD155), on dendritic cells: relevance for natural killer-dendritic cell interaction. Blood 107, 2030–2036. [DOI] [PubMed] [Google Scholar]
- Prod'homme, V., Griffin, C., Aicheler, R. J., Wang, E. C., McSharry, B. P., Rickards, C. R., Stanton, R. J., Borysiewicz, L. K., Lopez-Botet, M. & other authors (2007). The human cytomegalovirus MHC class I homolog UL18 inhibits LIR-1+ but activates LIR-1− NK cells. J Immunol 178, 4473–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond, N., Imbert, A. M., Devilard, E., Fabre, S., Chabannon, C., Xerri, L., Farnarier, C., Cantoni, C., Bottino, C. & other authors (2004). DNAM-1 and PVR regulate monocyte migration through endothelial junctions. J Exp Med 199, 1331–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, J. L., Dunn, S. M., Jin, B., Hillam, A. J., Walton, S., Berndt, M. C., Murray, A. W., Krissansen, G. W. & Burns, G. F. (1989). Characterization of a novel membrane glycoprotein involved in platelet activation. J Biol Chem 264, 13475–13482. [PubMed] [Google Scholar]
- Shibuya, A., Campbell, D., Hannum, C., Yssel, H., Franz-Bacon, K., McClanahan, T., Kitamura, T., Nicholl, J., Sutherland, G. R. & other authors (1996). DNAM-1, a novel adhesion molecule involved in the cytolytic function of T lymphocytes. Immunity 4, 573–581. [DOI] [PubMed] [Google Scholar]
- Shibuya, K., Lanier, L. L., Phillips, J. H., Ochs, H. D., Shimizu, K., Nakayama, E., Nakauchi, H. & Shibuya, A. (1999). Physical and functional association of LFA-1 with DNAM-1 adhesion molecule. Immunity 11, 615–623. [DOI] [PubMed] [Google Scholar]
- Shibuya, K., Shirakawa, J., Kameyama, T., Honda, S., Tahara-Hanaoka, S., Miyamoto, A., Onodera, M., Sumida, T., Nakauchi, H. & other authors (2003). CD226 (DNAM-1) is involved in lymphocyte function-associated antigen 1 costimulatory signal for naive T cell differentiation and proliferation. J Exp Med 198, 1829–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton, R. J., McSharry, B. P., Rickards, C. R., Wang, E. C., Tomasec, P. & Wilkinson, G. W. (2007). Cytomegalovirus destruction of focal adhesions revealed in a high-throughput Western blot analysis of cellular protein expression. J Virol 81, 7860–7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton, R. J., McSharry, B. P., Armstrong, M., Tomasec, P. & Wilkinson, G. W. (2008). Re-engineering adenovirus vector systems to enable high-throughput analyses of gene function. Biotechniques 45, 659–668. [DOI] [PubMed] [Google Scholar]
- Stern-Ginossar, N., Elefant, N., Zimmermann, A., Wolf, D. G., Saleh, N., Biton, M., Horwitz, E., Prokocimer, Z., Prichard, M. & other authors (2007). Host immune system gene targeting by a viral miRNA. Science 317, 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara-Hanaoka, S., Shibuya, K., Onoda, Y., Zhang, H., Yamazaki, S., Miyamoto, A., Honda, S., Lanier, L. L. & Shibuya, A. (2004). Functional characterization of DNAM-1 (CD226) interaction with its ligands PVR (CD155) and nectin-2 (PRR-2/CD112). Int Immunol 16, 533–538. [DOI] [PubMed] [Google Scholar]
- Takai, Y., Ikeda, W., Ogita, H. & Rikitake, Y. (2008). The immunoglobulin-like cell adhesion molecule nectin and its associated protein afadin. Annu Rev Cell Dev Biol 24, 309–342. [DOI] [PubMed] [Google Scholar]
- Tomasec, P., Braud, V. M., Rickards, C., Powell, M. B., McSharry, B. P., Gadola, S., Cerundolo, V., Borysiewicz, L. K., McMichael, A. J. & Wilkinson, G. W. (2000). Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 287, 1031. [DOI] [PubMed] [Google Scholar]
- Tomasec, P., Wang, E. C., Davison, A. J., Vojtesek, B., Armstrong, M., Griffin, C., McSharry, B. P., Morris, R. J., Llewellyn-Lacey, S. & other authors (2005). Downregulation of natural killer cell-activating ligand CD155 by human cytomegalovirus UL141. Nat Immunol 6, 181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trgovcich, J., Cebulla, C., Zimmerman, P. & Sedmak, D. D. (2006). Human cytomegalovirus protein pp71 disrupts major histocompatibility complex class I cell surface expression. J Virol 80, 951–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrecht, M., Martinozzi, S., Grzeschik, M., Hengel, H., Ellwart, J. W., Pla, M. & Weiss, E. H. (2000). Cutting edge: the human cytomegalovirus UL40 gene product contains a ligand for HLA-E and prevents NK cell-mediated lysis. J Immunol 164, 5019–5022. [DOI] [PubMed] [Google Scholar]
- Wang, E. C., McSharry, B., Retiere, C., Tomasec, P., Williams, S., Borysiewicz, L. K., Braud, V. M. & Wilkinson, G. W. (2002). UL40-mediated NK evasion during productive infection with human cytomegalovirus. Proc Natl Acad Sci U S A 99, 7570–7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiertz, E. J., Jones, T. R., Sun, L., Bogyo, M., Geuze, H. J. & Ploegh, H. L. (1996a). The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84, 769–779. [DOI] [PubMed] [Google Scholar]
- Wiertz, E. J., Tortorella, D., Bogyo, M., Yu, J., Mothes, W., Jones, T. R., Rapoport, T. A. & Ploegh, H. L. (1996b). Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384, 432–438. [DOI] [PubMed] [Google Scholar]
- Wilkinson, G. W., Tomasec, P., Stanton, R. J., Armstrong, M., Prod'homme, V., Aicheler, R., McSharry, B. P., Rickards, C. R., Cochrane, D. & other authors (2008). Modulation of natural killer cells by human cytomegalovirus. J Clin Virol 41, 206–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills, M. R., Ashiru, O., Reeves, M. B., Okecha, G., Trowsdale, J., Tomasec, P., Wilkinson, G. W., Sinclair, J. & Sissons, J. G. (2005). Human cytomegalovirus encodes an MHC class I-like molecule (UL142) that functions to inhibit NK cell lysis. J Immunol 175, 7457–7465. [DOI] [PubMed] [Google Scholar]
- Xu, Z. & Jin, B. (2010). A novel interface consisting of homologous immunoglobulin superfamily members with multiple functions. Cell Mol Immunol 7, 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]