Abstract
We have used the hematopoietic system as a model to investigate whether acute myeloid leukemia arises exclusively from self-renewing stem cells or also from short-lived myeloid progenitors. When transduced with a leukemogenic MLL fusion gene, prospectively isolated stem cells and myeloid progenitor populations with granulocyte/macrophage differentiation potential are efficiently immortalized in vitro and result in the rapid onset of acute myeloid leukemia with similar latencies following transplantation in vivo. Regardless of initiating cell, leukemias displayed immunophenotypes and gene expression profiles characteristic of maturation arrest at an identical late stage of myelomonocytic differentiation, putatively a monopotent monocytic progenitor stage. Our findings unequivocally establish the ability of transient repopulating progenitors to initiate myeloid leukemias in response to an MLL oncogene, and support the existence of cancer stem cells that do not necessarily overlap with multipotent stem cells of the tissue of origin.
Keywords: MLL, leukemia, stem cell, myeloid progenitors, cancer
Hematopoiesis takes place through the stepwise differentiation of multipotent stem cells to generate a hierarchy of progenitor populations with progressively restricted developmental potential, ultimately leading to the production of multiple lineages of mature effector cells (Morrison and Weissman 1994; Akashi et al. 2000; Orkin 2000). Importantly, long-term self-renewal is normally limited to long-term hematopoietic stem cells (HSC) in the hematopoietic stem and progenitor pool (for review, see Weissman et al. 2001). A unifying feature of all cancers is the capacity for unlimited self-renewal, which is also a defining characteristic of normal stem cells (for review, see Reya et al. 2001). Given these shared attributes, it has been proposed that human cancer may be initiated by transforming events that take place in rare tissue-specific stem cells. Alternatively, cancers may arise from more committed progenitors due to mutations and/or selective gene expression that enhance their otherwise limited self-renewal capabilities (for review, see Passegué et al. 2003).
Many of the transcription factors that control normal hematopoietic differentiation and proliferation are also targets for mutations associated with the development of acute leukemias (for review, see Tenen et al. 1997; Park et al. 2002), malignant diseases characterized by unregulated self-renewal. The MLL protein represents one such transcriptional regulator that is required for normal hematopoiesis (Hess et al. 1997; Yagi et al. 1998) and implicated in the maintenance of Hox gene expression (Breen and Harte 1993; Yu et al. 1995; Ayton and Cleary 2003), which is intimately tied to stem/progenitor cell expansion (Sauvageau et al. 1995; Lawrence et al. 1996). MLL fusion proteins, created by chromosomal translocation events, are frequently associated with the development of acute myeloid and lymphoid leukemias, and the oncogenic properties of a variety of MLL fusion proteins have been extensively studied in vitro and in vivo in the mouse (for review, see Ayton and Cleary 2001). Hence, retroviral-mediated gene transfer of MLL fusion proteins transforms stem and progenitor-enriched bone marrow cells and results in acute myeloid leukemia in mice (Lavau et al. 2000). However, it is still unclear as to which compartment, for example, self-renewing HSC or short-lived progenitor, is susceptible to each specific MLL fusion protein-mediated transformation.
This issue can now be addressed in the mouse because of recent advances that have facilitated the prospective isolation of phenotypically and functionally defined myeloid progenitors that lie developmentally downstream of the HSC (Akashi et al. 2000; Na Nakorn et al. 2002). These populations include the common myeloid progenitors (CMP), and the lineal descendent granulocytic/monocytic-restricted progenitors (GMP) and megakaryocytic/erythroid-restricted progenitors (MEP), respectively.
Results and Discussion
MLL-ENL transforms HSC and myeloid progenitors with granulocyte/macrophage differentiation potential in vitro
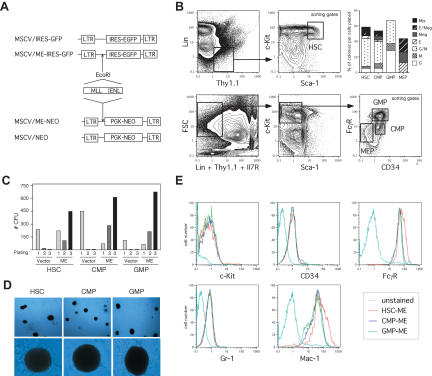
We first evaluated the in vitro transformation susceptibility of HSC, CMP, GMP, and MEP compartments by the representative MLL fusion protein MLL-ENL (ME; Tkachuk et al. 1992) using an in vitro serial replating assay (Fig. 1; Lavau et al. 1997). Transduction with either MSCV/MLL-ENL/Neo (ME) or control MSCV/neo retroviruses (Fig. 1A,B), and subsequent cultivation in methylcellulose supplemented with G-418 and myeloid differentiation-inducing cytokines, yielded numerous colonies in cultures initiated by ME-transduced HSC, CMP, and GMP populations (Fig. 1C). All ME-transduced progenitor populations generated colonies with identical compact, blast-like morphology (Fig. 1D), unlike the normal range of colony morphologies obtained in vector-transduced cultures (data not shown). Furthermore, colonies that arose from semisolid cultures initiated with ME-transduced HSC, CMP, or GMP readily adapted to growth in liquid culture in the presence of granulocyte/macrophage (GM)-CSF to generate immortalized factor-dependent cell lines. Interestingly, all ME-transduced HSC, CMP, and GMP cell lines exhibited a very similar immunophenotype characterized as Sca-1-, c-Kitlow/int., CD34+, FcγRhigh, Gr-1low, and Mac-1high (Fig 1E). In contrast, MEP and the progenitor-depleted c-kit- populations from BM did not retain clonogenic activity in the presence of myeloid-inducing cytokines (data not shown). Thus, both long-term self-renewing HSC and transient repopulating progenitors with GM differentiation potential (e.g., CMP and GMP) are susceptible to transformation by MLL-ENL in vitro and appeared to be arrested at an identical stage of differentiation regardless of the origin of the starting population.
Figure 1.
Enhanced in vitro self-renewal following transduction of purified myeloid progenitors with MLL-ENL. (A) Schematic representation of the retroviral control and oncogenic MLL-ENL constructs used for the in vitro and in vivo transduction experiments. (B) Example of FACS sorting conditions used to obtain highly purified populations of HSC (lin-/Thy1.1low/c-Kithigh/Sca-1high), CMP (lin-/Thy1.1-/Il7R-/c-Kithigh/Sca-1-/CD34+/FcγRlow), GMP (lin-/Thy1.1-/Il7R-/c-Kithigh/Sca-1-/CD34high/FcγRhigh), and MEP (lin-/Thy1.1-/Il7R-/c-Kithigh/Sca-1-/CD34-/FcγRlow). In methylcellulose CFU assay, both HSC and CMP gave rise to the entire range of myeloid and erythroid colonies including mix colonies (Mix), whereas GMP only gave rise to colonies composed of macrophages (M) and/or granulocytes (GM and G) and MEP to colonies composed of erythrocytes (E) and/or megakaryocytes (E/Meg and Meg), thereby attesting the purity of the starting populations. (C) In vitro immortalization of HSC and committed CMP and GMP progenitors. Bar graph indicates the numbers of colonies obtained after each round of plating in methylcellulose supplemented with IL-3, IL-6, SCF, GM-CSF, and G-418 for the first round only, and are representative of three independent experiments. Transduction efficiencies can be calculated from the first round of plating as the number of drug-resistant positively selected colonies divided by the numbers of starting cells (HSC: vector 30.3%, ME 22.7%; CMP: vector 80%, ME 26.3%; GMP: vector 77.1%, ME 50%). (D) Typical colonies showing blast morphology obtained after MLL-ENL-transduction of HSC, CMP, and GMP populations (upper panel: 40× magnification; lower panel: 200× magnification). (E) Immunophenotype characterization of the ME-transduced HSC, CMP, and GMP cell lines showing similar expression levels of the c-Kit, CD34, FcγR, Gr-1, and Mac-1 surface markers.
Similar onset of leukemias in mice transplanted with MLL-ENL-transduced HSC and myeloid progenitors with granulocyte/macrophage potential
We then tested the relative ability of HSC and each myeloid progenitor to induce acute myeloid leukemia (AML) in vivo. HSC and myeloid progenitors were prospectively isolated, retrovirally transduced with ME (MSCV/ME-IRES-GFP) or control (MSCV/IRES-GFP) retroviruses (Fig. 1A), and transplanted into lethally irradiated congenic recipients. The degree of engraftment and the development of AML were monitored by FACS analysis of blood. None of the animals transplanted with control-transduced HSC (n = 9) or progenitor populations (CMP, n = 3; GMP, n = 5; MEP, n =2) developed AML or displayed increased numbers of donor-derived GFP+ cells (Fig. 2A). In contrast, all of the animals transplanted with ME-transduced HSC (103 cells, n = 23), CMP (104 cells, n = 17), or GMP (104 cells n = 31) displayed considerable increases in the numbers of donor-derived GFP+ cells (Fig. 2A) and developed AML over an identical period of 90-100 d on average (Table 1). Titration studies revealed the transformation efficiency to be HSC > CMP > GMP, indicating that the more primitive or expandable population required fewer cells for transformation (Table 1). MEP were also efficiently transduced by the ME virus (Table 1), but no donor-derived GFP+ cells were detected in the peripheral blood of mice transplanted with ME-transduced MEP and none of the animals developed AML (104 cells, n = 8; Fig. 2A). Thus, in addition to HSC, all progenitors with GM differentiation potential could initiate MLL-associated acute myeloid leukemias in vivo.
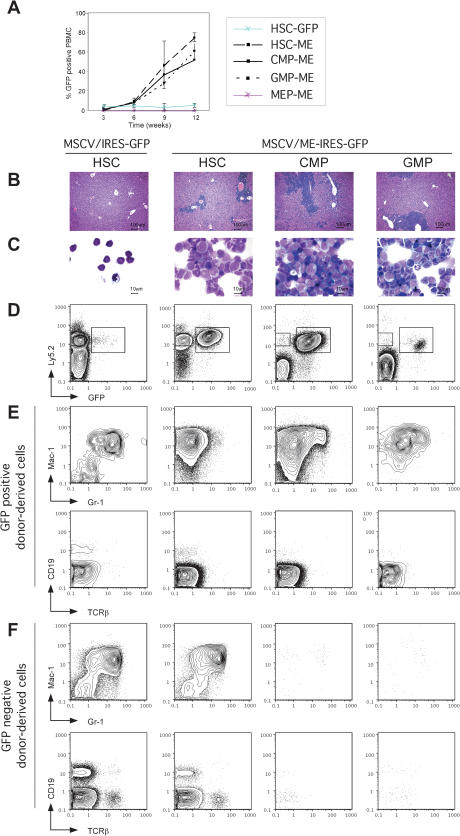
Figure 2.
Acute myeloid leukemias induced by MLL-ENL from stem and progenitor cells. (A) The extent of engraftment and development of AML was monitored by FACS analysis of GFP+ cells in peripheral blood mononuclear cells (PBMC). Mice (five per cohort) were transplanted with 1000 HSC transduced with control (blue line) or ME (dashed line) retroviruses, or with 10,000 ME-transduced CMP (black line), GMP (dotted line), or MEP (red line). Mice transplanted with control-transduced CMP, GMP, or MEP did not show any donor-derived Ly5.2+ cells after 6 to 7 wk posttransplantation (data not shown). (B-C) AML in preterminally leukemic mice. (B) Liver histology showing massive blast invasion in recipients of ME-transduced stem and progenitor cells. (C) May-Grünwald-Giemsa staining of splenocytes, indicating mostly blast and myelomonocytic morphologies. (D-E) Chimera analysis in mice 9 wk after transplantation of transduced stem and progenitor populations. ACK-treated bone marrow cells were analyzed for expression of GFP, Ly5.2 (donor-derived), and various myeloid (Mac-1, Gr-1) and lymphoid (CD19, TCRβ) markers. (D) An expanded population of ME-transduced donor-derived GFP+ cells is present in all leukemic mice. (E) ME-transduced GFP+ donor-derived cells only expressed myeloid markers (Mac-1high, Gr-1-/low) in contrast to multilineage readout of control-transduced HSC. (F) Multilineage reconstitution from untransduced GFP- donor-derived cells in ME-transduced HSC reconstituted mice. Consistent with their transient repopulating ability, untransduced GFP- donor-derived cells are not detected in CMP or GMP transplanted mice, confirming the absence of HSC contamination in the transplant.
Table 1.
MLL-ENL retroviral transduction/transplantation of purified hematopoietic stem cells and progenitors
| Starting population | Transduction efficiency (% GFP+) | Transplanted cells (#) | Estimated transduced cells (#) | Transplanted animals (#) | Incidence of AML (%) | Latency of AML (days ± SD) |
|---|---|---|---|---|---|---|
| HSC | 2.2-7.7 | 1,000 | 22-77 | 5 | 100 | 92 ± 12 |
| 600 | 13-46 | 5 | 100 | 91 ± 8 | ||
| 300 | 7-23 | 5 | 20 | 84 | ||
| CMP | 3.8-16.8 | 10,000 | 380-1680 | 5 | 100 | 98 ± 14 |
| 1,000 | 38-168 | 5 | 40 | 91 ± 10 | ||
| GMP | 8.0-24.9 | 10,000 | 800-2490 | 5 | 100 | 98 ± 8 |
| 1,000 | 80-249 | 5 | 20 | 102 | ||
| MEP | 5.0-9.7 | 10,000 | 500-970 | 5 | 0 | NA |
| 1,000 | 50-97 | 5 | 0 | NA |
Purified HSC, CMP, GMP, and MEP populations were transduced overnight with the MLL-ENL retrovirus. Lethally irradiated congenic recipient mice (five per cohort) were transplanted with the indicated numbers of cells. The percentage of ME-transduced GFP positive cells was measured after 2 to 5 d of in vitro culture and was used to estimate the number of transducer cells for each population. The given numbers represent the minimum and maximum transduction efficiency obtained from three independent experiments. The development of AML was monitored by FACS analysis of peripheral blood and confirmed by histological analysis of the sacrificed preterminally ill recipient mice. (NA) Not applicable.
All terminally ill leukemic mice presented with similar clinical manifestations, including infiltration of organ parenchyma (Fig. 2B) and presence of numerous blast cells in the spleen (Fig. 2C). Consistent with a myeloid derivation, all analyzed donor-derived GFP+ cells from ME-transduced HSC, CMP, and GMP origin expressed the myeloid markers Mac-1 and Gr-1, with only background staining for the lymphocytic markers CD19 and TCRβ (Fig. 2D,E). Although the spleens of animals transplanted with ME-transduced HSC retained a significant fraction of untransduced donor-derived GFP- cells that gave rise to multilineage readout (Fig. 2D,F), mice transplanted with ME-transduced CMP or GMP did not, consistent with the transient repopulating capability of untransformed myeloid progenitors. No donor-derived contribution to the B and T cell lineages was detected in any CMP or GMP transplanted animals (data not shown). These observations confirm the self-renewal and differentiation capacity of untransformed HSC, as well as the myeloid-restricted developmental potential and the absence of contaminating HSC in the starting CMP and GMP populations.
The MLL fusion genes are also known to cause multilineage leukemias (Ayton and Cleary 2001). Another MLL-associated gene, MLL-GAS7, can transform multipotent progenitor cells (MPP) within the stem cell pool (Morrison et al. 1997) and induces mixed-lineage leukemia in vivo (So et al. 2003). The capability of MLL-ENL to generate biphenotypic lympho-myeloid transformed cells in vitro has also been reported (Zeisig et al. 2003). To assess the ability of MLL-ENL to transform cells of lymphoid origin, we transduced highly purified populations of common lymphoid progenitor (CLP; Kondo et al. 1997) with control and ME retroviruses and cultivated them in vitro in the presence of Flt-3 ligand, stem cell factor (SCF), and interleukin-7 (IL-7), already used to generate biphenotypic lympho-myeloid transformed cells (So et al. 2003). Although control-transduced CLP produced numerous GFP+/B220+ cells after 8 d of culture, ME-transduced CLP failed to produce any GFP+ cells in these conditions (data not shown). These results indicate that MLL-ENL cannot transform CLP in vitro, although it does transform myeloid progenitors with GM differentiation potential both in vitro and in vivo.
Similar maturation arrest in leukemias originated from MLL-ENL-transduced HSC and myeloid progenitors with GM potential
Because a major feature of AML is a severe block to terminal myeloid differentiation associated with the subsequent expansion of immature myeloid precursors, we determined whether leukemias arising from transduced HSC and progenitors with GM developmental potential were arrested at specific stages of myeloid differentiation dependent on the progenitor compartment used to initiate the leukemias. None of the leukemias exhibited the immunophenotype reminiscent of the starting populations. All analyzed GFP+ cells from ME-transduced HSC, CMP, and GMP origin exhibited a similar immunophenotype characterized as c-Kitlow/int., Sca-1-, CD34+/high, Mac-1high, and Gr-1low (Figs. 3B,C, 2E), reminiscent of the common immunophenotype of the ME-transduced HSC, CMP, and GMP immortalized cell lines (Fig. 1E). As expected (Lavau et al. 2000), purified GFP+ cells from ME-transduced HSC, CMP, and GMP mice serially transplanted the AML disease in lethally irradiated recipient mice, hence demonstrating their leukemic potential. However, transplantation of fractionated GFP+ populations based on their c-Kit expression levels (c-Kitlow and c-Kitint.) similarly propagated the disease, indicating that the leukemic populations are heterogeneous in terms of c-Kit expression (data not shown).
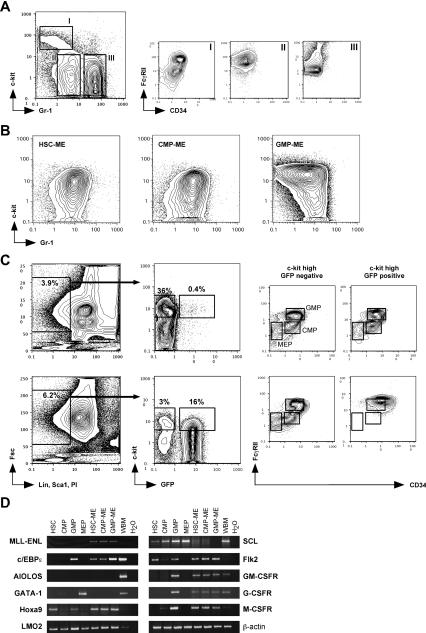
Figure 3.
Characterization of the leukemic cells. (A) Normal bone marrow stained with c-Kit and Gr-1 shows three distinct populations: a c-Kitint./high/Gr-1-/low population (I), which is CD34+/FcγR+ and contains the myeloid CMP and GMP progenitors (Akashi et al. 2000); a c-Kit-/Gr-1low population (II), which is CD34-/FcγRhigh and contains the myelomonocytic cells; and a c-Kit-/Gr-1high population (III), which is CD34-/FcγRint. and contains the neutrophilic granulocytes (Lagasse and Weissman 1996). (B) In leukemic mice, all ME-transduced GFP+ donor-derived cells from HSC, CMP, and GMP origin do not show the immunophenotypes of the starting populations, but are consistently c-Kitlow-int./Gr-1int.. (C) Mice transplanted with control-(upper panel) or ME-(lower panel) transduced HSC were analyzed for GFP expression in myeloid progenitor compartments at 9 wk posttransplantation. In control-transduced mice, c-Kithigh/GFP+ donor-derived cells contributed to all myeloid progenitor populations, whereas in leukemic mice the same population only yielded leukemic cells with a FcγRIIhigh/CD34+ phenotype. (D) Gene expression analyses comparing HSC and myeloid progenitors to their ME-transduced leukemic counterparts. RT-PCR was performed on 100 sorted cells or 5 × 105 total cells for control whole bone marrow (WBM).
In normal marrow, oligolineage myeloid progenitors are enriched in the Lin-, Sca-1-, c-Kithigh, CD34+, FcγRII+ population (Fig. 3A, population I). With differentiation, c-Kit and CD34 are down-regulated, and myelomonocytes become Mac1high, Gr-1low, FcγRIIhigh (Fig. 3A, population II), whereas neutrophilic granulocytes become Mac-1low-high, Gr-1high, FcγRIIlow (Fig. 3A, population III; Lagasse and Weissman 1996). Within this scheme, all the ME-induced leukemias appeared to be arrested at an intermediate point between stages I and II (Fig. 3B), with a Mac-1high, Gr-1low expression pattern (Fig. 2E) and high expression for FcγRII and the macrophage marker F4/80 (data not shown) typical for cells of the monocytic lineage (Hume et al. 1983). Cytospin and histology analysis further confirmed their myelomonocytic morphology (Fig. 2C and Lavau et al. 1997). This immunologic phenotype places the most abundant progeny of the leukemic stem cells within the monocytic lineage in close vicinity, but downstream of the GMP.
This immunocharacterization was further supported by analysis of mRNA expression profiles for various lineage and stage-specific genes (Fig. 3C). The expression of the myeloid transcript c/EBPε, in conjunction with the lack of expression for the B-cell transcript AIOLOS and for the erythroid transcript GATA-1 (except for the MEP population), confirmed the purity of the starting HSC and progenitor populations (Akashi et al. 2000). Recently, we have shown that transformation by MLL-ENL depends on the expression of Hoxa9 and Hoxa7 (Ayton and Cleary 2003). Interestingly, MLL-ENL was associated with high expression levels of Hoxa9 not only in leukemic cells derived from HSC, but also in leukemic cells derived from CMP and GMP, which normally expressed Hoxa9 at much lower levels than HSC. Hence, Hoxa9 up-regulation in ME-transduced transient repopulating CMP and GMP populations may contribute to their acquisition of leukemic self-renewal activity. Furthermore, all ME-transduced leukemic cells displayed no or greatly reduced expression of the specific stem cell transcript SCL but highly expressed transcripts for the stem and progenitor marker Flk2 (Christensen and Weissman 2001), as recently reported (Armstrong et al. 2002, 2003). Southern blot analysis showed that all ME-induced leukemias, regardless of their origin, were oligoclonal (data not shown), indicating that they were initiated by several transformed cells. To exclude the possibility that transformation of the committed CMP and GMP populations resulted from insertional oncogenesis of the ME retrovirus, we analyzed the expression of transcriptionally active loci, such as the LMO2 locus, which did not show any changes in expression levels. Interestingly, all ME-transduced leukemic cells expressed transcripts of the different myeloid growth factor receptors (GM-CSF, G-CSF, and M-CSF), which normally are only present in GMP and downstream myelomonocytic populations. Thus, despite the use of distinct HSC and myeloid progenitor populations, the gene expression profiles and the immunophenotypes of the resultant leukemias suggest that transformation-associated maturation arrest occurred at an identical late stage of myelomonocytic differentiation positioned developmentally downstream of the GMP but prior to terminal differentiation, putatively at a monopotent monocytic progenitor stage.
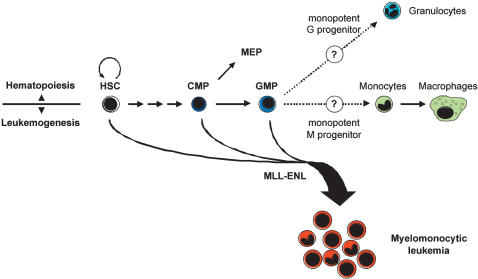
Despite the massive expansion of the GFP+ myelomonocytic leukemic progeny, GFP expression was not detected in the HSC compartment (data not shown) nor in the CMP, GMP, or MEP compartments of mice transplanted with ME-transformed HSC, in contrast to mice transplanted with control-transduced HSC (Fig. 3C). This indicates that the ME oncogene does not appreciably function to expand these progenitor populations, but selectively imposes a further downstream arrest regardless of which upstream progenitor initiates the leukemic process following acquisition of the activated oncogene by retroviral transduction. This result also suggests that the MLL-ENL oncogene may directly instruct differentiation to the leukemic stage from HSC, CMP, or GMP (Fig. 4).
Figure 4.
Proposed model for MLL-ENL initiated leukemogenesis. In normal hematopoiesis, only HSC possess unlimited self-renewal potential. Differentiation occurs through a phenotypically and functionally defined series of progenitor populations with restricted half-lives and lineage potentials. Myelomonocytic leukemias induced by MLL-ENL can arise in both self-renewing HSC and short-lived progenitor cells with GM differentiation potential. Introduction of MLL-ENL uniformly leads to the accumulation of leukemic cells with the phenotype of cells immediately downstream of GMP, putatively a monopotent monocytic progenitor stage.
Conclusions
The MLL-ENL oncogene is unique in our experience in its ability to initiate the oncogenic process rapidly and effectively in CMP and GMP, cell populations with very limited life spans in vivo (Akashi et al. 2000). Normally, the progeny of the most mature subset (GMP) transiently persist in transplanted recipients for only 2-3 wk, and at no time can self-renewing GMP be detected, although limited early expansion is possible and likely (Na Nakorn et al. 2002). Yet, ME-transduced GMP gave leukemic myelomonocytic lineage progeny with the same latency as the self-renewing ME-transduced HSC. It seems likely, therefore, that one effect of MLL-ENL expression is the rapid induction of self-renewal at a very restricted stage of hematopoiesis downstream of the GMP compartment (Fig. 4). However, the MLL-ENL oncogene cannot sustain unregulated self-renewal in the more primitive long- or short-term HSC and MPP, or in the oligolineage precursors CMP and GMP. Although previous studies have reported that MLL fusion proteins can antagonize MPP differentiation in vitro, the resultant in vivo leukemias generally displayed more differentiated precursor phenotypes (Schulte et al. 2002; So et al. 2003). The identification of developmentally upstream transformation-initiating events leading to subsequent downstream tumorigenic and transplantable phenotypes lends further support to the existence of the cancer and leukemic stem cell (Reya et al. 2001; Passegué et al. 2003). Moreover, our studies establish that a cancer stem cell does not necessarily overlap with the multipotent stem cell of the tissue in which the tumor arose (Bonnet and Dick 1997). The use of developmentally restricted, prospectively sorted, phenotypically monomorphic bone marrow populations for transformation experiments may allow for the analysis of the critical gene expression profiles of early transformation events or the critical tumor antigens against which effective immunity must be directed. Future identification of such target genes will be helpful in the analysis of the molecular mechanisms that regulate both normal and leukemogenic stem cell and progenitor self-renewal.
Materials and methods
Plasmid constructions and retroviral production
The MLL-ENL cDNA (Tkachuk et al. 1992) was subcloned into the pMSCV/IRES-GFP or pMSCV/neo vectors. Retroviral stocks were produced from the Phoenix packaging cell line by transient transfection using FuGENE 6 (Roche Molecular Biochemicals). Virus containing supernatant medium was collected 2-3 d following transfections, concentrated by ultracentrifugation, and used for infection of bone marrow cells.
Stem/progenitor cell purification and transplantation
HSC, CMP, GMP, MEP, and CLP were purified from C57BL/6-Ly5.2 mice by FACS sorting as previously described (Morrison and Weissman 1994; Kondo et al. 1997; Akashi et al. 2000). For retroviral transduction, progenitors were cultured in 200 μL in 96-well plates with 20%-50% virus stock in Iscove's Modified Dulbecco's Media (IMDM) containing 10% FCS, 50 mM 2-mercaptoethanol, 4 μg/mL polybrene, SCF (20 ng/mL), Flt3L (20 ng/mL), and IL-11 (10 ng/mL) for 12-18 h at 32°C. Transduced progenitors were mixed with a radioprotective dose of 2.5 × 105 congenic C57BL/6-Ly5.1 bone marrow cells and transplanted via retro-orbital injection into lethally irradiated (960 rad) congenic C57BL/6-Ly5.1 recipients. Transduction efficiency of progenitor populations was assessed by FACS analysis of GFP-expressing cells after expansion in IMDM containing 10% FCS, SCF (20 ng/mL), Flt3L (20 ng/mL), IL-11 (10 ng/mL), IL-3 (20 ng/mL), and Tpo (20 ng/mL) for 2 to 5 d after infection.
In vitro myeloid assays and generation of cytokine-dependent cell lines
Methylcellulose replating assays and derivation of cytokine-dependent myeloid progenitor cell lines were carried out as previously described (Lavau et al. 1997) using 1000 HSC, 10,000 CMP, or 10,000 GMP per experiment. Immortalized cell lines were maintained in RPMI supplemented with 10% FCS and 1 ng/mL GM-CSF.
RT-PCR
Total RNA was isolated from double-sorted progenitors and leukemic cells or from unfractionated whole bone marrow using Trizol (Invitrogen). RNA was reverse transcribed using the SuperScript First-Strand Synthesis kit (Invitrogen) and subjected to PCRamplification. Details for all primer sequences and conditions are available on request.
Immunophenotype analyses
Single-cell suspensions from bone marrow and spleen were ACK-treated for 3 min on ice to lyses erythrocytes, washed twice in staining medium (HBSS containing 2% FCS), incubated with 20 μg/mL rat IgG for 10 min to prevent nonspecific binding, and stained with the indicated fluorochrome-conjugated antibodies for 30 min on ice. Cells were then washed twice in staining medium and resuspended in 1 μg/mL propidium iodide (PI) before analysis using a FACS Vantage (modified dual 488-nm argon and 599-nm dye lasers, Becton Dickinson Immunocytometry System). Dead cells were gated out by high PI staining and forward light scatter.
Acknowledgments
We are grateful to L. Jerabek for excellent lab management and to A. Baiker for help with the retrovirus production. We thank L. Hidalgo and B. Lavarro for animal care and the Stanford Shared FACS Facility. A. Cozzio is a fellow of the Swiss National Foundation, E. Passegué of the Jose Carreras Leukemia Foundation (FIJC-01/EDTHOMAS), and H. Karsunky of the Ernst Schering Research Foundation. The National Institutes of Health (CA55209 and CA86017), the Children's Health Initiative (MLC), and a De Villier Award from the Leukemia and Lymphoma Society (ILW) supported this work.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1143403.
References
- Akashi K., Traver, D., Miyamoto, T., and Weissman, I.L. 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404: 193-197. [DOI] [PubMed] [Google Scholar]
- Armstrong S.A., Staunton, J.E., Silverman, L.B., Pieters, R., den Boer, M.L., Minden, M.D., Sallan, S.E., Lander, E.S., Golub, T.R., and Korsmeyer, S.J. 2002. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat. Genet. 30: 41-47. [DOI] [PubMed] [Google Scholar]
- Armstrong S.A., Kung, A.L., Mabon, M.E., Silverman, L.B., Stam, R.W., Den Boer, M.L., Pieters, R., Kersey, J.H., Sallan, S.E., Fletcher, J.A., et al. 2003. Inhibition of FLT3 in MLL. Validation of a therapeutic target identified by gene expression based classification. Cancer Cell 3: 173-183. [DOI] [PubMed] [Google Scholar]
- Ayton P.M. and Cleary, M.L. 2001. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene 20: 5695-5707. [DOI] [PubMed] [Google Scholar]
- ____. 2003. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes & Dev. 17: 2298-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D. and Dick, J.E. 1997. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 3: 730-737. [DOI] [PubMed] [Google Scholar]
- Breen T.R. and Harte, P.J. 1993. Trithorax regulates multiple homeotic genes in the bithorax and antennapedia complexes and exerts different tissue-specific, parasegment-specific and promoter-specific effects on each. Development 117: 119-134. [DOI] [PubMed] [Google Scholar]
- Christensen J.L. and Weissman, I.L. 2001. Flk-2 is a marker in hematopoietic stem cell differentiation: A simple method to isolate long-term stem cells. Proc. Natl. Acad. Sci. 98: 14541-14546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess J.L., Yu, B.D., Li, B., Hanson, R., and Korsmeyer, S.J. 1997. Defects in yolk sac hematopoiesis in Mll-null embryos. Blood 90: 1799-1806. [PubMed] [Google Scholar]
- Hume D.A., Robinson, A.P., MacPherson, G.G., and Gordon, S. 1983. The mononuclear phagocyte system of the mouse defined by immunohistochemical localization of antigen F4/80. Relationship between macrophages, Langerhans cells, reticular cells, and dendritic cells in lymphoid and hematopoietic organs. J. Exp. Med. 158: 1522-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M., Weissman, I.L., and Akashi, K. 1997. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell 91: 661-672. [DOI] [PubMed] [Google Scholar]
- Lagasse E. and Weissman, I.L. 1996. Flow cytometric identification of murine neutrophils and monocytes. J. Immunol. Methods 197: 139-150. [DOI] [PubMed] [Google Scholar]
- Lavau C., Szilvassy, S.J., Slany, R., and Cleary, M.L. 1997. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J. 16: 4226-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavau C., Luo, R.T., Du, C., and Thirman, M.J. 2000. Retrovirus-mediated gene transfer of MLL-ELL transforms primary myeloid progenitors and causes acute myeloid leukemias in mice. Proc. Natl. Acad. Sci. 97: 10984-10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence H.J., Sauvageau, G., Humphries, R.K., and Largman, C. 1996. The role of HOX homeobox genes in normal and leukemic hematopoiesis. Stem Cells 14: 281-291. [DOI] [PubMed] [Google Scholar]
- Morrison S.J. and Weissman, I.L. 1994. The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity 1: 661-673. [DOI] [PubMed] [Google Scholar]
- Morrison S.J., Wandycz, A.M., Hemmati, H.D., Wright, D.E., and Weissman, I.L. 1997. Identification of a lineage of multipotent hematopoietic progenitors. Development 124: 1929-1939. [DOI] [PubMed] [Google Scholar]
- Na Nakorn T., Traver, D., Weissman, I.L., and Akashi, K. 2002. Myeloerythroid-restricted progenitors are sufficient to confer radioprotection and provide the majority of day 8 CFU-S. J. Clin. Invest. 199: 1579-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin S.H. 2000. Diversification of hematopoietic stem cells to specific lineages. Nat. Rev. Genet. 1: 57-64. [DOI] [PubMed] [Google Scholar]
- Park I.K., He, Y., Lin, F., Laerum, O.D., Tian, Q., Bumgarner, R., Klug, C.A., Li, K., Kuhr, C., Doyle, M.J., et al. 2002. Differential gene expression profiling of adult murine hematopoietic stem cells. Blood 99: 488-498. [DOI] [PubMed] [Google Scholar]
- Passegué E., Jamieson, C.H.M., Ailles, L.E., and Weissman, I.L. 2003. Normal and leukemic hematopoiesis: Are leukemias a stem cell disorder or a reacquisition of stem cell characteristics? Proc. Nat. Acad. Sci. (Suppl. 1) 100: 11842-11849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T., Morrison, S.J., Clarke, M.F., and Weissman, I.L. 2001. Stem cells, cancer, and cancer stem cells. Nature 414: 105-111. [DOI] [PubMed] [Google Scholar]
- Sauvageau G., Thorsteinsdottir, U., Eaves, C.J., Lawrence, H.J., Largman, C., Lansdorp, P.M., and Humphries, R.K. 1995. Overexpression of HOXB4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. Genes & Dev. 9: 1753-1765. [DOI] [PubMed] [Google Scholar]
- Schulte C.E., von Lindern, M., Steinlein, P., Beug, H., and Wiedemann, L.M. 2002. MLL-ENL cooperates with SCF to transform primary avian multipotent cells. EMBO J. 21: 4297-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So C.W., Karsunky, H., Passegué, E., Cozzio, A., Weissman, I.L., and Cleary, M.L. 2003. MLL-GAS7 transforms multipotent hematopoietic progenitors and induces mixed lineage leukemias in mice. Cancer Cell 3: 161-171. [DOI] [PubMed] [Google Scholar]
- Tenen D.G., Hromas, R., Licht, J.D., and Zhang, D.-E. 1997. Transcription factors, normal myeloid development, and leukemia. Blood 90: 489-519. [PubMed] [Google Scholar]
- Tkachuk D.C., Kohler, S., and Cleary, M.L. 1992. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell 71: 691-700. [DOI] [PubMed] [Google Scholar]
- Weissman I.L., Anderson, D.J., and Gage, F. 2001. Stem and progenitor cells: Origins, phenotypes, lineage commitments, and transdifferentiations. Annu. Rev. Cell Dev. Biol. 17: 387-403. [DOI] [PubMed] [Google Scholar]
- Yagi H., Deguchi, K., Aono, A., Tani, Y., Kishimoto, T., and Komori, T. 1998. Growth disturbance in fetal liver hematopoiesis of Mll-mutant mice. Blood 92: 108-117. [PubMed] [Google Scholar]
- Yu B.D., Hess, J.L., Horning, S.E., Brown, G.A., and Korsmeyer, S.J. 1995. Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378: 505-508. [DOI] [PubMed] [Google Scholar]
- Zeisig B.B., Garcia-Cuellar, M.P., Winkler, T.H., and Slany, R.K. 2003. The oncoprotein MLL-ENL disturbs hematopoietic lineage determination and transforms a biphenotypic lymphoid/myeloid cell. Oncogene 22: 1629-1637. [DOI] [PubMed] [Google Scholar]