Abstract
Murine cytomegalovirus (MCMV) immediate-early protein 3 (IE3) is essential for successful viral infection. This study developed MCMVs with an EGFP-fused IE3 gene in order to study IE3 gene expression, subnuclear distribution and biological function, as well as to examine the interaction of IE3 with cellular and viral proteins. The generated viruses included MCMVIE3gfp, in which IE1 was completely removed by the in-frame fusion of exons 3 and 5 and the C terminus of IE3 was tagged with EGFP, and MCMVIE1/3gfp, in which IE1 was kept intact and EGFP was also fused to the C terminus of IE3. Unlike human CMV (HCMV), whose growth was significantly reduced when IE2 (the HCMV homologue of IE3 in MCMV) was tagged with EGFP, MCMVs with IE3–EGFP presented an unchanged replication profile. Using these new constructs, the distribution of IE3 was revealed as well as its interaction with viral and cellular proteins, especially proteins pertaining to DNA replication (M44 and E1) and cellular intrinsic defence [promyelocytic leukemia protein and histone deacetylases (HDACs)]. It was also shown that IE3 domains co-localize with DNA replication domains, and IE3 attracted other required proteins into IE3 domains via protein–protein interactions. In addition, IE3 was shown to interact with HDAC2 and to eliminate the inhibitory effect of HDAC2 on early viral gene production. Together, these results suggest that IE3 acts as a key protein for viral DNA replication by establishing pre-replication domains via recruitment of the required viral and cellular proteins, and by reducing host defences.
INTRODUCTION
Cytomegaloviruses (CMVs) are large, dsDNA viruses with genomes of ∼230 kb, and are highly adapted to their respective hosts. CMV replication in host cells is a well-defined, sequential process: entry into cells, immediate-early (IE) gene expression, early gene expression, DNA replication, late gene expression and viral production (Mocarski et al., 2006). Many viral gene products function through interaction with cellular proteins or other viral proteins at different stages of viral infection. Studies of the essential gene products of CMV may lead to novel therapeutic strategies against CMV-associated diseases.
Murine CMV (MCMV) infection in the mouse is the most used and, so far, the best small animal model for studies of human CMV (HCMV) as the viruses have similar genomic structures, infectious cycles and methods of pathogenesis (Reddehase et al., 2008; Stinski & Isomura, 2008; Stinski & Petrik, 2008). MCMV and HCMV present a similar biological interaction with host cells at the IE stage. In this stage, the major IE (MIE) gene products such as IE3 of MCMV, or the HCMV homologue IE2, are needed to shut off host gene expression and DNA replication and cause cell-cycle arrest (Salvant et al., 1998; Wiebusch & Hagemeier, 1999; Wiebusch et al., 2008). Both HCMV and MCMV MIE genes are expressed independently of other de novo viral gene expression (Keil et al., 1987). The IE gene transcription unit is activated by tegument proteins, possibly through sequestration of Daxx and histone deacetylases (HDACs) (Tang & Maul, 2003; Taylor & Bresnahan, 2005), and the transcript is differentially spliced. Translation of the major spliced MIE transcripts produces the two most abundant viral proteins: IE1 and IE2 in HCMV and IE1 and IE3 in MCMV (Angulo et al., 2000; Keil et al., 1987; Messerle et al., 1992; Stenberg, 1996). For both CMVs, IE1 is formed by splicing exons 1–4, and MCMV IE3 (or HCMV IE2) by splicing exons 1–3 plus exon 5 (Keil et al., 1987). These spliced products appear to be necessary for the activation of early promoters in the tightly regulated transcription cascade (Angulo et al., 2000; Ghazal et al., 2005; Mendelson et al., 1996; Messerle et al., 1992).
HCMV IE2 has been shown to interact with several cellular proteins that are involved in transcription, such as the TATA-binding protein (Caswell et al., 1993) and the transcription-associated factor TFIID (Hagemeier et al., 1992). In fact, HCMV IE2 itself appears to function much like transcription-associated factors (Lukac et al., 1997). The MCMV IE3 or HCMV IE2 protein is essential to activate early gene promoters (Angulo et al., 2000; Buhler et al., 1990; Cherrington & Mocarski, 1989; McElroy et al., 2000; Scully et al., 1995; Stenberg, 1996; Stenberg & Stinski, 1985; Stenberg et al., 1990), whereas IE1 has been shown to be non-essential for virus replication for both MCMV and HCMV at high m.o.i. (Ghazal et al., 2005; Greaves & Mocarski, 1998). The biological functions of HCMV IE2 and the importance of its post-translational modifications and functional regions have been widely investigated and were recently reviewed by Stinski & Petrik (2008). However, IE3 of MCMV remains uninvestigated in many aspects, largely due to the lack of a specific antibody.
In MCMV, deletion of IE1 does not affect virus replication in cell culture, even though it is important for virus replication in vivo (Ghazal et al., 2005), suggesting that IE3 function precedes IE1 effects. A recent paper showed that IE3 can arrest MCMV-infected cells in the G1 and G2 phases (Wiebusch et al., 2008), which is similar to HCMV IE2, except that HCMV IE2 arrests HCMV-infected cells in the G1 phase only (Jault et al., 1995; Wiebusch & Hagemeier, 1999). Previously, it has been shown that MCMV IE3 (but not IE1) can activate early gene expression and interact with several cellular proteins (Messerle et al., 1992; Tang et al., 2005); this finding resulted from experiments using a transfection and co-transfection system. It is important to know whether this protein exhibits the same or different functions in a cell-culture infection system. In addition, we were curious as to whether MCMV IE3 could also interact with HDACs that have been demonstrated to exert regulatory effects on viral gene expression and replication. In this paper, we showed that IE3 can interact with several key viral and cellular proteins during viral infection and may be the key protein in the formation of viral DNA replication domains.
RESULTS
Generation and confirmation of MCMVIE3gfp and MCMVIE1/3gfp
The MCMV IE3 gene is essential for virus replication (Angulo et al., 2000). Although there is a high level of amino acid sequence similarity between MCMV IE3 and its HCMV homologue IE2, especially in terms of the C-terminal acidic sequences (Wiebusch et al., 2008), there are differences between MCMV and HCMV regarding their interactions with their respective hosts at the IE stage of infection [reviewed by Maul & Negorev (2008)]. Therefore, we cannot infer the function of IE3 simply from studying HCMV IE2; however, new findings regarding one counterpart may be instructive about the other.
In this study, we focused on the role of MCMV IE3 in the formation of DNA replication compartments (RCs) by using new viral constructs that could be followed by using a fluorescent tag. A galactokinase (galK)-positive/counterselection bacterial artificial chromosome (BAC) system was used to tag EGFP to the C terminus of exon 5, thus leaving IE1 functional and tagging IE3 (MCMVIE1/3gfp; Fig. 1). We also fused exon 3 with exon 5 (thus excluding exon 4, which is required to produce IE1) and tagged exon 5 to create MCMVIE3gfp, which produces functional tagged IE3, but no transcript for IE1. This BAC system used Escherichia coli SW102 to recombine DNA with 50 bp homology, thus making the resulting DNA mutation, deletion or fusion exactly as desired; this is also called a seamless BAC system. The DNA sequences of the homologous DNA and primers used for recombination are shown in Supplementary Fig. S1 (available in JGV Online).
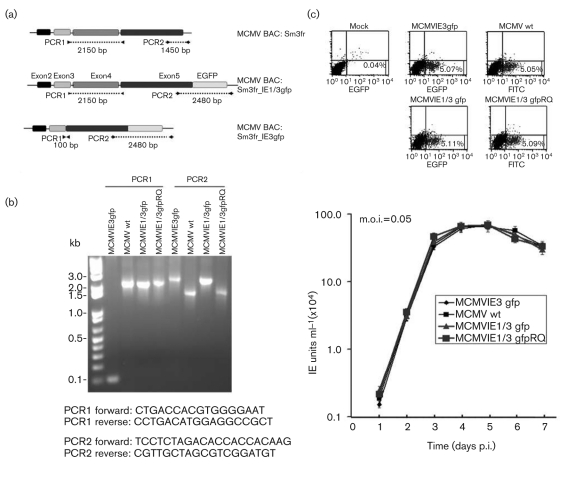
Fig. 1.
Construction and confirmation of MCMV mutants. (a) Diagram of the MIE gene structure of MCMV and its mutants: Sm3fr is the original BAC in which IE1/3 is the wild-type (wt) MIE gene; IE1/3gfp has an in-frame fusion of EGFP with exon 5 at its C terminus; IE3gfp resulted from the fusion of exons 3 and 5 and has an in-frame fusion of EGFP with exon 5 at its C terminus. PCR1 and PCR2 sites are shown and the sizes of the PCR products are labelled. (b) PCR to identify the resulting viruses using viral DNA isolated from infected NIH3T3 cells with the Hirt isolation protocol (Eizuru et al., 1984). The DNA sequences of the primers indicated in (a) are shown below the gel. (c) Viral growth curve in NIH3T3 cells. NIH3T3 cells seeded in 24-well plates were infected with MCMV (wt), mutants and RQ (rescued) as indicated (graph). Samples of cells with medium were collected at the indicated time and used for viral IE unit detection with a FACSCalibur (using two lasers and four channels; see Methods). The infection rate of each virus was detected by FACSCalibur (upper panels) to ensure the viral infections were at the same rate so that the IE unit assay was comparable.
The resultant BAC DNAs (BACmids MCMVIE3gfp, MCMVIE1/3gfp and the rescued MCMVIE1/3-gfpRQ; Fig. 1a) were purified and transfected into NIH3T3 cells, and the resultant viruses were amplified and purified. First, we performed PCR (to show the size of the changed DNAs) and sequenced the viral DNAs around the following mutation sites: the site of the fusion of exon 3 and exon 5 for MCMVIE3gfp, the egfp gene sequences at the C terminus of exon 5, and around the mutation sites. The results showed the sizes of the PCR products to be correct (Fig. 1b).
Virus replication curve in NIH3T3 cells
HCMV IE2 was recently tagged with EGFP to show its dynamic interactions with host cells during infection; however, even though the generated virus with EGFP-tagged IE2 could replicate, it showed significantly lower virus production compared with wild type (Sourvinos et al., 2007). To determine whether MCMVs with EGFP-tagged IE3 had a changed replication profile, we used a FACSCalibur flow cytometer to detect IE units and measure viral growth. As shown in Fig. 1(c), the same amounts of different viruses were used to infect NIH3T3 cells and viral growth logarithmic curves were determined. As can be seen, all three constructed viruses showed the same growth pattern as wild-type MCMV.
Viral protein production
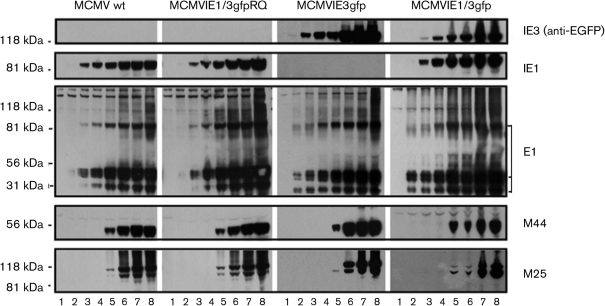
After infecting NIH3T3 cells with the viruses at an m.o.i. of 1, we utilized Western blotting (WB) to detect the production of several viral proteins at different times of infection. Cellular lysis samples from infections with MCMV mutants or mock-infected cells were collected at seven time points post-infection (p.i.), ranging from 3 to 72 h (Fig. 2). Lysed samples were run on WBs and probed with antibodies against IE1, EGFP, E1 (also called M112/113), M44 (viral DNA polymerase-associated protein) and M25. IE1 could be detected in wild-type, MCMVIE1/3gfp and the rescued virus infection; anti-EGFP antibody detected a band with a size of ∼108 kDa (fused IE3–EGFP) in the MCMVIE3gfp and MCMVIE1/3gfp infections. Other viral proteins detected included E1 and M44, both of which are essential for viral DNA replication. We also detected M25 95 and 105 kDa proteins (Wu et al., 1999), which are both late-early proteins of unknown function.
Fig. 2.
Confirmation of the resultant MCMVs with EGFP-fused IE3 gene by WB. NIH3T3 cells were mock-infected (lane 1) or infected with MCMV (indicated on the top) for 3 h (lane 2), 6 h (lane 3), 12 h (lane 4), 24 h (lane 5), 36 h (lane 6), 48 h (lane 7) and 72 h (lane 8). The whole-cell lysis samples were separated by PAGE and transferred to nitrocellulose membranes for WB. Blots were first probed with anti-EGFP to detect ∼108 kDa IE3–EGFP (top), then stripped and reprobed with antibodies against the proteins indicated on the right. MCMVIE3gfp did not produce IE1 protein. Note that the revertant (MCMVIE1/3gfpRQ) produced IE1, but did not produce IE3–EGFP. Also note that the presence of IE1 protein appeared to delay the production of IE3–EGFP (top right panel), which in turn may have delayed the production of M25 protein (bottom right panel).
Viral proteins and ND10
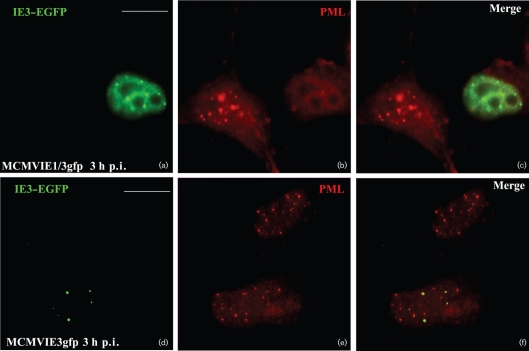
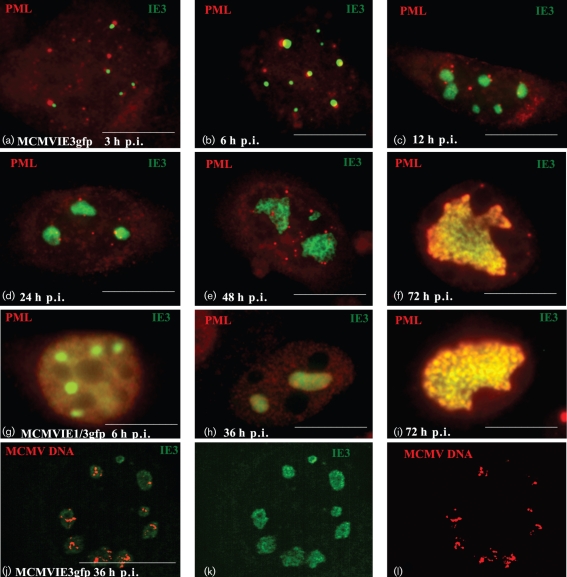
We also characterized MIE protein distribution and detected its relationship to nuclear domain 10 [ND10; also called promyelocytic leukemia protein (PML) oncogenic domain (POD), or a PML body] after infection of our MCMVs with or without IE1 in NIH3T3 cells. At 3 h p.i. in NIH3T3 cells grown on coverslips, the cells were fixed, permeabilized and then probed with anti-PML antibody to demonstrate the position of ND10 (Fig. 3b, e). IE3–EGFP (Fig. 3a, d) was seen only in infected cells. Additionally, we found that IE3–EGFP was distributed adjacent to ND10 at this time point of infection (Fig. 3f), which is consistent with our previous report that IE3 is distributed at ND10 after transfection of IE3–GFP-expressing plasmid (Tang et al., 2005). In MCMVIE1/3gfp-infected cells, ND10 was dispersed (Fig. 3b), which is consistent with a previously described action of IE1 (Tang & Maul, 2003).
Fig. 3.
EGFP-fused IE3 gene products and ND10 after viral infection. NIH3T3 cells were infected with MCMVIE3gfp (a–c) and MCMVIE1/3gfp (d–f) for 3 h, fixed and permeabilized, and stained with anti-PML antibody to detect ND10, followed by Texas Red-labelled secondary antibody. The results show PML alone (red; b and e), IE3–GFP alone (green; a and d) and the merged images (c and f). Bars, 10 μm.
Viral protein production sequence
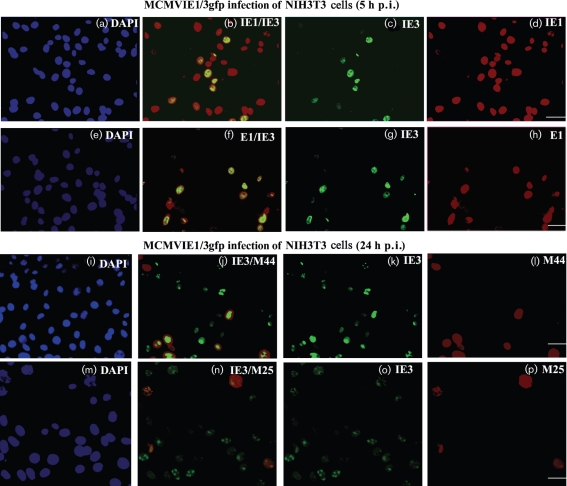
After confirming IE3–EGFP production and its associations with ND10, we were next able to use immunofluorescent methods to follow the production sequence of viral proteins after infection. At 5 or 24 h after MCMVIE1/3gfp infection in NIH3T3 cells (m.o.i. of 1), cells were fixed and immunostained for IE1, E1, M44 or M25, as shown in Fig. 4 (red staining). DAPI staining of nuclei indicated the total number of cells (infected and uninfected). Fig. 4(a–d) and Table 1 showed that IE3–EGFP was not detectable in many IE1-positive cells, suggesting that IE1 is produced ahead of IE3, consistent with in vivo studies (Kurz & Reddehase, 1999). IE3 is the activator of the E1 promoter (Messerle et al., 1992; Tang & Maul, 2005); therefore, we expected the production of IE3 to be significantly earlier than E1. Surprisingly, E1 could be detected at almost the same time as IE3 (Fig. 4e–h), which was consistent with the results of the WB assay (Fig. 2). Furthermore, we randomly selected 1000 cells (both infected and uninfected) by counting DAPI-stained cells and counted IE3-positive cells (green due to EGFP expression) with IE1- or E1-positive (red) cells. Table 1 showed a similar percentage of IE3- and E1-positive cells but a significant difference between IE3- and IE1-positive cells. In NIH3T3 cells infected with MCMVIE1/3gfp, M44 was not detectable by either WB or immunofluorescence (IF) at 5 h p.i. (data not shown), so IF was performed at 24 h p.i. (Fig. 4i–l). There were significantly fewer M44-positive cells than IE3-positive cells at 24 h p.i.; we also noted that M44 appeared to be ‘attracted’ to the IE3 domain at the same time that M44 could be detected. As M44 is an essential protein for viral DNA replication, this observation suggested that IE3 and E1 might form pre-replication domains. In addition, viral membrane protein M25 could be detected at later times after infection as shown in Fig. 4(m–p). This, combined with the data shown by WB in Fig. 2, indicated that IE3 is produced after IE1 and at the almost same time as E1; however, M44 and M25 are early-late proteins that are detectable significantly later than either E1 or IE3.
Fig. 4.
Immunofluorescent assay for detection of viral proteins after infection. NIH3T3 cells on coverslips were infected with MCMVIE1/3gfp for 5 h (a–h) or 24 h (i–p) and fixed. DAPI staining (panels a, e, i and m) was used to show the total number of cells (infected and uninfected). IE3 was detected as green fluorescence due to the fused EGFP (c, g, k, and o). Other viral proteins (IE1 and E1, early proteins; M44 and M25, early-late proteins) were stained in red (d, h, l and p). The merged images are shown in (b), (f), (j) and (n). Bars, 20 μm.
Table 1.
Numbers of cells with different viral proteins detected at 5 h after infection of NIH3T3 cells with MCMVIE1/3gfp
At 5 h p.i. after MCMVIE1/3gfp infection of NIH3T3 cells (m.o.i. of 1), cells were fixed and immunostained for IE1 (left) or E1 (right). IE3–EGFP signals were also counted. A total of 1000 randomly selected cells (infected and uninfected) was counted by DAPI staining. The difference between IE1- and IE3-positive cells was statistically significant (Student's t-test, P<0.01), whilst there no significant difference between IE3- and E1-positive cells.
| IE3/IE1 | IE3/E1 | ||||
|---|---|---|---|---|---|
| Total cells | IE1 positive | IE3 positive | Total cells | IE3 positive | E1 positive |
| 1000 | 688 (68.8 %) | 288 (28.8 %) | 1000 | 298 (29.8 %) | 286 (28.6 %) |
IE3–EGFP visualizes viral DNA replication domain formation
Previously, we reported that IE3 interacts with E1 and forms domains in the nucleus after co-transfection (Tang et al., 2005). Because IE3 co-localizes with E1, and E1 has been determined to be a DNA replication protein (Tang et al., 2005), we asked whether IE3 could be used to visualize viral DNA replication domain formation. An expansion of the IE3 domain in size during the time course of infection would support our hypothesis that IE3 is essential for the formation of DNA RCs in MCMV-infected cells.
We infected NIH3T3 cells with MCMVIE3gfp (m.o.i. of 1), and cells were fixed, permeabilized and probed with anti-PML antibody at various time points p.i. (Fig. 5a–f). At 3 and 6 h p.i. (Fig. 5a, b), there were several green IE3–EGFP dots adjacent to ND10; at 12, 24, 48 and 72 h p.i., the IE3–EGFP regions became noticeably larger (Fig. 5c–f). By 24 h p.i., when MCMV DNA replication is supposed to begin, the IE3 domains formed different shapes, and ND10 surrounded these domains (Fig. 5e). Later, at 48 and 72 h p.i., ND10 protein co-localized with the IE3 domains (Fig. 5e, f), at which time IE3 was seen to form ‘sausage-like’ compartments (Fig. 5f).
Fig. 5.
Development of IE3 domains. NIH3T3 cells were infected with MCMVIE3gfp for different lengths of time as indicated, fixed and permeabilized, and stained with anti-PML antibody and reacted with Texas Red-labelled secondary antibody (a–i). IE3–EGFP (green, a–k) was used to detect IE3 domain development in terms of both size and variation of shape. FISH was used to detect viral DNA replication in cells that had been infected with MCMVIE3gfp for 36 h (l); the replicating MCMV DNA is shown in red, whilst IE3–EGFP is shown in green (k) and the merged image in (j). Bars, 10 μm.
We wondered whether the formation of IE3 domains was affected by the IE1 that was detectable prior to IE3. To find out, we infected NIH3T3 cells with MCMVIE1/3gfp, and the cells were fixed and probed at 6, 36 and 72 h p.i. (Fig. 5g–i). We observed similar expansion of an IE3 domain, as with MCMVIE3gfp infection, after which the IE3 domain again formed a large domain that had a ‘sausage-like’ structure. By 72 h p.i., ND10 strongly co-localized with the IE3 domains. The only notable difference between the infection of MCMVIE1/3gfp (with IE1) and that of MCMVIE3gfp (without IE1) was that ND10 appeared more dispersed and less punctate in the MCMVIE1/3gfp infection.
We then tested whether the IE3 domains co-localized with replicating DNA using a fluorescent in situ hybridization (FISH) assay. Fig. 5(j–l) shows the results for MCMVIE3gfp-infected NIH3T3 cells at 36 h p.i. After infection, the cells were fixed and treated with RNase to remove RNA. Viral DNA was hybridized with a biotin-labelled probe prepared from MCMV BAC DNA and visualized with Texas Red-conjugated avidin. As seen in the merged panel (Fig. 5j), replicating viral DNA (red) only appeared in IE3 domains (green). Because IE3–EGFP can be produced in the very early stages of infection and forms domains before DNA replication, we propose that our IE3–EGFP probe can be used to localize viral pre-RCs and DNA replication domains.
Viral DNA replication proteins are attracted to IE3 domains through protein–protein interaction
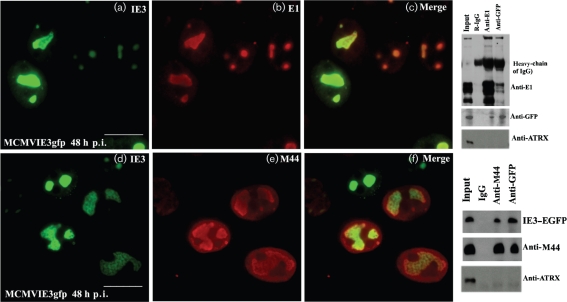
Two viral proteins, E1 and M44, have previously been shown to act as viral DNA replication proteins (Ciocco-Schmitt et al., 2002; Loh et al., 1999; Tang et al., 2005). After MCMVIE3gfp infection in NIH3T3 cells for 48 h (Fig. 6), we fixed cells and performed IF with anti-E1 or anti-M44 antibodies. As can be seen, IE3 domains co-localized well with E1 in the nucleus (Fig. 6c), and this occurred in almost all cells observed, consistent with previous findings (Tang et al., 2005). Fig. 6 showed cells that were unequally (or not simultaneously) infected, suggested by the sizes of the IE3 dots or domains. We noted that M44 was detectable and co-localized with IE3 only in the cells with large IE3 domains (Fig. 6f).
Fig. 6.
Interactions of IE3 with E1 and M44. Left: NIH3T3 cells were infected with MCMVIE3gfp for 48 h, fixed and permeabilized, stained with anti-E1 antibody (upper panels) or M44 (lower panels) and reacted with Texas Red-labelled secondary antibody. The IE3–EGFP signal (green) was merged with that of the viral DNA replication proteins to show co-localization. Bars, 10 μm. Right: co-IP was performed to determine the interactions of the viral proteins. Upper panels show the interaction of IE3 and E1. Anti-E1 (lane 3) and anti-EGFP (lane 4) rabbit antibodies were used, and rabbit IgG was used as a control (lane 2). The blot was probed with anti-E1, anti-EGFP and anti-ATRX antibodies to detect E1, IE3 and IE1, respectively. Lower panels show the interaction of IE3 and M44. Anti-M44 (lane 3) and anti-EGFP (lane 4) antibodies were from mouse, and mouse IgG (lane 2) was used as a control. The blot was probed with anti-EGFP, anti-M44 and anti-ATRX antibodies. As M44 is very close to the heavy chain of IgG, the TrueBlot system was used to eliminate the heavy chain.
To determine whether the overlapping of IE3 domains with either E1 or M44 could be due to protein–protein interactions, we performed a co-immunoprecipitation (co-IP) assay. A nuclear extract was prepared at 36 h after infection of NIH3T3 cells with MCMVIE3gfp (m.o.i. of 1) and reacted with anti-EGFP (mouse or rabbit), anti-E1 (rabbit) and anti-M44 (mouse) antibodies. Mouse and rabbit pre-immune sera were used as IgG controls. As shown in the blots in Fig. 6 (right panels), both M44 and E1 could be pulled down with anti-EGFP (IE3) antibody, whilst anti-E1 or anti-M44 antibody could pull down IE3. We also probed a nuclear protein (ATRX) in both co-IP systems and found that ATRX was not pulled down by anti-EGFP (IE3), anti-E1 or anti-M44 antibodies. Therefore, we concluded that IE3 interacts with these virus replication proteins in the RC. The same results were also found for MCMVIE1/3gfp infection in NIH3T3 cells (data not shown); thus, the interactions of IE3 with E1 and M44 appeared to be independent of IE1.
IE3 interacts with cellular proteins and eliminates the repressive effects of HDAC on E1 production
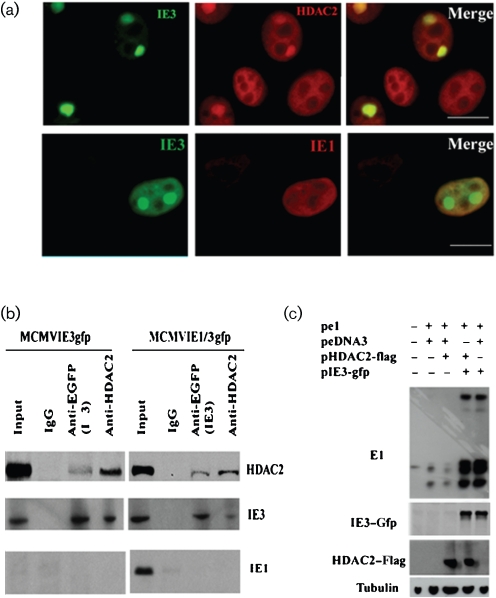
One of the most important gene silencers involved in effectively suppressing virus replication is HDAC. HDAC2 and -3 both interact with viral proteins, and CMV needs to counter their defensive silencing for successful replication (Michaelis et al., 2005; Nevels et al., 2004; Park et al., 2007; Wright et al., 2005). Using IE3–EGFP, we also showed a relationship between IE3 and HDAC2 (Fig. 7). We infected NIH3T3 cells with MCMVIE3gfp and fixed the cells at 24 h p.i. for IF to detect HDAC2 and IE3–EGFP. As can be seen in the merged image, some of the HDAC2 was drawn to the IE3 domains, although most remained diffused in the nucleus (Fig. 7a, upper panels). We reported previously that MCMV IE1 is able to interact with HDAC2 and reduce its activity at 2 h p.i. (Tang & Maul, 2003), so we wanted to know whether IE1 could be present in the IE3 domains and thus be responsible for any HDAC2 activity reduction. However, in this study, we observed that IE1 diffused in the nucleus of MCMVIE1/3gfp-infected cells (24 h p.i.) and was not associated with the IE3 domains (Fig. 7a, lower panels), suggesting that IE1 only interacts with HDAC2 at a very early stage of infection and that IE3 takes over interference with HDAC2 at later stages.
Fig. 7.
Interaction of IE3 with HDAC2. (a) NIH3T3 cells were infected with MCMVIE3gfp for 24 h, fixed and permeabilized, and stained with anti-HDAC2 (upper panels) or anti-IE1 (lower panels) antibodies and detected with Texas Red-conjugated secondary antibody (red). IE3–EGFP (green) shows the IE3 domains. Co-localization is shown in the merged panels. Bars, 10 μm. (b) Mouse anti-EGFP antibodies, anti-HDAC2 antibodies and mouse IgG were bound to protein G beads and incubated with nuclear extracts prepared from NIH3T3 cells infected with MCMVIE3gfp or MCMVIE1/3gfp for 24 h. In the WB of the co-IP, we used the TrueBlot system in order to eliminate the heavy and light chains of IgG (which are usually not easy to separate from HDAC2). Note that IE3–EGFP interacted with HDAC2 reciprocally, but IE1 interacted with neither. (c) Plasmids expressing E1, HDAC2 and IE3 (pe1, pHDAC2-flag and pIE3-gfp, respectively) were co-transfected into NIH3T3 cells as indicated for 24 h (pcDNA3 was added as necessary to make the total DNA amount equal in each sample). Total cell lysates were separated by PAGE for subsequent WB and probed with the antibodies indicated on the left. Tubulin production was used as a sample loading control.
Unlike HCMV IE1, MCMV IE1 appears to have only a weak effect on virus replication (Tang & Maul, 2003). Therefore, we wanted to know whether IE3 might substitute as the major protein responsible for countering cellular intrinsic defence in MCMV. To test first for protein–protein interactions between HDAC2 and IE3, we performed a co-IP assay using mouse anti-EGFP (against IE3–EGFP) and anti-HDAC2 antibodies to react with nuclear extracts prepared from NIH3T3 cells infected with MCMVIE3gfp for 24 h. As shown in Fig. 7(b), IE3–EGFP interacted with HDAC2. In contrast, IE1 was not pulled down with either IE3–EGFP or HDAC2, suggesting little or no direct interaction of these proteins with IE1. In addition, the interactions of IE3 and HDAC2 could be detected in both MCMVIE3gfp- and MCMVIE1/3gfp-infected cells, suggesting that IE1 does not interfere with the interaction of IE3 with other proteins, nor, in all probability, with its function.
It has been reported previously that IE3 activates the E1 gene promoter and increases E1 production (Messerle et al., 1992; Tang et al., 2005). Therefore, we wondered whether E1 production could also be affected by HDAC2. When we co-transfected E1-expressing plasmid (Fig. 7c; pe1) with HDAC2, E1 production was clearly reduced (Fig. 7c; compare lanes 3 and 2, with and without HDAC2, respectively). We then co-transfected an IE3-expressing plasmid with the E1 and HDAC2 plasmids (Fig. 7c, lane 4), which showed that IE3 could eliminate the repressive effect of HDAC2 on E1 gene expression. Therefore, IE3 not only interacts with HDAC2 but also inhibits HDAC2's repressive effect on gene expression.
DISCUSSION
The interactions of CMV and host cells at the molecular level have been studied in recent years. These interactions start once the virus contacts the cells and involve many processes, including cell signal transduction, cell-cycle control and cellular defensive responses; in addition, they usurp the cellular functional machineries leading to replicative success (Fortunato et al., 2000). It is believed that the interactions between CMV and host cells at different stages determine the fate of infected viruses: productive replication, latency or being cleared out of cells. Chemical arrays have been used to screen for small molecules that interfere with virus replication by targeting the interaction between viruses and cells (Loregian & Coen, 2006). Therefore, understanding the complicated interplay between viruses and host cells is critical not only for elucidating the pathogenesis of viruses but also for developing strategies against viral infections.
One of the consequences of virus–host interactions is the formation of virus RCs. Whether the formation of RCs is viral DNA dependent remains a subject of controversy. As in the current studies, it has been demonstrated elsewhere that, in both HCMV and herpes simplex virus type 1 infections, large globular domains are formed before DNA replication (Wilkinson & Weller, 2004; Zhong & Hayward, 1997), designated pre-RCs. Apparently, pre-RCs contain both viral and cellular proteins that are needed for later viral DNA replication. We are interested in the key viral proteins of MCMV that are important for the assembly of pre-RCs. Regarding MCMV, we reported previously that E1 can itself form large irregularly shaped structures in the nucleus after plasmid transfection in mouse cells (Tang et al., 2005). However, E1 gene expression depends on transactivation of viral IE gene products and IE3 is required for activating the E1 promoter. Therefore, we hypothesized that IE3 is important to the formation of viral DNA replication domains.
To test our hypothesis, we first revealed that IE3 forms speckles in the nucleus at the very beginning of infection, and these speckles expanded in volume with increased infection time (Fig. 5). Furthermore, the IE3 domains were demonstrated to co-localize with DNA replication domains, as shown by FISH. We then found that some viral proteins (such as E1) interact with IE3 and co-localize with IE3 throughout the time course of infection (Figs 4 and 6). Therefore, our observations suggest that IE3 is important in the formation of pre-RCs. During the time of pre-RC formation, different viral proteins are attracted into the domains by IE3 through protein–protein interactions. That E1 can be detected and is present at virtually the same time as IE3 seems to contradict the fact that IE3 activates the E1 promoter and that E1 production relies on IE3 activation (this activation is what defines E1 as an early gene as opposed to an IE gene) (Ciocco-Schmitt et al., 2002). However, it is still possible that only a small amount of IE3 protein is required to activate the E1 promoter. Finally, we found that IE3's activities of attracting viral and cellular proteins to the pre-RCs were not affected by IE1, as similar results were observed with both MCMVIE3gfp- and MCMVIE1/3gfp-infected cells.
Host cells try to silence or eliminate viruses using cellular intrinsic defensive proteins such as HDACs. In this study, we observed that HDAC2 inhibited E1 gene expression. The observation was obtained in a co-transfection system: E1 production was clearly reduced when HDAC2-expressing plasmid was co-transfected with E1-expressing plasmid (Fig. 7c). When an IE3-expressing plasmid was added to the co-transfection system, the repressive effects of HDAC2 on E1 production were eliminated (Fig. 7c), indicating that the activating effect of IE3 on the E1 promoter might be through segregation and inhibition of HDAC2. This idea was also supported by the observations that IE3 interacts with HDAC2 (Fig. 7b) and co-localizes with HDAC2 in DNA replication domains (Fig. 7a).
Both IE1 and IE3 of MCMV can aggregate in the nucleus as granules at the beginning of infection (Tang & Maul, 2006; Tang et al., 2005). Some IE1 dots co-localize with ND10, and later IE1 diffuses and disperses ND10 (Tang & Maul, 2003). Previously, we showed that IE3 can form dense dots and co-localize with E1, so E1 could segregate IE3 and inhibit its repressive function on the MIE promoter (Tang et al., 2005). In this study, we constructed the viruses with EGFP-tagged MIE genes and showed that IE3 could attract HDAC2 into DNA RCs (Fig. 7a) and that this procedure was independent of IE1 because IE1 was not in the IE3 domains (Fig. 7a). These results also suggest that IE1 and IE3 might play different roles in virus replication, as IE1 is not important for MCMV replication in cell culture but significantly enhances MCMV infection in vivo (Ghazal et al., 2005).
In summary, we fused EGFP with the major MIE gene IE3 in order to visualize DNA RC formation and demonstrated that IE3 recruits cellular and viral proteins into the RC through protein–protein interactions. Our results suggest that MCMV IE3 is a multifunctional protein. Future studies will include the use of MCMV with an EGFP-tagged MIE gene in a time-lapse study in order to directly visualize RC formation and the interaction of IE3 with other proteins. The constructed virus can also be used for MCMV studies in vivo.
METHODS
Tissue culture.
NIH3T3 cells (ATCC) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10 % FCS and 1 % penicillin/streptomycin. For immunohistochemical staining, cells were grown on round coverslips (Corning) in 24-well plates (Falcon). Plasmids were transfected into cells using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions.
Antibodies.
The antibodies used for WB and IF and their dilutions were as follows: rabbit anti-HDAC2 and anti-HDAC3 (Abcam; 1 : 250 for IF, 1 : 1000 for WB); mAb against tubulin (Sigma-Aldrich; 1 : 1000 for WB); polyclonal antibodies against PML and ATRX and monoclonal anti-EGFP (Santa Cruz Biotechnology; 1 : 100 for IF to show PML and 1 : 1000 for WB to probe EGFP); mAbs against MCMV IE1 and E1 (from Dr S. Jonjic; 1 : 50 for IF and 1 : 200 for WB; Hengel et al., 1994; Tang et al., 2005); and mAbs against M44 and M25 (from Dr J. D. Shanley; 1 : 50 for IF and 1 : 250 for WB; Loh et al., 1999; Wu et al., 1999).
Generation of MCMVs with EGFP-fused IE3.
The MCMV BACmid DNA Sm3fr (Wagner et al., 1999) was transformed into Escherichia coli SW102 containing a galK-positive/counterselection cassette (Warming et al., 2005) by electroporation using a Gene Pulser Xcell Electroporation System (Bio-Rad). E. coli SW102/Sm3fr harbouring MCMV BACmid DNA was obtained and identified, kept in Luria broth with chloramphenicol and used as template for mutagenesis.
A seamless BAC system was used to construct MCMV tagged with EGFP at the C terminus of exon 5, for either MCMVIE1/3gfp or MCMVIE3gfp. In MCMV, IE1 is spliced from exons 1–4 in the MIE gene, whereas IE3 is spliced from exons 1–3 plus exon 5. To make MCMVIE3gfp, we fused exon 3 with exon 5, eliminating exon 4 and therefore IE1 (Fig. 1a). First, we utilized recombination to replace the DNA fragment between exons 3 and 5 by electroporation with a galK gene PCR product. The galK gene was flanked by homologous sequences H3 (on the left) and H4 (on the right) (DNA sequences are provided in Supplementary Fig. S1). The galK gene was then replaced with a short sequence (H3+H4; see Supplementary Fig. S1).
To add an EGFP tag at the C terminus of exon 5, we first needed to insert the galK gene between the last amino acid and the stop codon of exon 5. The galK gene was produced by PCR and flanked with a 50 bp homology sequence on left side (H1; Supplementary Fig. S1) and another on the right side (H2; Supplementary Fig. S1). Finally, the galK gene was replaced with an egfp gene produced by PCR and flanked by H1 and H2 (Supplementary Fig. S1).
Lastly, we constructed two BACmids: SM3fr_IE1/3gfp, in which exon 5 of IE1/3 was tagged with EGFP, and SM3fr_IE3gfp, in which IE1 was removed by the fusion of exons 3 and 5, and exon 5 of IE3 was tagged with EGFP (Fig. 1a). The BACmid DNAs were further confirmed by sequencing the sites of insertions or deletions. We subsequently transfected the BACmid DNAs into NIH3T3 cells by electroporation with a Bio-Rad Gene Pulser Xcell Electroporation System. The transfected cells were cultured back and the resulting viruses were purified: MCMVIE1/3gfp and MCMVIE3gfp, respectively.
To make revertant MCMV from the MCMVIE1/3gfp, the egfp gene was replaced with the galK gene using the method described above, after which the galK gene was replaced by the short fragment of H1+H2 (Supplementary Fig. S1). Finally, the resultant BACmid DNA was transfected into NIH3T3 cells and the viral particles were purified, resulting in MCMVIE1/3gfpRQ.
Co-immunoprecipitation assay.
Nuclear extracts were obtained essentially as described before (Tang et al., 2003). Antibodies were coupled to protein G–Sepharose beads (Amersham Pharmacia Biotech), according to the manufacturer's instructions. After blocking with PBS containing 0.1 % BSA, the beads were incubated overnight at 4 °C with clarified nuclear extracts, washed repeatedly in 0.1 % BSA in buffered saline and resuspended in a mixture of PBS and 2× Laemmli buffer. After being heated at 95 °C for 5 min, the beads were isolated by centrifugation; proteins were separated by SDS-PAGE and analysed by WB, as described below.
Immunoblot analysis.
Proteins were separated by 7.5 % SDS-PAGE (10–20 μg in each lane), transferred to nitrocellulose membranes (Amersham), and blocked with 5 % non-fat milk for 60 min at room temperature. Membranes were incubated overnight at 4 °C with primary antibody, followed by incubation with horseradish peroxidase-coupled secondary antibody [for regular WB, we used secondary antibody from Amersham; for the detection of protein in the immunoprecipitation, we used mouse or rabbit secondary antibodies from TrueBlot ULTRA (eBioscience)]. Detection was accomplished with ECL (Pierce), according to standard methods. Membranes were stripped with stripping buffer [100 mM β-mercaptoethanol, 2 % SDS, 62.5 mM Tris/HCl (pH 6.8)], washed with PBS containing 0.1 % Tween 20 and reprobed with new primary antibodies to detect additional proteins.
Nick translation.
dsDNA probes for in situ hybridization were labelled by nick translation, as described previously (Tang et al., 2000). Briefly, 1 μg pSM3fr plasmid DNA, 10× nick translation buffer, 0.05 mM dNTPs (dATP, dCTP and dGTP), 0.01 mM dTTP, 0.04 mM biotinylated UTP, 1 U DNA polymerase I and appropriate concentrations of DNase I were incubated at 15 °C for 50 min. Labelled fragments obtained from the protocol were 200–500 nt as determined on 2 % agarose gels.
Immunocytochemistry and FISH.
Immunostaining was performed on cells grown on coverslips fixed in 1 % paraformaldehyde (10 min at room temperature) and permeabilized in 0.2 % Triton (20 min on ice), followed by sequential incubation with primary antibodies and Texas Red- or FITC-labelled secondary antibodies (Vector Laboratories) for 30 min each (all solutions in PBS). For simultaneous detection of viral protein and specific DNA sequences, cells were first immunostained for viral proteins and then treated for 1 h at 37 °C with RNase (Roche; 100 μg ml−1 in PBS) for the detection of DNA. After refixing in 4 % paraformaldehyde (10 min at room temperature), samples were equilibrated in 2× SSC (1× SSC: 0.15 M NaCl plus 0.015 M sodium citrate), dehydrated in ethanol (70, 80 and 100 % ethanol for 3 min each at 20 °C), air dried and incubated overnight at 37 °C with the hybridization mixture.
For DNA detection, the probe and cells were simultaneously heated at 94 °C for 4 min to denature the DNA. After hybridization, samples were washed at 37 °C with 55 % formamide in 2× SSC (twice for 15 min each), 2× SSC (10 min) and 0.25× SSC (twice for 5 min each). Hybridized probes were labelled with Texas Red–avidin (Vector Laboratories; 1 : 500 in 4× SSC with 0.5 % BSA) and signals were amplified using biotinylated anti-avidin antibody (Vector Laboratories; 1 : 250), followed by another round of Texas Red–avidin staining. Finally, cells were equilibrated in PBS, stained for DNA with DAPI and mounted in Fluoromount G (Fisher Scientific).
Confocal microscopy.
Cells were examined with a Leica TCS SP II confocal laser-scanning system. Two channels were recorded simultaneously or sequentially and controlled for possible breakthrough between the green and red channels.
Flow cytometry-based quantitative virus replication assays.
MIE gene expression can only be detected after infection of live viral particles; in addition, MIE gene expression can be detected very soon (before DNA replication) after infection. Therefore, the number of infected cells expressing MIE gene products (IE unit) can be used to determine the number of infectious viral particles. Compared with conventional plaque assays, detection of the IE unit is more time-efficient and less labour-intensive. Flow cytometry was used to detect IE gene expression and determine the viral growth curve.
Viral infection and sample collection.
NIH3T3 cells were prepared in 12-well plates and infected with virus (wild-type MCMV, MCMVIE3gfp, MCMVIE1/3gfp and its rescued MCMVIE1/3gfpRQ) at an m.o.i. of 0.05 when cells reached ∼80 % confluency. Eight wells of cells were infected for each virus in six-well dishes (Corning). At 5 h p.i., the medium was discarded and replaced with 1 ml fresh MEM medium containing 5 % FCS. To ensure that the input viruses were at the same level, one well of cells for each viral infection was collected at 5 h p.i. to detect EGFP expression or IE1 expression by immunofluorescent assay using anti-IE1 antibody (followed by FITC-conjugated secondary antibody to show IE1 in green) and to determine the infection efficiency. One well of cells (together with the medium) for each viral infection was collected every day for 7 days. All samples were stored at −80 °C. The viral particles were released from cells by three freeze–thaw cycles and centrifuged at 13 253 g for 15 min at 4 °C to remove cellular debris. The supernatants became the viral samples for the next infection in NIH3T3 cells for the IE unit assay.
IE unit assay.
The number of IE units ml−1 could be shown by EGFP expression, as the MIE gene was fused to EGFP (or IE1–FITC for wild-type MCMV) and could only be expressed after being infected with live viral particles. NIH3T3 cells at ∼80 % confluency were infected with viral samples prepared as above (wild-type MCMV, MCMVIE3gfp, MCMVIE1/3gfp and its rescued MCMVIE1/3gfpRQ) with different dilutions for 6 h; the medium was discarded and cells were trypsinized and washed with PBS. The cells were then assayed by FACSCalibur (using two lasers and four channels; Becton Dickinson) in order to ascertain the total number of cells as well as the total number of EGFP-positive (or FITC-positive) cells. Mock-infected cells were prepared at the same time in order to have background control for the FACS. The real IE cell number was calculated as: [EGFP (or FITC) cell number of infected cells − EGFP (or FITC) cell number (false EGFP or FITC) of mock-infected cells]. The real IE cell number in 1 ml viral solution was defined as the number of IE units ml−1.
Supplementary Material
Acknowledgments
We would like to thank Drs S. Jonjic, M. Messerle, J. Shanley and H. Zhu for donating reagents. This study was supported by a Pilot Grant from the Research Center for Minority Institutes (RCMI) program of NIH-NCRR (2G12RR003050-24), an American Cancer Society grant RSG-090289-01-MPC, an ACS-IRG grant (IRG-92-032-13, subaward # 60-14599-01-01-S6) and a start-up fund from the Ponce School of Medicine (all to Q. T.). We also thank Pablo Lopez Colon and Omayra De Jesus Matos for technical assistance. We acknowledge Bob Ritchie for English editing and Dr Andrew Boileau for critical reading.
Footnotes
The sequences of the primers used in this study are available with the online version of this paper.
References
- Angulo, A., Ghazal, P. & Messerle, M. (2000). The major immediate-early gene ie3 of mouse cytomegalovirus is essential for viral growth. J Virol 74, 11129–11136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler, B., Keil, G. M., Weiland, F. & Koszinowski, U. H. (1990). Characterization of the murine cytomegalovirus early transcription unit e1 that is induced by immediate-early proteins. J Virol 64, 1907–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell, R., Hagemeier, C., Chiou, C. J., Hayward, G., Kouzarides, T. & Sinclair, J. (1993). The human cytomegalovirus 86K immediate early (IE) 2 protein requires the basic region of the TATA-box binding protein (TBP) for binding, and interacts with TBP and transcription factor TFIIB via regions of IE2 required for transcriptional regulation. J Gen Virol 74, 2691–2698. [DOI] [PubMed] [Google Scholar]
- Cherrington, J. M. & Mocarski, E. S. (1989). Human cytomegalovirus ie1 transactivates the α promoter-enhancer via an 18-base-pair repeat element. J Virol 63, 1435–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciocco-Schmitt, G. M., Karabekian, Z., Godfrey, E. W., Stenberg, R. M., Campbell, A. E. & Kerry, J. A. (2002). Identification and characterization of novel murine cytomegalovirus M112–113 (e1) gene products. Virology 294, 199–208. [DOI] [PubMed] [Google Scholar]
- Eizuru, Y., Inagawa, S. & Minamishima, Y. (1984). Application of “Hirt supernatant” DNA to the molecular epidemiology of cytomegalovirus infections. J Clin Microbiol 20, 1012–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunato, E. A., McElroy, A. K., Sanchez, I. & Spector, D. H. (2000). Exploitation of cellular signaling and regulatory pathways by human cytomegalovirus. Trends Microbiol 8, 111–119. [DOI] [PubMed] [Google Scholar]
- Ghazal, P., Visser, A. E., Gustems, M., García, R., Borst, E. M., Sullivan, K., Messerle, M. & Angulo, A. (2005). Elimination of ie1 significantly attenuates murine cytomegalovirus virulence but does not alter replicative capacity in cell culture. J Virol 79, 7182–7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves, R. F. & Mocarski, E. S. (1998). Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J Virol 72, 366–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeier, C., Walker, S., Caswell, R., Kouzarides, T. & Sinclair, J. (1992). The human cytomegalovirus 80-kilodalton but not the 72-kilodalton immediate-early protein transactivates heterologous promoters in a TATA box-dependent mechanism and interacts directly with TFIID. J Virol 66, 4452–4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengel, H., Lucin, P., Jonjic, S., Ruppert, T. & Koszinowski, U. H. (1994). Restoration of cytomegalovirus antigen presentation by gamma interferon combats viral escape. J Virol 68, 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jault, F. M., Jault, J. M., Ruchti, F., Fortunato, E. A., Clark, C., Corbeil, J., Richman, D. D. & Spector, D. H. (1995). Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J Virol 69, 6697–6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil, G. M., Ebeling-Keil, A. & Koszinowski, U. H. (1987). Immediate-early genes of murine cytomegalovirus: location, transcripts, and translation products. J Virol 61, 526–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz, S. K. & Reddehase, M. J. (1999). Patchwork pattern of transcriptional reactivation in the lungs indicates sequential checkpoints in the transition from murine cytomegalovirus latency to recurrence. J Virol 73, 8612–8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh, L. C., Keeler, V. D. & Shanley, J. D. (1999). Sequence requirements for the nuclear localization of the murine cytomegalovirus M44 gene product pp50. Virology 259, 43–59. [DOI] [PubMed] [Google Scholar]
- Loregian, A. & Coen, D. M. (2006). Selective anti-cytomegalovirus compounds discovered by screening for inhibitors of subunit interactions of the viral polymerase. Chem Biol 13, 191–200. [DOI] [PubMed] [Google Scholar]
- Lukac, D. M., Harel, N. Y., Tanese, N. & Alwine, J. C. (1997). TAF-like functions of human cytomegalovirus immediate-early proteins. J Virol 71, 7227–7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul, G. G. & Negorev, D. (2008). Differences between mouse and human cytomegalovirus interactions with their respective hosts at immediate early times of the replication cycle. Med Microbiol Immunol (Berl) 197, 241–249. [DOI] [PubMed] [Google Scholar]
- McElroy, A. K., Dwarakanath, R. S. & Spector, D. H. (2000). Dysregulation of cyclin E gene expression in human cytomegalovirus-infected cells requires viral early gene expression and is associated with changes in the Rb-related protein p130. J Virol 74, 4192–4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelson, M., Monard, S., Sissons, P. & Sinclair, J. (1996). Detection of endogenous human cytomegalovirus in CD34+ bone marrow progenitors. J Gen Virol 77, 3099–3102. [DOI] [PubMed] [Google Scholar]
- Messerle, M., Buhler, B., Keil, G. M. & Koszinowski, U. H. (1992). Structural organization, expression, and functional characterization of the murine cytomegalovirus immediate-early gene 3. J Virol 66, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis, M., Suhan, T., Reinisch, A., Reisenauer, A., Fleckenstein, C., Eikel, D., Gumbel, H., Doerr, H. W., Nau, H. & Cinatl, J., Jr (2005). Increased replication of human cytomegalovirus in retinal pigment epithelial cells by valproic acid depends on histone deacetylase inhibition. Invest Ophthalmol Vis Sci 46, 3451–3457. [DOI] [PubMed] [Google Scholar]
- Mocarski, E. S., Jr, Shenk, T. & Pass, R. F. (2006). Cytomegaloviruses. Philadelphia. : Lippincott Williams &Wilkins.
- Nevels, M., Paulus, C. & Shenk, T. (2004). Human cytomegalovirus immediate-early 1 protein facilitates viral replication by antagonizing histone deacetylation. Proc Natl Acad Sci U S A 101, 17234–17239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J. J., Kim, Y. E., Pham, H. T., Kim, E. T., Chung, Y. H. & Ahn, J. H. (2007). Functional interaction of the human cytomegalovirus IE2 protein with histone deacetylase 2 in infected human fibroblasts. J Gen Virol 88, 3214–3223. [DOI] [PubMed] [Google Scholar]
- Reddehase, M. J., Simon, C. O., Seckert, C. K., Lemmermann, N. & Grzimek, N. K. (2008). Murine model of cytomegalovirus latency and reactivation. Curr Top Microbiol Immunol 325, 315–331. [DOI] [PubMed] [Google Scholar]
- Salvant, B. S., Fortunato, E. A. & Spector, D. H. (1998). Cell cycle dysregulation by human cytomegalovirus: influence of the cell cycle phase at the time of infection and effects on cyclin transcription. J Virol 72, 3729–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully, A. L., Sommer, M. H., Schwartz, R. & Spector, D. H. (1995). The human cytomegalovirus IE2 86-kilodalton protein interacts with an early gene promoter via site-specific DNA binding and protein–protein associations. J Virol 69, 6533–6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourvinos, G., Tavalai, N., Berndt, A., Spandidos, D. A. & Stamminger, T. (2007). Recruitment of human cytomegalovirus immediate-early 2 protein onto parental viral genomes in association with ND10 in live-infected cells. J Virol 81, 10123–10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg, R. M. (1996). The human cytomegalovirus major immediate-early gene. Intervirology 39, 343–349. [DOI] [PubMed] [Google Scholar]
- Stenberg, R. M. & Stinski, M. F. (1985). Autoregulation of the human cytomegalovirus major immediate-early gene. J Virol 56, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg, R. M., Fortney, J., Barlow, S. W., Magrane, B. P., Nelson, J. A. & Ghazal, P. (1990). Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J Virol 64, 1556–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinski, M. F. & Isomura, H. (2008). Role of the cytomegalovirus major immediate early enhancer in acute infection and reactivation from latency. Med Microbiol Immunol (Berl) 197, 223–231. [DOI] [PubMed] [Google Scholar]
- Stinski, M. F. & Petrik, D. T. (2008). Functional roles of the human cytomegalovirus essential IE86 protein. Curr Top Microbiol Immunol 325, 133–152. [DOI] [PubMed] [Google Scholar]
- Tang, Q. & Maul, G. G. (2003). Mouse cytomegalovirus immediate-early protein 1 binds with host cell repressors to relieve suppressive effects on viral transcription and replication during lytic infection. J Virol 77, 1357–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Q. & Maul, G. (2005). Immediate early interactions and epigenetic defense mechanisms. In Cytomegaloviruses: Molecular Biology and Immunology. Edited by Reddehase, M. J.. Norwich, UK. : Horizon Scientific Press.
- Tang, Q. & Maul, G. G. (2006). Mouse cytomegalovirus crosses the species barrier with help from a few human cytomegalovirus proteins. J Virol 80, 7510–7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Q., Bell, P., Tegtmeyer, P. & Maul, G. G. (2000). Replication but not transcription of simian virus 40 DNA is dependent on nuclear domain 10. J Virol 74, 9694–9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Q., Li, L., Ishov, A. M., Revol, V., Epstein, A. L. & Maul, G. G. (2003). Determination of minimum herpes simplex virus type 1 components necessary to localize transcriptionally active DNA to ND10. J Virol 77, 5821–5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Q., Li, L. & Maul, G. G. (2005). Mouse cytomegalovirus early M112/113 proteins control the repressive effect of IE3 on the major immediate-early promoter. J Virol 79, 257–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, R. T. & Bresnahan, W. A. (2005). Human cytomegalovirus immediate-early 2 gene expression blocks virus-induced beta interferon production. J Virol 79, 3873–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, M., Jonjic, S., Koszinowski, U. H. & Messerle, M. (1999). Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J Virol 73, 7056–7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming, S., Costantino, N., Court, D. L., Jenkins, N. A. & Copeland, N. G. (2005). Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res 33, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebusch, L. & Hagemeier, C. (1999). Human cytomegalovirus 86-kilodalton IE2 protein blocks cell cycle progression in G1. J Virol 73, 9274–9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebusch, L., Neuwirth, A., Grabenhenrich, L., Voigt, S. & Hagemeier, C. (2008). Cell cycle-independent expression of immediate-early gene 3 results in G1 and G2 arrest in murine cytomegalovirus-infected cells. J Virol 82, 10188–10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, D. E. & Weller, S. K. (2004). Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J Virol 78, 4783–4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, E., Bain, M., Teague, L., Murphy, J. & Sinclair, J. (2005). Ets-2 repressor factor recruits histone deacetylase to silence human cytomegalovirus immediate-early gene expression in non-permissive cells. J Gen Virol 86, 535–544. [DOI] [PubMed] [Google Scholar]
- Wu, C. A., Carlson, M. E., Henry, S. C. & Shanley, J. D. (1999). The murine cytomegalovirus M25 open reading frame encodes a component of the tegument. Virology 262, 265–276. [DOI] [PubMed] [Google Scholar]
- Zhong, L. & Hayward, G. S. (1997). Assembly of complete, functionally active herpes simplex virus DNA replication compartments and recruitment of associated viral and cellular proteins in transient cotransfection assays. J Virol 71, 3146–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.