Abstract
Powassan virus (POW) is a tick-borne flavivirus distributed in Canada, the northern USA and the Primorsky region of Russia. POW is the only tick-borne flavivirus endemic to the western hemisphere, where it is transmitted mainly between Ixodes cookei and groundhogs (Marmota monax). Deer tick virus (DTV), a genotype of POW that has been frequently isolated from deer ticks (Ixodes scapularis), appears to be maintained in an enzootic cycle between these ticks and white-footed mice (Peromyscus leucopus). DTV has been isolated from ticks in several regions of North America, including the upper Midwest and the eastern seaboard. The incidence of human disease due to POW is apparently increasing. Previous analysis of tick-borne flaviviruses endemic to North America have been limited to relatively short genome fragments. We therefore assessed the evolutionary dynamics of POW using newly generated complete and partial genome sequences. Maximum-likelihood and Bayesian phylogenetic inferences showed two well-supported, reciprocally monophyletic lineages corresponding to POW and DTV. Bayesian skyline plots based on year-of-sampling data indicated no significant population size change for either virus lineage. Statistical model-based selection analyses showed evidence of purifying selection in both lineages. Positive selection was detected in NS-5 sequences for both lineages and envelope sequences for POW. Our findings confirm that POW and DTV sequences are relatively stable over time, which suggests strong evolutionary constraint, and support field observations that suggest that tick-borne flavivirus populations are extremely stable in enzootic foci.
INTRODUCTION
Powassan virus (POW; family Flaviviridae, genus Flavivirus) is the sole North-American representative of the tick-borne encephalitis serological complex of the flaviviruses. POW comprises two genetic lineages including the prototype lineage (POW, lineage I) and a second lineage that was first described in the late 1990s [deer tick virus (DTV), lineage II] (Telford et al., 1997). In nature POW and DTV appear to perpetuate in relatively distinct transmission cycles, with the former maintained by Ixodes cookei and/or Ixodes marxi and medium-sized mammals, and the latter maintained by Ixodes scapularis and white-footed mice (Artsob, 1988; Ebel et al., 2000). Whereas POW has been considered to be a minor public-health concern due to the relative host-specificity of its arthropod vector, DTV could be a greater threat since it is mainly associated with the tick vectors that have driven the ongoing epidemics of Lyme disease, human babesiosis and human granulocytic anaplasmosis (Rodgers & Mather, 2007; Daniels et al., 1998; Falco et al., 1995). DTV has been isolated from ticks in several regions of North America (Brackney et al., 2008; Ebel et al., 1999; Telford et al., 1997). It is considered a distinct lineage of POW because of high nucleotide and amino acid sequence identity (84 and 93 %, respectively) (Kuno et al., 2001) and strong serological cross-reactivity (Beasley et al., 2001). POW has been isolated from ticks and/or seropositive animals in Canada, the USA and the Primorsky region of Russia (Artsob, 1988; Leonova et al., 2009). Although data from field-based studies of POW and DTV are scant, two recent reports demonstrated that infection rates in ticks in a DTV focus in northern Wisconsin were remarkably stable over a 10-year period (Brackney et al., 2008; Ebel et al., 1999), and that infections within ticks were surprisingly genetically homogeneous, exhibiting little or no quasispecies structure (Brackney et al., 2010). Therefore, POW and DTV appear to be similar to many other tick-borne agents in that they are nidal infections that are maintained in highly stable enzootic foci for long periods of time. Paradoxically, human disease due to infection by POW and/or DTV appears to be increasing. Human cases have increased markedly between the periods 1958–1998 and 1999–2007 (Hinten et al., 2008), and a recent fatality was unequivocally attributed to DTV (Tavakoli et al., 2009). These observations raise the possibility that these agents may be an emerging burden to public health in regions of North America where they are enzootic. Accordingly, we sought to determine whether the apparent emergence of tick-borne flavivirus disease in North America may be driven by increases in the virus population size, reasoning that the well-documented expansion in the population of deer ticks that has occurred in recent decades may have consequently increased the population size, or sizes, of their pathogens. In particular, full genome sequences were determined and the phylogenetic relationships of POW and DTV from North America and Russia were reconstructed. We then estimated the divergence time between lineages and tested for evidence of positive selection. Finally, population dynamics were inferred under a Bayesian framework, thus shedding light on the mechanisms influencing the epidemiology of POW in North America.
RESULTS
Phylogenetic analyses
To assess the phylogenetic relationships of POW, we used 11 full-genome, 23 envelope and 22 NS-5 viral sequences. Kyasanur forest disease virus (KFDV) and Alkhurma virus were included as out-groups. Maximum-likelihood topologies were congruent in full-genome, NS-5 and envelope partitions in recovering two supported monophyletic groups that correspond to lineage I POW and DTV (lineage II POW) (Fig. 1a, b). Full genome topology showed that, within lineage I POW, samples were subdivided into Russia–Ontario [maximum-likelihood bootstrap (MLB)=100] and the sample from New York was basal. DTV showed two subclades clustering all samples from Wisconsin (MLB=99) and Massachusetts with Connecticut (MLB=100). Envelope and NS-5 topologies supported these divisions, with DTV from Colorado being basal to the Wisconsin strains, and the isolate of DTV from West Virginia basal to the New England strains.
Fig. 1.
Maximum-likelihood tree obtained from full coding sequences (10 245 nt) (a), envelope sequences (1491 nt) (b) and NS-5 partial sequences (2709 nt) (c) of POW and DTV strains. Numbers above branches indicate pseudoreplicate bootstrap values.
Substitution rates, divergence times and population size changes
Bayesian skyline plots (BSPs) were generated for envelope (Fig. 2a) and NS-5-encoding sequences (Fig. 2b), using relaxed (uncorrelated log normal) molecular-clock models. Overall, results indicate that the effective number of POW and DTV infections through time was stable over the distant past (years 1100–1950) with recent declines, apparently beginning around 1950. BSPs for envelope and NS-5 partitions were congruent in placing the time to the most recent common ancestor (MRCA) for DTV and POW at around 1000 years before the present (ybp) (Fig. 2). Estimated nucleotide substitution rates were 2.2×10−4 substitutions site−1 year−1 using envelope sequences and 3.9×10−5 substitutions site−1 year−1 using NS-5 sequences.
Fig. 2.
Population dynamics of POW and DTV depicted using BSPs derived from envelope-encoding sequences (a) and NS-5-encoding sequences (b). The plots show changes in relative genetic diversity, depicted as the effective number of infections (Neτ; Ne is the effective population size and τ the generation time from infected host to infected host) through time. The thick, solid line is the median estimate of Neτ, while the 95 % HPD intervals are shown in grey.
Using concatenated envelope and NS-5 sequences, we estimated divergence times of POW and DTV lineages under a Bayesian–Markov chain Monte Carlo (BMCMC) framework (Fig. 3). The time to the MRCA of DTV isolates was estimated at 195 ybp [highest posterior density (HPD)=73–382] and for lineage I POW, at 196 ybp (HPD=67–382); the analysis placed the time for the MRCA between the two lineages at approximately 484 ybp (HPD=114–1048).
Fig. 3.
Maximum clade credibility tree generated from concatenated envelope and NS-5 sequences (4200 nt) of POW and DTV strains. The median node ages and their 95 % HPD values are shown on the major nodes, with the 95 % HPD values shown in parentheses.
Selection analyses in POW strains
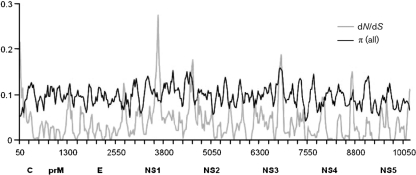
Sliding window analysis of dN/dS and π, conducted with complete genomic coding sequences of both POW and DTV, indicated purifying selection throughout the genome, with values distributed below 0.3 for dN/dS throughout (Fig. 4). Likelihood-based approaches mostly agreed, showing that most substitutions were silent and probably reflect strong purifying selection acting over the POW genome. The single-likelihood ancestor counting (SLAC), random effects likelihood (REL) and partitioning approach for robust inference of selection (PARRIS) models detected purifying selection throughout the sequences, while the fixed effects likelihood (FEL) model found evidence for positive selection at two different sites in two different NS-5 partitions (one in POW lineage I only, the other evident in comparisons of the two lineages) (Table 1). Using the selecton server (http://selecton.tau.ac.il) we found evidence for positive selection in the envelope and NS-5 sequences using the MEC model and in the DTV NS-5 sequences using the M8 model (Table 1). McDonald–Kreitman (M–K) tests showed significant values for partitions of full-genome sequences (P=0.035) and NS-5 sequences (P=0.001), but did not conclusively show significant differences from neutral evolution for envelope sequences (P=0.077) (Table 1).
Fig. 4.
Estimates of dN/dS and genetic diversity (π) across POW and DTV full-genome sequences.
Table 1.
Results from selection analyses
Models to test for selection were conducted on the datamonkey and selecton servers as indicated. CDS, Coding sequence; NS, not significant.
| Partition | datamonkey | selecton | M–K test | |||||
|---|---|---|---|---|---|---|---|---|
| SLAC | FEL | REL | PARRIS | MEC (vs M8a) | M8 (vs M8a) | M7 | Neutrality index (P-value) | |
| NS5 POW | Purifying | Positive (735) | Purifying | No + | Significant | NS | No + | |
| NS5 DTV | Purifying | Purifying | Purifying | No + | NS | Significant | No + | |
| NS5 All | Purifying | Positive (383) | Purifying | No + | Significant | NS | No + | 2.089 (0.00107) |
| ENV POW | Purifying | Purifying | Purifying | No + | Significant | NS | No + | |
| ENV DTV | Purifying | Purifying | Purifying | No + | NS | NS | No + | |
| ENV All | Purifying | Purifying | Purifying | No + | Significant | NS | No + | 1.820 (0.0753) |
| POW CDS | Purifying | Purifying | Purifying | No + | NS | NS | No + | |
| DTV CDS | Purifying | Purifying | Purifying | No + | NS | NS | No + | |
| CDS All | Purifying | Purifying | Purifying | No + | NS | No + | No + | 0.753 (0.03539) |
DISCUSSION
Our phylogenetic analyses using both partial and whole-genome sequences found support for two distinct lineages of POW, similar to previous findings by Ebel et al. (2001) and Kuno et al. (2001). The clustering of Russian samples with the prototypic 1958 LB strain from Ontario supports assertions that a strain similar to this was recently imported into Russia, most probably in the past 100 years, potentially from importation of mink for fur farms in the area (Leonova et al. 2009). Envelope and NS-5 topologies showed DTV from Colorado basal to the Wisconsin strains and the isolate of DTV from West Virginia basal to the New England strains. Isolates taken from overlapping geographical areas in both Ontario and New England were placed into the two separate lineages (POW and DTV), suggesting that these two partially sympatric lineages exploit different ecological niches, and thus supporting the association of each lineage with a distinct transmission cycle (Ebel et al., 1999).
BSPs, which measure population dynamics based on genetic diversity, show a long, stable history for both POW and DTV, with some evidence of recent declines in these populations. These results support field data that suggest that a stable, persistent virus population is present in the upper Midwest USA (Brackney et al., 2008; Ebel et al., 2000). Although the BSPs show a recent decline in populations of POW, an increase in human cases of POW encephalitis has been reported over the past 10 years (Hinten et al., 2008). This may be the result of more human contact with areas where the virus is present (Lloyd-Smith et al., 2009). In addition or alternatively, there could be an enhanced awareness of arthropod-borne flaviviruses due to the introduction of West Nile virus (WNV) to the USA in 1999, which lead to increased testing and better diagnosis (Hinten et al., 2008). Genetic diversity of flaviviruses is very low within tick hosts (Brackney et al., 2010), especially compared with mosquito-borne flaviviruses, where intrahost populations have greater genetic diversity (Jerzak et al., 2005). The lack of diversity within POW populations may limit the number of unique genomes available and consequently the adaptability of POW to new ecological niches. Ultimately, this could produce the patterns of strong genetic conservation and population stability that we document in this study. Other factors that may influence population size, as estimated by genetic diversity, could be: the low levels of replication achieved in ticks, resulting in smaller populations generated at this stage of the life cycle; transstadial or transovarial transmission that may occur as part of the viral life cycle, providing a bottleneck for virus transmission; and the potential for co-feeding transmission, which would separate the need for replication in vertebrate hosts from the population dynamics of the virus overall (Ebel & Kramer, 2004; Nuttall et al., 1994). Finally, our sampling effort might not be sufficient to detect very recent (i.e. in the past 30 years) changes in virus population size that would be associated with the well-documented expansion in the populations of their main vector, I. scapularis. In sum, our results are concordant with previously published data which suggest that populations of POW (and DTV), like tick-borne encephalitis and other tick-borne pathogens, are very stable in nature (Blaskovic & Nosek, 1972; Goethert & Telford, 2009; Gresikova & Calisher, 1989).
Estimated nucleotide substitution rates ranged from 2×10−4 to 4×10−5 substitutions site−1 year−1, with NS-5-derived estimates approximately one log10 lower than those derived from envelope sequences. Nucleotide substitution estimates for the segmented negative-sense RNA virus transmitted by ticks, Crimean–Congo haemorrhagic fever virus (family Bunyaviridae, genus Nairovirus), found a rate of 5.8×10−5 for the L segment, which encodes the RNA-dependent RNA polymerase, and 1.09×10−4 for the S segment, which encodes the structural nucleocapsid protein, although the authors concluded this could be a result of greater taxon sampling for the S segment, which is not true of our dataset (Carroll et al., 2010). Nucleotide substitution rates for other vector-borne flaviviruses, such as KFDV and tick-borne encephalitis, tick-borne flaviviruses related to POW, or the mosquito-borne flaviviruses yellow fever virus, St Louis encephalitis virus, and WNV fall within this range (Baillie et al., 2008; Bryant et al., 2007; Mehla et al., 2009; Snapinn et al., 2007). The nucleotide substitution rates estimated for POW and DTV are lower than the typical estimate of approximately 10−3 substitutions site−1 year−1 found for RNA viruses (Jenkins et al. 2002). However, vector-borne RNA viruses have been found, overall, to display lower mutation rates than other RNA viruses, probably because of the requirement for replication in two distinct cell types (vertebrate and invertebrate), which may lead to stronger evolutionary constraint and lower overall accumulation of mutant types (Jenkins et al., 2002). Although the mutation rates are lower than expected for RNA viruses, they are 20-fold higher than those found in a recent study of POW using only the Russian and LB sequence data (Leonova et al., 2009). This difference may be due to a restricted focus of Russian transmission, or because the strain of POW in Russia is closely related to the LB strain because it was only recently introduced there from North America, thus leading to lower genetic diversity in those samples (Leonova et al., 2009). The apparently non-native Russian focus may prove a useful laboratory, over time, for studying the actual rate of divergence in a new viral focus in nature, and provide an interesting contrast to literature concerning the similar importation of mosquito-borne WNV into North America (Ebel & Kramer, 2009; Beasley et al., 2003; Bertolotti et al., 2007). In any case, our data show that nucleotide substitution rates for POW are similar to those of other flaviviruses and are variable along the length of the genome.
Divergence times were estimated to assess the likelihood that DTV and POW diverged recently as a result of expanding deer-tick populations. Using the BMCMC approach and concatenated complete NS-5 and envelope sequences (approximately 4000 nt total), we estimated the MRCA for POW from DTV at approximately 484 ybp (95 %, HPD=114–1048). This estimate agrees overall with previous studies based on an approximately 2000 nt sequence comprising portions of envelope, NS-3 and NS-5 sequences (Charrel et al., 2005). Previously, greater genetic diversity was found within the DTV lineage, leading researchers to speculate that this may be the older lineage (Ebel et al., 2001). Our divergence estimates show a nearly simultaneous radiation of DTV and POW approximately 200 years ago, with no indication that one clade is ancestral to the other. Divergence of DTV and POW may have occurred well before the current expansion of deer-tick populations, and must have been driven by other factors.
We observed strong purifying selection, and some evidence of positive selection, in the sequences we studied. An indicator of positive selection has previously been detected in partial POW-envelope alignments, but not in NS-5 alignments, using an M–K test (Ebel et al., 2001). We found significant signals for positive selection occurring in full-genome sequences and NS-5-encoding sequences using M–K tests, and approaching significance for envelope-encoding sequences. The likelihood models we employed were also able to identify positive selection in certain sites. However, we were unable to consistently detect specific codons that appeared to be subject to positive selection using multiple programs. All methods that were capable of doing so detected purifying selection at multiple sites. Our results indicate that most substitutions in POW and DTV genomes were synonymous, indicating that strong purifying selection limits the accumulation of genetic changes in POW virus populations. Similar results have been found in other arthropod-borne flaviviruses (Baillie et al., 2008). Despite some incongruence in the methods identifying positive selection, we suggest that this process is acting on POW and DTV genomes.
METHODS
Virus strains.
RNA samples were generated from three isolates of ticks collected in Wisconsin in 2008 (Brackney et al., 2008), one isolate from the Nantucket field station in 2000 (Ebel et al., 2000), wicf9901 from Wisconsin in 1999 (Ebel et al., 1999) and 64-7062 in 1964 (Whitney & Jamnback, 1965) (Table 2).
Table 2.
Isolates of POW used in this study
All locations are in the USA unless indicated otherwise.
| Code | Strain | Collection location | Year | Source | Genbank accession no. |
|---|---|---|---|---|---|
| DTVWiA08 | Spooner, WI | 2008 | I. scapularis | HM440560 | |
| DTVWiB08 | Spooner, WI | 2008 | I. scapularis | HM440561 | |
| DTVWiC08 | Spooner, WI | 2008 | I. scapularis | HM440562 | |
| POWPa06 | Part/2006 | Partizansk, Russia | 2006 | Human | EU543649 |
| DTVWi99 | wicf9901 | Chippewa Falls, WI | 1999 | I. scapularis | HM440558 |
| DTVWi97 | DTV-SPO | Spooner, WI | 1997 | I. scapularis | AF310921, AF310938 |
| DTVMa96 | NFS001 | Nantucket, MA | 1996 | I. scapularis | AF310918, AF310947, HM440559 |
| DTVCt94 | DTV-CT | Connecticut | 1994 | I. scapularis | AF310919, AF311056 |
| DTVMa94 | DTV-IPS | Ipswich, MA | 1994 | I. scapularis | AF310947, AF310918 |
| POWNad91 | Nad-1991 | Nadezdinsk, Russia | 1991 | Human | EU670438 |
| POWOn81 | t18-23-81 | Ontario, Canada | 1981 | I. cookei on Marmota sp. | AF310909, AF310943 |
| DTVWV77 | 12542 | West Virginia | 1977 | Vulpes sp. | AF310920, AF310949 |
| POWSp75 | Sp-9 | Spassk, Russia | 1975 | Dermacentor silvarum | EU770575 |
| POWOn65 | m11665 | Ontario, Canada | 1965 | Tick | AF310910, AF310937 |
| POWNYA64 | 64-7062 | New York | 1964 | Ticks on Marmota sp. | AF310915, AF310944, HM440563 |
| POWNYB64 | 64-7483 | New York | 1964 | Unknown | AF310916, AF310945 |
| POWOn64A | M8998 | Ontario, Canada | 1964 | Unknown | AF310911, no NS-5 |
| POWOn64B | 1982-64 | Ontario, Canada | 1964 | Marmota monax | AF310913, AF310939 |
| POWOn62 | 1247-62 | Ontario, Canada | 1962 | Tamiasciurus hudsonicus | AF310914, AF310942 |
| POWOn60s | m1409 | Ontario, Canada | 1960s | Tick | AF310912, AF310940 |
| POWOn58 | LB | Powassan, Ontario, Canada | 1958 | Human brain | NC003687 |
| DTVCo52 | 791A-52 | Colorado | 1952 | Dermacentor andersonii | AF310922, AF310950 |
RNA extraction, RT-PCR and sequencing.
RNA was extracted using an RNeasy Protect Mini-kit (Qiagen), according to the manufacturer's protocol, from 100 μl viral stock and eluted into 100 μl double-distilled H2O. RT-PCR was conducted using Superscript III One-Step RT-PCR with Platinum Taq (Life Technologies) using the following conditions: 55 °C for 30 min (reverse transcription), 95 °C for 15 min (initial denaturation) and 40 cycles of 94 °C for 15 s, 55 °C for 30 s and 72 °C for 30 s, followed by a final extension at 72 °C for 10 min. Primers amplifying approximately 1 kb segments were used (sequences available on request). PCR products were purified using StrataPrep PCR purification kits (Stratagene), and sequenced with the primers used for RT-PCR, yielding greater than twofold coverage, by the University of New Mexico DNA Research Services. Sequences were assembled using SeqMan (dnastar), and aligned with clustal w in BioEdit (Hall, 1999). The full genome sequences from the POW and DTV strains sequenced for this study have been deposited in GenBank and have been assigned accession numbers HM440558–HM440563. Other previously published sequences used in the study were obtained from GenBank. Sequences were tested for evidence of recombination using gard (genetic algorithm for recombination assessment) which is available at http://datamonkey.org.
Phylogenetic analysis.
Maximum-likelihood trees were generated in paup* version 4 (Swofford, 2002), considering all characters as unordered with four possible states (A, C, T, G). We employed heuristic searches with ten random stepwise additions of sequences and tree bisection–reconnection branch swapping. Node support was evaluated with 1000 non-parametric bootstrap pseudoreplicates (Felsenstein, 1985), using the search conditions described above. Analysis was performed with 11 full-genome sequences.
Divergence times, substitution rates and population size changes.
Bayesian phylogenetic analyses were performed to investigate the timescale of diversification of POW and DTV lineages using beast version 1.5.1 (Drummond & Rambaut, 2007). For these analyses, 24 sequences or partial sequences including ENV and NS-5 (4000 nt) were included. To assess the extent of temporal structure in the sequence data, we performed a regression analysis of tree root-to-tip genetic distance against sampling date using the program Path-O-Gen, (http://tree.bio.ed.ac.uk/software/pathogen/) based on a neighbour-joining tree. Regression analysis showed sufficient temporal structure in the viral sequences of this study (having the range 1958–2008) to estimate rates and dates. Analyses were performed by comparing uniform rates across branches (strict clock) and uncorrelated relaxed-clock assumptions. We used the uncorrelated log-normal assumption after comparing the marginal posterior distribution with the prior distribution of the standard deviation of the uncorrelated log-normal distribution (Suchard et al., 2001).
The Hasegawa, Kishino and Yano model of nucleotide substitution was used with γ-rate heterogeneity among sites as selected by comparison of Akaike information criterion scores. Two independent MCMC chains were run for 50 000 000 steps with sampling every 5000 steps, following a discarded burn-in of 5 000 000 steps of the posterior samples. The samples from the two runs were combined, and the convergence of the chain, sampling and mixing was confirmed by inspection of the MCMC samples using the program tracer version 1.5 (http://beast.bio.ed.ac.uk/Tracer).
Selection and recombination detection.
Alignments of POW sequences were tested for possible breakpoints using gard (Kosakovsky Pond et al., 2006), which is available at http://datamonkey.org (Kosakovsky Pond & Frost, 2005a). Models of nucleotide substitution were calculated using modeltest (Posada & Crandall, 1998). Non-synonymous to synonymous rate ratios were estimated in HyPhy (www.hyphy.org), using the maximum-likelihood phylogeny and the general time-reversible model of nucleotide substitution. Tests for positive selection were conducted on the datamonkey server using the following methods: SLAC, FEL, REL and PARRIS. SLAC counts the number of non-synonymous changes per non-synonymous site (dN) and tests whether it is significantly different from the number of synonymous changes per synonymous site (dS). FEL and REL estimate ratios of non-synonymous to synonymous changes for each site in an alignment, but FEL fixes estimates of branch lengths and substitution rate parameters for each site independently, whereas REL allows rate variation in both synonymous and non-synonymous rates and the nucleotide substitution model (Kosakovsky Pond & Frost, 2005b). PARRIS is able to take both recombination and differences in synonymous evolution rate into account (Scheffler et al., 2006). Additionally, we tested for selection using three models from the selecton server: MEC, M7+β and M8. DNAsp (Librado & Rozas, 2009) was used to calculate M–K tests, the ratio of the mean number of non-synonymous (dN) to synonymous (dS) nucleotide substitutions (dN/dS) and the genetic diversity (π). Analyses were performed using full coding sequences.
Acknowledgments
This work was supported in part by funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health under grant number AI067380 and by the University of New Mexico School of Medicine, Department of Pathology. F. T. P. was supported by the Fogarty Actions for Building Capacity Award 5D43 TW01133.
Footnotes
The GenBank/EMBL/DDBJ accession numbers for the sequences reported in this manuscript are: HM440558–HM440563.
References
- Artsob, H. (1988). Powassan encephalitis. In The Arboviruses, pp. 29–50. Edited by Monath, T. P.. Boca Raton, FL. : CRC Press.
- Baillie, G. J., Kolokotronis, S., Waltari, E., Maffei, J. G., Kramer, L. D. & Perkins, S. L. (2008). Phylogenetic and evolutionary analyses of St. Louis encephalitis virus genomes. Mol Phylogenet Evol 47, 717–728. [DOI] [PubMed] [Google Scholar]
- Beasley, D. W. C., Suderman, M. T., Holbrook, M. R. & Barrett, A. D. T. (2001). Nucleotide sequencing and serological evidence that the recently recognized deer tick virus is a genotype of Powassan virus. Virus Res 79, 81–89. [DOI] [PubMed] [Google Scholar]
- Beasley, D. W. C., Davis, C. T., Guzman, H., Vanlandingham, D. L., Travassos da Rosa, A., Parsons, R. E., Higgs, S., Tesh, R. B. & Barrett, A. D. T. (2003). Limited evolution of West Nile virus has occurred during its southwesterly spread in the United States. Virology 309, 190–195. [DOI] [PubMed] [Google Scholar]
- Bertolotti, L., Kitron, U. & Goldberg, T. L. (2007). Diversity and evolution of West Nile virus in Illinois and the United States, 2002–2005. Virology 360, 143–149. [DOI] [PubMed] [Google Scholar]
- Blaskovic, D. & Nosek, J. (1972). The ecological approach to the study of tick-borne encephalitis. Prog Med Virol 14, 275–320. [PubMed] [Google Scholar]
- Brackney, D. E., Nofchissey, R. A., Fitzpatrick, K. A., Brown, I. K. & Ebel, G. D. (2008). Stable prevalence of Powassan virus in Ixodes scapularis in a northern Wisconsin focus. Am J Trop Med Hyg 79, 971–973. [PMC free article] [PubMed] [Google Scholar]
- Brackney, D. E., Brown, I. K., Nofchissey, R. A., Fitzpatrick, K. A. & Ebel, G. D. (2010). Homogeneity of Powassan virus populations in naturally infected Ixodes scapularis. Virology 402, 366–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant, J. E., Holmes, E. C. & Barrett, A. D. T. (2007). Out of Africa: a molecular perspective on the introduction of yellow fever virus into the Americas. PLoS Pathog 3, e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, S. A., Bird, B. H., Rollin, P. E. & Nichol, S. T. (2010). Ancient common ancestry of Crimean–Congo hemorrhagic fever virus. Mol Phylogenet Evol 55, 1103–1110. [DOI] [PubMed] [Google Scholar]
- Charrel, R. N., Zaki, A. M., Fakeeh, M., Yousef, A. I., de Chesse, R., Attoui, H. & de Lamballerie, X. (2005). Low diversity of Alkhurma hemorrhagic fever virus, Saudi Arabia, 1994–1999. Emerg Infect Dis 11, 683–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels, T. J., Boccia, T. M., Varde, S., Marcus, J., Le, J., Bucher, D. J., Falco, R. C. & Schwartz, I. (1998). Geographic risk for Lyme disease and human granulocytic ehrlichiosis in southern New York state. Appl Environ Microbiol 64, 4663–4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond, A. J. & Rambaut, A. (2007). beast: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel, G. D. & Kramer, L. D. (2004). Short report: duration of tick attachment required for transmission of Powassan virus by deer ticks. Am J Trop Med Hyg 71, 268–271. [PubMed] [Google Scholar]
- Ebel, G. D. & Kramer, L. D. (2009). West Nile virus: molecular epidemiology and diversity. In West Nile Encephalitis Virus Infection, pp. 25–43. Edited by Diamond, M. S.. New York. : Springer ScienceBusiness Media.
- Ebel, G. D., Foppa, I., Spielman, A. & Telford, S. R., III (1999). A focus of deer tick virus transmission in the northcentral United States. Emerg Infect Dis 5, 570–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel, G. D., Campbell, E. N., Goethert, H. K., Spielman, A. & Telford, S. R., III (2000). Enzootic transmission of deer tick virus in New England and Wisconsin sites. Am J Trop Med Hyg 63, 36–42. [DOI] [PubMed] [Google Scholar]
- Ebel, G. D., Spielman, A. & Telford, S. R. (2001). Phylogeny of North American Powassan virus. J Gen Virol 82, 1657–1665. [DOI] [PubMed] [Google Scholar]
- Falco, R. C., Daniels, T. J. & Fish, D. (1995). Increase in abundance of immature Ixodes scapularis (Acari: Ixodidae) in an emergent Lyme disease endemic area. J Med Entomol 32, 522–526. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. [DOI] [PubMed] [Google Scholar]
- Goethert, H. K. & Telford, S. R., III (2009). Nonrandom distribution of vector ticks (Dermacentor variabilis) infected by Francisella tularensis. PLoS Pathog 5, e1000319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresikova, M. & Calisher, C. (1989). Tick-borne encephalitis. In The Arboviruses, pp. 177–202. Edited by Monath, T. P.. Boca Raton, FL. : CRC Press.
- Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 41, 95–98. [Google Scholar]
- Hinten, S. R., Beckett, G. A., Gensheimer, K. F., Pritchard, E., Courtney, T. M., Sears, S. D., Woytowicz, J. M., Preston, D. G., Smith, R. P., Jr & other authors (2008). Increased recognition of Powassan encephalitis in the United States, 1999–2005. Vector Borne Zoonotic Dis 8, 733–740. [DOI] [PubMed] [Google Scholar]
- Jenkins, G. M., Rambaut, A., Pybus, O. G. & Holmes, E. C. (2002). Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J Mol Evol 54, 156–165. [DOI] [PubMed] [Google Scholar]
- Jerzak, G., Bernard, K. A., Kramer, L. D. & Ebel, G. D. (2005). Genetic variation in West Nile virus from naturally infected mosquitoes and birds suggests quasispecies structure and strong purifying selection. J Gen Virol 86, 2175–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond, S. L & Frost, S. D. (2005a). Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21, 2531–2533. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond, S. L. K. & Frost, S. D. W. (2005b). Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol 22, 1208–1222. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond, S. L. K., Posada, D., Gravenor, M. B., Woelk, C. H. & Frost, S. D. W. (2006). Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol 23, 1891–1901. [DOI] [PubMed] [Google Scholar]
- Kuno, G., Artsob, H., Karabatsos, N., Tsuchiya, K. R. & Chang, G. J. (2001). Genomic sequencing of deer tick virus and phylogeny of Powassan-related viruses of North America. Am J Trop Med Hyg 65, 671–676. [DOI] [PubMed] [Google Scholar]
- Leonova, G. N., Kondratov, I. G., Ternovoi, V. A., Romanova, E. V., Protopopova, E. V., Chausov, E. V., Pavlenko, E. V., Ryabchikova, E. I., Belikov, S. I. & Loktev, V. B. (2009). Characterization of Powassan viruses from far eastern Russia. Arch Virol 154, 811–820. [DOI] [PubMed] [Google Scholar]
- Librado, P. & Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. [DOI] [PubMed] [Google Scholar]
- Lloyd-Smith, J. O., George, D., Pepin, K. M., Pitzer, V. E., Pulliam, J. R. C., Dobson, A. P., Hudson, P. J. & Grenfell, B. T. (2009). Epidemic dynamics at the human–animal interface. Science 326, 1362–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehla, R., Kumar, S. R. P., Yadav, P., Barde, P. V., Yergolkar, P. N., Erickson, B. R., Carroll, S. A., Mishra, A. C., Nichol, S. T. & Mourya, D. T. (2009). Recent ancestry of Kyasanur forest disease virus. Emerg Infect Dis 15, 1431–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuttall, P. A., Jones, L. D., Labuda, M. & Kaufman, W. R. (1994). Adaptations of arboviruses to ticks. J Med Entomol 31, 1–9. [DOI] [PubMed] [Google Scholar]
- Posada, D. & Crandall, K. (1998). modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818. [DOI] [PubMed] [Google Scholar]
- Rodgers, S. E. & Mather, T. N. (2007). Human Babesia microti incidence and Ixodes scapularis distribution, Rhode Island, 1998–2004. Emerg Infect Dis 13, 633–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffler, K., Martin, D. P. & Seoighe, C. (2006). Robust inference of positive selection from recombining coding sequences. Bioinformatics 22, 2493–2499. [DOI] [PubMed] [Google Scholar]
- Snapinn, K. W., Holmes, E. C., Young, D. S., Bernard, K. A., Kramer, L. D. & Ebel, G. D. (2007). Declining growth rate of West Nile virus in North America. J Virol 81, 2531–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard, M. A., Weiss, R. E. & Sinsheimer, J. S. (2001). Bayesian selection of continuous-time Markov chain evolutionary models. Mol Biol Evol 18, 1001–1013. [DOI] [PubMed] [Google Scholar]
- Swofford, D. L. (2002). paup*: Phylogenetic analysis using parsimony (and other methods), version 4. Sunderland, MA: Sinauer Associates.
- Tavakoli, N. P., Wang, H., Dupuis, M., Hull, R., Ebel, G. D., Gilmore, E. J. & Faust, P. L. (2009). Fatal case of deer tick virus encephalitis. N Engl J Med 360, 2099–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford, S. R., Armstrong, P. M., Katavolos, P., Foppa, I., Garcia, A. S., Wilson, M. L. & Spielman, A. (1997). A new tick-borne encephalitis-like virus infecting New England deer ticks, Ixodes dammini. Emerg Infect Dis 3, 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney, E. & Jamnback, H. (1965). The first isolations of Powassan virus in New York state. Proc Soc Exp Biol Med 119, 432–435. [DOI] [PubMed] [Google Scholar]