Abstract
Progressive multifocal leukoencephalopathy (PML) is an often fatal demyelinating disease caused by lytic infection of oligodendrocytes with JC virus (JCV). The development of PML in non-immunosuppressed individuals is a growing concern with reports of mortality in patients treated with mAb therapies. JCV can persist in the kidneys, lymphoid tissue and bone marrow. JCV gene expression is restricted by non-coding viral regulatory region sequence variation and cellular transcription factors. Because JCV latency has been associated with cells undergoing haematopoietic development, transcription factors previously reported as lymphoid specific may regulate JCV gene expression. This study demonstrates that one such transcription factor, Spi-B, binds to sequences present in the JCV promoter/enhancer and may affect early virus gene expression in cells obtained from human brain tissue. We identified four potential Spi-B-binding sites present in the promoter/enhancer elements of JCV sequences from PML variants and the non-pathogenic archetype. Spi-B sites present in the promoter/enhancers of PML variants alone bound protein expressed in JCV susceptible brain and lymphoid-derived cell lines by electromobility shift assays. Expression of exogenous Spi-B in semi- and non-permissive cells increased early viral gene expression. Strikingly, mutation of the Spi-B core in a binding site unique to the Mad-4 variant was sufficient to abrogate viral activity in progenitor-derived astrocytes. These results suggest that Spi-B could regulate JCV gene expression in susceptible cells, and may play an important role in JCV activity in the immune and nervous systems.
INTRODUCTION
The human polyomavirus JC is the aetiological agent of the fatal demyelinating disease, progressive multifocal leukoencephalopathy (PML), that occurs in patients in states of immune suppression or modulation. PML is an AIDS-defining illness that occurs in approximately 3 % of HIV-infected patients (Major, 2010). Recently, PML has been reported in patients undergoing immuno-modulatory therapies (Major, 2010; Marshall & Major, 2010). PML is caused by lytic multiplication of the JC virus (JCV) in oligodendrocytes, which results in the loss of myelination of neurons and loss of neuronal function. Despite its lytic capability, JCV can persist in a variety of cell types including CD34+ haematopoietic precursors and B cells present in bone marrow (Houff et al., 1988; Marzocchetti et al., 2008; Tan et al., 2009), brain, tonsil and in circulation (Sabath & Major, 2002). Detection of JCV DNA in the absence of viral capsid protein in these cell types from patients prior to the development of PML suggests that such cells may act as a reservoir for latent virus (Houff et al., 1988; Tan et al., 2009). Trafficking of JCV-infected B cells, or haematopoietic precursors, between the bone marrow and brain is a possible method of viral dissemination.
The JCV genome is a closed, circular, supercoiled DNA chromosome that permits temporal expression of early and late genes that are physically separated by the non-coding viral regulatory region (RR) (Frisque et al., 1984). RR is the most highly variable sequence among JCV isolates but always contains the origin of replication, one or more TATA boxes and a variety of enhancer elements (Ault & Stoner, 1993; Frisque, 1983; Frisque et al., 1984; Iida et al., 1993). The RR from the original isolate of JCV, Mad-1, contains a promoter/enhancer that exists as 98 bp tandem repeats containing two TATA boxes and multiple cellular transcription factor-binding sites (Frisque, 1983). TATA boxes in the tandem repeat sequence are essential for transcription of early and late viral genes (Daniel & Frisque, 1993; Kenney et al., 1986a, b; Khalili et al., 1986; Krebs et al., 1995; Vacante et al., 1989). RR variants containing tandem repeats have been isolated from tissues of patients with PML (Martin et al., 1985). The RR of a naturally occurring variant of JCV shed in urine, referred to as ‘archetype’, contains a single 98 bp unit with internal 23 and 66 bp insertions (Yogo et al., 1990). Archetype is not associated with PML and is replication incompetent in tissue culture (Daniel et al., 1996). Consistent isolation of tandem repeat containing sequences in tissues obtained from PML patients strongly suggests the importance of these sequences in viral pathogenesis (Frisque et al., 1984; Jensen & Major, 1999; Martin et al., 1985; Marzocchetti et al., 2008; Vaz et al., 2000).
Interestingly, JCV multiplication in susceptible cells is restricted by activation of viral gene expression by host transcription factors such as Oct-6/tst-1/SCIP (Wegner et al., 1993), pur α (Chen & Khalili, 1995), YB-1 (Chen et al., 1995c; Kerr et al., 1994) and NF-1 (Amemiya et al., 1989, 1992; Messam et al., 2003; Monaco et al., 2001; Ravichandran & Major, 2008; Shivakumar & Das, 1994; Sumner et al., 1996; Tamura et al., 1988). However, NF-1X is the only factor that has been shown to be important for JCV activity in glial (Kumar et al., 1993, 1996) and immune cells (Monaco et al., 2001). Because JCV latency is associated with cells undergoing haematopoietic development, it is probable that transcription factors previously reported as lymphoid specific regulate JCV gene expression. One such factor, Spi-B, activates gene expression from a lymphotrophic variant of SV40 (Petterson & Schaffner, 1987) and lymphotrophic papovavirus (LPV) (Erselius et al., 1990), both of which share genetic architecture and promoter regulation with JCV. In addition, Spi-B is upregulated in peripheral blood mononuclear cells in response to treatment with Natalizumab (Lindberg et al., 2008), an immuno-modulatory therapy that has been associated with the development of PML (Major, 2010). Spi-B is an Ets transcription factor that is required for normal B-cell receptor signalling and formation of germinal centres in the spleen (Garrett-Sinha et al., 1999). Spi-B binds target sequences containing a 5′-GGAA/T-3′ core (Araki et al., 1988; Dorn et al., 1988; Erselius et al., 1990; Laux et al., 1994; Petterson & Schaffner, 1987; Wasylyk et al., 1993) and can cooperate with the retinoblastoma protein (pRB) to alter expression of proteins involved in B-cell maturation (Hagemeier et al., 1993; Mao et al., 1996; Rao et al., 1999b; Weintraub et al., 1995). Spi-B is involved in differentiation and maturation of B cells and is expressed at high levels in developing and mature B cells (Chen et al., 1995a, b; Ray et al., 1992; Su et al., 1996).
RESULTS
Spi-B-binding sites are present in JCV promoter/enhancer sequences
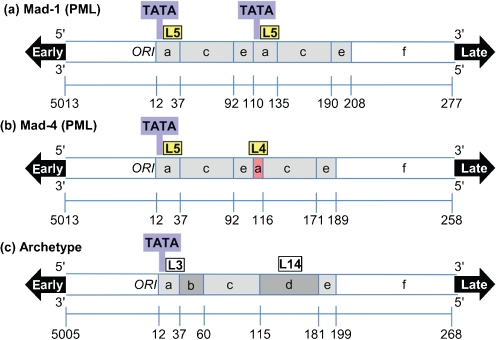
The Spi-B-binding site consensus sequence was identified as 5′-WWWRRRGAASNDR-3′ where the internal RGAA core is conserved (Laux et al., 1994). Promoter/enhancer sequences from PML-associated Mad-1 (Frisque et al., 1984), Mad-4 (Martin et al., 1985) and the non-pathogenic archetype variants (Yogo et al., 1990) were analysed for Spi-B-binding sites using the criteria of a 5′-NGAA-3′ core. Four potential Spi-B-binding sites (Fig. 1) were identified in the JCV promoter/enhancer sequences and are listed in Table 1. Mad-1 (Fig. 1a) and Mad-4 (Fig. 1b) promoter/enhancers contain a binding site, labelled L5, adjacent to the TATA box in the first repeat. Mad-1 contains a second L5 site in the second repeat. A naturally occurring 19 bp deletion in the Mad-4 promoter/enhancer results in the loss of the second TATA box and a unique Spi-B-binding site labelled L4. The archetype promoter/enhancer (Fig. 1c) contains 23 and 66 bp insertions within a single 98 bp unit, resulting in the loss of the L5/L4-binding sites. The 23 bp insertion results in a single base pair change, 5′-AAAAGGGAAGGGA-3′, at the 3′ end of the L5 site to 5′-AAAAGGGAAGGTA-3′, labelled L3. The 66 bp insertion resulted in a unique site labelled L14.
Fig. 1.
Diagram of potential Spi-B-binding sites in JCV promoter/enhancer sequences. The non-coding viral RRs from the PML-associated Mad-1 (a), Mad-4 (b) and the non-pathogenic archetype (c) JCV variants are represented. The PML-associated variants contain 98 bp tandem repeats (light grey). A 19 bp deletion in the second repeat of Mad-4 (red) results in the loss of the second TATA box (blue). The archetype sequence contains two inserts (dark grey) in a single 98 bp unit. Sites that bind Spi-B protein in EMSA assays are shaded in yellow and sites that did not bind protein are white.
Table 1.
Potential Spi-B-binding site sequences present in the promoter/enhancers of JC virus variants
| Spi-B-binding site | Viral variant | Sequence (5′–3′) |
|---|---|---|
| SV40 | SV40 | CTGAAAGAGGAACTTG |
| SV40 mutant | – | CTGAAAGACCAACTTG |
| L4 | Mad-4 | CAAGGGGAAGGGA |
| L4 mutant | – | CAAGGCCAAGGGA |
| L5 | Mad-1/Mad-4 | AAAAGGGAAGGGA |
| L5 mutant | – | AAAAGCCAAGGGA |
| L3 | Archetype | AAAAGGGAAGGTA |
| L14 | Archetype | TATAGTGAAACCC |
Spi-B is expressed in JCV susceptible cells
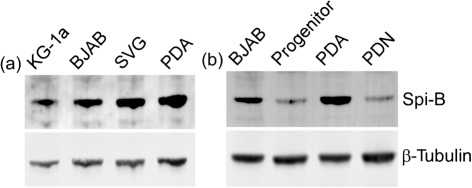
JCV infects a variety of different cell types including CD34+ haematopoietic precursors, B cells, and astrocytes in culture. However, Spi-B gene expression has been described exclusively in the B cell lineage (Dahl et al., 2002; Erselius et al., 1990; Garrett-Sinha et al., 2005; Kim et al., 2003; Ray et al., 1992; Schmidlin et al., 2008; Schweitzer & DeKoter, 2004; Su et al., 1996, 1997). Northern blot analysis of Spi-B mRNA in brain tissue from mice showed no message present (Ray et al., 1992; Su et al., 1996). Using a sensitive method of quantitative real-time RT-PCR (qRT-PCR), Spi-B mRNA expression was measured in the various cell types susceptible to JCV (Table 2). Spi-B mRNA was detected above the non-template control for background detection in all cell types tested. Spi-B mRNA expression was measured at low levels in two cell types, HeLa cells and primary CD3+ T lymphocytes, in which Spi-B protein has not been detected. Immortalized B cells (Raji and BJAB) and primary CD19+ B cells expressed 100-fold and greater levels of Spi-B mRNA in comparison. A CD34+ cell line (KG-1a) and primary CD34+ haematopoietic precursors expressed less Spi-B mRNA than B cells, which is consistent with published reports (Su et al., 1996). The human fetal brain-derived SVG cell line, which supports robust JCV multiplication, expressed Spi-B mRNA comparable to CD34+ haematopoietic precursors. Human fetal brain-derived cells (progenitors, astrocytes and neurons) expressed Spi-B mRNA at levels similar to HeLa and primary T lymphocytes. Spi-B protein expression was measured by protein blotting using an antibody that detects a single 43 kDa band for Spi-B (Arguello et al., 2003). Spi-B protein was detected in BJAB cells and slightly lower in KG-1a cells (Fig. 2a) consistent with mRNA levels. Spi-B protein was detected in SVG cells and in progenitor-derived astrocytes (PDA), but not progenitor cells or progenitor-derived neurons (PDN) (Fig. 2b). Spi-B protein expression in the absence of high levels of mRNA in PDAs may be due to instability and/or turnover of the mRNA. Stability of the highly related Spi-1 mRNA has been demonstrated to be extensively regulated at the post-transcriptional level (Hensold et al., 1996), which may also be the case for Spi-B.
Table 2.
Spi-B mRNA expression in immune- and brain-derived cells types
| Category | Cell type | Spi-B mRNA, relative level | Standard deviation |
|---|---|---|---|
| Control | HeLa | 0.01 | 0.0002 |
| CD3+ lymphocytes | 0.01* | 0.0061 | |
| B cells | Raji | 2.17 | 0.1262 |
| BJAB | 1.66 | 0.0014 | |
| CD19+ lymphocytes | 1.16* | 0.1616 | |
| Haematopoietic precursor | KG-1a | 0.31 | 0.0082 |
| CD34+ lymphocytes | 0.75* | 0.3661 | |
| Brain-derived cells | SVG | 0.30 | 0.0273 |
| Human fetal brain progenitor cells | 0.02 | 0.0001 | |
| PDA | 0.01 | 0.0013 | |
| PDN | 0.10 | 0.0036 |
*Spi-B mRNA value is a mean of cell purified from multiple blood donors.
Fig. 2.
Spi-B is expressed in JCV susceptible cells. Spi-B protein was detected in JCV susceptible KG-1a, BJAB, SVG and PDA (a) and cells derived from human fetal brain progenitors, PDA and PDN (b) by protein blotting. β-Tubulin protein levels were detected to demonstrate equal loading.
Spi-B binds unique sites on JCV enhancers
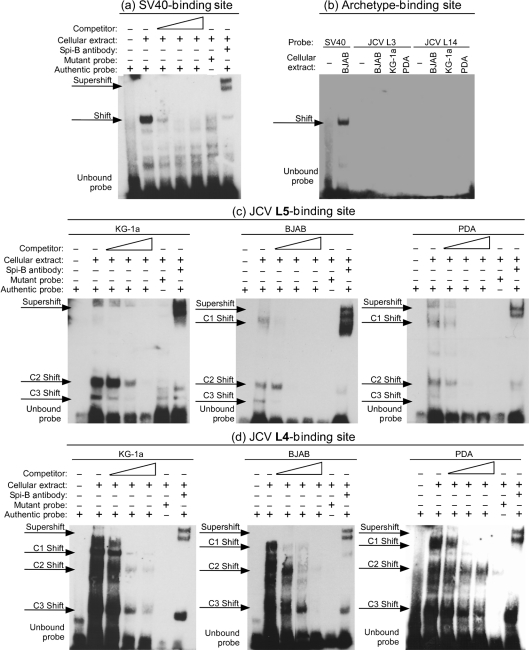
Because potential Spi-B-binding sites are present in JCV promoter/enhancer sequences and cells susceptible to JCV infection express Spi-B, the association of Spi-B with potential binding sites was measured by electromobility shift assay (EMSA) using whole-cell extracts from KG-1a cells, BJAB cells and PDAs (Fig. 3). Cell extracts were incubated with double-stranded oligonucleotide probes encoding the Spi-B-binding sites described in Table 1, or the SV40 Spi-B site described previously (Petterson & Schaffner, 1987) as a positive control. Fig. 3(a) illustrates that protein present in BJAB cell extract binds the SV40 probe and causes a shift that was competed by a 5-, 100- and 400-fold excess of unlabelled probe. Alteration of the Spi-B site core from 5′-GGAA-3′ to 5′-CCAA-3′ abrogated protein binding to the mutant probe, indicating that complex formation was specific for the Spi-B core. Addition of Spi-B antiserum, suitable for distinguishing Spi-B from other related proteins in an EMSA assay (Laux et al., 1994), caused a supershift in the original complex, which indicates that protein bound to the SV40 probe in the shifted complex is Spi-B.
Fig. 3.
Spi-B binds unique sites present on JCV promoter/enhancers. An EMSA was performed using BJAB cell extract and biotin-labelled oligonucleotides for the SV40 Spi-B site as a positive control for Spi-B-DNA complex formation (a). Authentic-binding site probe was incubated with cell extract alone or in combination with 5-, 100- or 400-fold excess unlabelled oligonucleotide competitor, or Spi-B antiserum. Mutant probe was incubated with cell extract to demonstrate specificity for the Spi-B-binding site core. Identical EMSAs were performed using BJAB, KG-1a, and/or PDA cell extract and biotin-labelled oligonucleotides for the archetype JCV L3 and L14 (b) and JCV L5 (c) and L4 (d) Spi-B sites as probes.
The same EMSAs were carried out for the potential JCV Spi-B-binding sites. Probes for Spi-B sites present in archetype promoter/enhancers, L3 and L14, did not bind protein in any cell type tested (Fig. 3b). Probes for Spi-B sites present in PML-associated promoter/enhancers, L5 and L4, bound protein expressed in KG-1a, BJAB and PDAs shown in Fig. 3(c) and (d), respectively. Incubation of both the L5 and L4 probes with cellular extracts resulted in the formation of multiple protein–DNA complexes labelled shifts C1–C3 that were consistent between cell types. Formation of multiple protein–DNA complexes on Spi-B-binding sites has been described using EMSA analysis for many promoters in published reports (Dekoninck et al., 2003; Erselius et al., 1990; Garrett-Sinha et al., 2005; Laux et al., 1994; Petterson & Schaffner, 1987; Zhao & Sample, 2000). In the case of JCV L5 and L4, each complex was competed by excess of unlabelled oligonucleotide probe and alteration of the Spi-B site core to 5′-CCAA-3′ resulted in abrogation of formation of all complexes. Addition of Spi-B antiserum caused supershifts of the original complexes, similar to that observed for SV40. The C2 shift for the L5 site and C3 shift for the L4 site required higher excess unlabelled probe for competition and were not completely supershifted upon addition of Spi-B antiserum, suggesting an abundance of these complexes over others. These results suggest that Spi-B is a component of multiple complexes at these locations on the viral promoter/enhancer. Spi-B is known to bind multiple cofactors that cooperate to affect gene expression such as cellular proteins TBP and pRB (Rao et al., 1999b), OBF-1 (Bartholdy et al., 2006) and CBP (Yamamoto et al., 2002), and the viral proteins EBNA-2 and EBNA-3C for Epstein–Barr virus (Zhao & Sample, 2000). It is possible that the multiple shifts represent separate complexes that may include cofactors necessary for activity of Spi-B. Importantly, competition with varying levels of unlabelled probe and Spi-B antiserum supershifts demonstrate that each of the complexes formed for both L5 and L4 are Spi-B specific.
Expression of exogenous Spi-B in semi- and non-permissive cells increases T-antigen expression
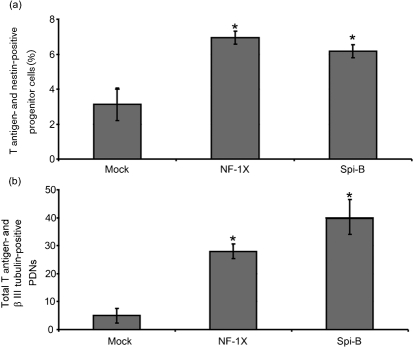
Because Spi-B is expressed in cells that are susceptible to JCV infection, we sought to determine if Spi-B overexpression in non-permissive cells could support viral activity. Previous studies showed that exogenous expression of NF-1X in non-permissive cells by transfection prior to JCV exposure increased T-antigen expression (Messam et al., 2003). NF-1X and Spi-B were expressed by transient transfection in semi-permissive progenitor cells and non-permissive PDNs for 24 h followed by exposure to Mad-4 JCV. Five days after JCV exposure, cells were analysed for expression of T antigen, as well as the cell-specific marker for progenitor cells (nestin) or neurons (β III tubulin). Double-positive cells for T antigen and the appropriate cell-specific marker were quantified from three separate experiments. Pre-expression of NF-1X and Spi-B resulted in a statistically significant increase in T-antigen expression in progenitors (Fig. 4a) and PDNs (Fig. 4b). NF-1X protein expression was confirmed 3 days post-transfection by immunofluorescence (progenitors: 5–10 %; PDN: 1 %). An increase in Spi-B mRNA expression was confirmed 3 days post-transfection by qRT-PCR. Spi-B gene expression was increased from 0.0005 to 1.7 in progenitor cells and from 0.05 to 1.7 in PDNs. These results demonstrate that expression of Spi-B, like NF-1X, supports early viral gene expression.
Fig. 4.
Expression of exogenous Spi-B in semi- and non-permissive cells increases T-antigen expression. The fraction of T antigen and nestin double-positive progenitor cells (a) or β III tubulin double-positive PDN (b) is represented along with the sd. An asterisk denotes the following statistically significant change in value. Progenitors: NF-1X P=0.0006, Spi-B P=0.0063; PDN: NF-1X P=0.0004, Spi-B P=0.0012.
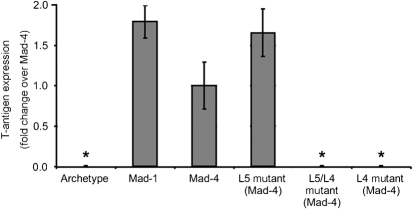
Mutation of the L4 Spi-B-binding site in the Mad-4 promoter/enhancer is sufficient to abrogate JCV activity in PDAs
To determine if Spi-B sites in the JCV promoter/enhancer that bind Spi-B are important for viral activity, site-directed mutagenesis was used to generate plasmids encoding mutations in the Spi-B site core of the L5 and L4 sites in Mad-4 (Table 1). Plasmids encoding archetype, Mad-1, Mad-4, L5 mutant Mad-4, L4 mutant Mad-4 or L5/L4 double-mutant Mad-4 were introduced into PDAs via nucleofection. T-antigen expression was quantified on a per cell basis by immunofluorescence, 6 days after nucleofection. Fig. 5 demonstrates that T-antigen expression occurred at similar levels from the Mad-1 and Mad-4 plasmids, while expression from the archetype plasmid was rarely detected. Mutation of the L5 site resulted in a slight, but not statistically significant, increase in T-antigen expression comparable to Mad-1. Mutation of both the L5 and the L4 sites resulted in abrogation of T-antigen expression. Importantly, mutation of the L4 site alone also resulted in abrogation of T-antigen expression. These results demonstrate that abrogation of viral activity due to the L4 site mutation is dominant in the presence of a wild-type L5 site.
Fig. 5.
Mutation of the L4 Spi-B-binding site in the Mad-4 promoter/enhancer is sufficient to abrogate JCV activity in PDA. The fraction of T antigen-positive cells as a fold change over the wild-type Mad-4 plasmid for archetype, Mad-1, Mad-4, L5 mutant Mad-4, L4 mutant Mad-4 or L5/L4 mutant Mad-4 plasmids is represented along with sd. An asterisk denotes the following statistically significant change in value. Archetype, P=0.039 ; L5/L4 mutant, P=0.039 ; L4 mutant, P=0.039.
DISCUSSION
The promoter/enhancer of JCV is considered to be the portion of the RR that confers specific tissue tropism and supports development of PML. PML-associated variants Mad-1 and Mad-4 contain promoter/enhancers that exist as tandem repeats, which contain TATA boxes and duplications of essential transcription factor-binding sites including Spi-B. In this study, Spi-B-binding sites that actively bound Spi-B protein expressed in JCV susceptible cell types are present in promoter/enhancer sequences from Mad-1 and Mad-4, but not the non-pathogenic archetype (Table 2, Figs 2 and 3). Mad-1 contains two identical Spi-B-binding sites (L5) within each of the 98 bp repeats, while Mad-4 contains the same site as Mad-1 (L5) in the first repeat followed by a unique site (L4) in the second repeat that results from a naturally occurring 19 bp deletion. Importantly, mutation of the unique Mad-4 L4 Spi-B site and not L5 resulted in abrogation of T-antigen expression in PDAs. The locations of the L5- and L4-binding sites are of particular interest in relation to JCV activity.
Abrogation of T-antigen gene expression in response to mutation of the L4 site within the viral promoter/enhancer suggests that an Spi-B site in the second tandem repeat is important for early viral gene expression (Fig. 5). Strikingly, the presence of a functional Spi-B site in the first repeat (L5) is incapable of compensating for the loss of the Spi-B site in the second repeat (L4). Mutation of the L5 site alone did not significantly alter the ability of the virus to express T antigen; however, this could be due to compensation by the L4 site. The L4 site may be dominant over an initial L5 site in the context of the tandem repeat. Due to its deletion in a variety of RR sequences from PML patients including Mad-4, Her-1, Mad-7, Mad-8, Mad-9 and Mad-11 (Major et al., 1987; Martin et al., 1985; Matsuda et al., 1987) the second TATA box was not thought to be necessary for virus multiplication (Lynch & Frisque, 1990; Martin et al., 1985). In fact, molecular studies on the function of the Mad-1 RR often utilize JCV sequences that only contain a single 98 bp unit and, therefore, a single TATA box and Spi-B site (Kerr et al., 1994; Sunden et al., 2007). However, the maintenance of tandem repeat nucleotide sequences throughout PML-associated variants illustrates their importance in the pathogenesis of JCV (Gosert et al., 2010). Utilizing RRs with a single 98 bp unit may miss the importance of measuring the relationship between tandem repeats. Future studies should include full-length RR to include the effects of a functional second repeat in the context of JCV multiplication and pathogenesis.
Spi-B-binding sites within the first repeat of the promoter/enhancers of PML-associated JCV variants are situated directly downstream of the viral origin and TATA box elements, which are essential for T-antigen binding and activation of viral gene expression (Daniel & Frisque, 1993; Kenney et al., 1986a, b; Khalili et al., 1986; Krebs et al., 1995; Vacante et al., 1989). Activation of gene expression from the basal promoter is an essential event for JCV multiplication and reactivation from latency. SV40 T antigen interacts with proteins that recruit the basal transcriptional apparatus (TFIID) to the viral promoter/enhancer including TATA-binding protein (TBP) and transcription-enhancing factor 1 (Damania & Alwine, 1996; Gruda et al., 1993; Zhai et al., 1997) and some evidence suggests similar interactions for JCV T antigen (Rekvig, 1997). Importantly, initial events of JCV infection occur in the absence of T-antigen protein, suggesting that other factors regulate this process. The amino-terminal portion of Spi-B binds TBP and is capable of recruiting the TFIID complex to promoters (Rao et al., 1999b). Therefore, a potential mechanism for activation of JCV early gene expression in the absence of T protein could involve Spi-B binding to TBP on the JCV promoter/enhancer and recruitment of the TFIID complex. It has been demonstrated that TBP containing transcription factor complexes form on TATA-less promoters and are required for transcription (Pugh & Tjian, 1991). Therefore, Spi-B bound to the L4 site in the TATA-less second repeat of Mad-4 could be capable of recruiting TBP and TFIID to initiate transcriptional activation in the absence of a TATA box. In this case the L4 site is positioned to compensate for the lack of the TATA box in the Mad-4 variant.
In addition to TATA boxes, these areas are bound heavily by other host factors that regulate viral activity. Oct-6/tst-1/SCIP-binding sites are located adjacent to both the L5 and L4 Spi-B sites on their 5′ ends and NF-1-binding sites are located adjacent to both Spi-B sites on their 3′ ends. Importantly, neither the Oct-6/tst-1/SCIP nor the NF-1-binding sites directly overlap the Spi-B-binding sites. Therefore, these factors would not affect Spi-B binding in the EMSA analysis presented in Fig. 3 and would not be affected by the mutations introduced in the Mad-4 L5, L4 or L5/L4 double mutants presented in Fig. 5. Pur α and YB-1 bind opposite strands of the single-stranded JCV promoter/enhancer sequence, which overlap the L5 Spi-B-binding site, and regulate early and late viral gene expression, respectively, in cooperation with the large T antigen (Chen et al., 1995c; Chen & Khalili, 1995; Kerr et al., 1994). Nucleotide changes equivalent to the mutations in the Mad-4 L5, L4 or L5/L4 double mutants were introduced into the YB-1/pur α-binding site reported by Chang et al. (1996) and did not have any significant effect on the ability of protein from BJAB cells or PDAs to bind the authentic probe in competition experiments (data not shown). Importantly, studies on YB-1/pur α-binding sites focus on sequences present in the first tandem repeat and emphasize the importance of T-antigen binding to sites present in the origin of replication (Chen et al., 1995c; Chen & Khalili, 1995; Kerr et al., 1994). In the context of a Mad-4 JCV promoter/enhancer, the naturally occurring 19 bp deletion results in alteration of the YB-1/pur α-binding site in the second tandem repeat. In addition, YB-1/pur α-binding site in the second tandem repeat of both Mad-1 and Mad-4 would not contain the important T-antigen binding sites in proximity because they occur directly after the first repeat. Taken together these points suggest that the YB-1/pur α-binding site would not play a role in the abrogation of early gene expression demonstrated in Fig. 5 for the L4 mutant plasmid.
Spi-B is largely considered a lymphotrophic transcriptional activator (Rao et al., 1999a; Yamamoto et al., 2002) and is known to transactivate the λ2–4 enhancer (Su et al., 1996), the LMP/TP2 promoter of Epstein–Barr virus (Laux et al., 1994), the SV40 enhancer (Petterson & Schaffner, 1987) and the LPV promoter (Erselius et al., 1990). JCV is capable of infecting a variety of cells of the lymphoid system. Detection of tandem repeat containing JCV DNA sequence in the absence of viral capsid protein in the bone marrow in patients prior to the development of PML suggests that these cells may act as a reservoir for latent virus (Houff et al., 1988; Tan et al., 2009). Spi-B expression in developing B cells correlates with reactivation of JCV in immune cells. Studies in mice demonstrate that Spi-B is expressed at a low level in pro-B cells, increases in pre-B cells and is highest in mature B cells (Su et al., 1996). Spi-B expression during B-cell development was confirmed in human using prototypic cell lines (Ray et al., 1992). If Spi-B is indeed a transcriptional activator of JCV gene expression, a minimal level of Spi-B expression in haematopoietic progenitors and pro-B cells may support latency of JCV, while upregulation of Spi-B expression in pre-B and mature B cells may lead to reactivation of JCV gene expression and production of viral progeny.
Because the number of patients undergoing immuno-modulatory therapies that develop PML continues to rise (Major, 2009), understanding the role of Spi-B during JCV latency and reactivation is increasingly important. These therapies cause mobilization and expansion of cells that have the potential to harbour latent JCV infection. Natalizumab has also been shown to upregulate genes involved in B-cell differentiation, including Spi-B (Lindberg et al., 2008). If Spi-B is an activator of JCV gene expression it could be a contributing factor in the development of PML in patients undergoing these immuno-modulatory therapies. In addition, PML-associated Mad-4 contains a duplication of the sites due to the tandem repeat nature of its promoter/enhancer, one of which (L4) was essential for T-antigen expression demonstrated by mutational analysis in Fig. 5. Accumulation of Spi-B sequence motifs in promoters can confer tissue specificity (Gerster et al., 1987; Ondek et al., 1987; Schirm et al., 1987) as illustrated in the case of a lymphotrophic SV40 variant (Petterson & Schaffner, 1987). Accumulation of additional, or unique, Spi-B-binding sites in the promoter/enhancers of JCV RRs may contribute to higher levels of replication in lymphoid tissues that in turn lead to development of PML. Further investigation into the role of Spi-B during JCV infection and the presence of duplicated, and or unique, Spi-B-binding sites in the RR sequences derived from PML patients will offer insight into the molecular mechanism of JCV reactivation and the development of PML.
METHODS
Cells, plasmids and viruses.
Human growth factors for cell culture were obtained from Peprotech. KG-1a cells were maintained in RPMI 1640 medium (Cellgro) supplemented with 20 % FBS (Atlanta Biologics) and 2 mM l-glutamine (Quality Biologics). BJAB and Raji cells were maintained in RPMI 1640 medium supplemented with 10 % FBS and 2 mM l-glutamine. SVG cells were maintained in minimal essential medium (MEM; Cellgro) supplemented with 10 % FBS and 2 mM l-glutamine. Human central nervous system progenitors were isolated and differentiated into astrocytes and neurons as described previously (Messam et al., 2003).
A plasmid encoding the full-length human Spi-B cDNA, pΔEB-Spi-B, was obtained from Francoise Moreau-Gauchelin at the Institut Curie in Paris, France (Ray et al., 1992). A plasmid encoding the full-length human NF-1X cDNA with an amino-terminal haemagglutinin (HA) tag, pCHA-NF1X, was described previously (Monaco et al., 2001). A plasmid encoding the full-length JCV Mad-4 genome, pMad4(586), was described previously (Martin et al., 1985). A plasmid encoding the full-length JCV Mad-4 genome, pM1TC, was described previously (Frisque et al., 1984). A plasmid encoding the full-length JCV archetype genome, CY, was described previously (Yogo et al., 1990).
Isolation of primary lymphocytes.
Mononuclear cells were isolated from the peripheral blood of normal donors provided by the NIH clinical centre blood bank by centrifugation on Ficoll-Hypaque gradients. CD34+ haematopoietic precursors, CD19+ B cell and CD3+ T-cell populations were purified using the RoboSep brand of immunomagnetic cell separation (StemCell Technologies).
Antibodies.
A mouse mAb for SV40 T antigen, which cross-reacts with JCV T antigen (EMD Calbiochem) was used at 5 μg ml−1. The mouse mAb GA5 for human glial fibrillary acid protein (GFAP) (Cell Signaling Technology) was used at a 1 : 300 dilution. The rabbit polyclonal antibody for GFAP (Covance) was used at a 1 : 1000 dilution. The rabbit polyclonal antibody TUJ1 for neuronal class III β-tubulin (Covance) was used at a 1 : 2000 dilution. HA-tagged human NF-1X was visualized with the rat mAb 3F10 for HA (Roche Diagnostics) at 200 ng ml−1. The rabbit polyclonal antibody 331B for human nestin was used at a 1 : 200 dilution (Messam et al., 2000). The goat polyclonal antibody for human Spi-B (Santa Cruz Biotechnology Inc) was used at a 1 : 200 dilution. The mouse mAbs for β actin AC-15 and for β-tubulin TUB 2.1 (Sigma) were used at 1 : 1000 dilutions. Fluorescent-labelled antibodies qualified for multiple labelling experiments were obtained from Invitrogen and used at 1–2 μg ml−1.
Preparation of Spi-B-binding site mutants in a Mad-4 background.
Site-directed mutagenesis was performed on the pMad4(586) plasmid to alter the L5 and L4 Spi-B site cores from 5′-GGAA-3′ to 5′-CCAA-3′ using the QuikChange II XL site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. Sense oligonucleotides (L4: 5′-GTAAACAAAGCACAAGGCCAAGGGATGGCTGCCAGC-3′; L5: 5′-TCCTGTATATATAAAAAAAAGCCAAGGGATGGCTGCCAGCCA-3′) and antisense oligonucleotides (L4: 5′-GCTGGCAGCCATCCCTTGGCCTTGTGCTTTGTTTAC-3′; L5: 5′-TGGCTGGCAGCCATCCCTTGGCTTTTTTTTATATATACAGGA-3′) containing the 5′-GG-3′ to 5′-CC-3′ mutations were synthesized by Integrated DNA Technologies (IDT). To create the L5/L4 double-mutant plasmid, site-directed mutagenesis was performed on the L4 mutant plasmid using the L5 sense and antisense oligonucleotides. DNA sequencing was performed to confirm the presence of each desired mutation as well as the fidelity of Mad-4 by the NINDS DNA sequencing facility.
Isolation of total RNA.
Total RNA was isolated from cell lines and primary cells using the RNeasy plus mini kit (Qiagen) according to the manufacturer's instructions. RNA was eluted from the column in nuclease-free water and quantified using a Nanodrop 8000 (Thermo Scientific).
Preparation of whole-cell extracts.
Whole-cell extracts were prepared by using a modification of the method of Andrews & Faller (1991) as described previously (Monaco et al., 2001).
Protein blotting.
Fifteen micrograms of protein was separated by electrophoresis in a 4–12 % Bis-Tris gel followed by transfer to PVDF membrane. Membranes were blocked for 1 h in 5 % non-fat dry milk in Tris-buffered saline containing 50 mg BSA ml−1, 10 mg glycine ml−1, 0.05 % Tween-20 (TBS-BGT). Membranes were incubated with primary antibodies diluted in TBS-BGT for 3 h. Unbound antibody was removed by washing in TBS-BGT followed by 1 h incubation in secondary fluorescent-conjugated antibodies (Invitrogen). Unbound antibody was removed by washing in TBS-BGT. The antibody–antigen complex was visualized using a FluorChem Q imager (Alpha Innotech).
Immunofluorescence.
Immunofluorescence was conducted on Lipofectamine 2000 transfected, nucleofected and JCV-infected cell cultures. Cells were fixed with 4 % paraformaldehyde and permeabilized with 0.2 % Triton X-100 before indirect antibody labelling. Samples were mounted with a glycerol-based mounting medium containing the DNA dye, 4′,6-diamidino-2-phenylindole (DAPI) and analysed by fluorescence microscopy using a Zeiss Axiovert 200M microscope fitted with filters appropriate for DAPI, Alexa Fluor 488 and Alexa Fluor 568 excitation.
qRT-PCR.
Spi-B mRNA present in cells was measured by reverse transcription followed by qRT-PCR using the TaqMan gene expression assay (Applied Biosystems) for human Spi-B (Hs00162150_m1), human β actin (Hs99999903_m1) and human PUM1 (Hs00206469_m1). Reverse transcription (RT) of 1 μg of total RNA was performed using qScript cDNA supermix (Quanta Biosciences) according to the manufacturer's instructions. Six 1 : 10 serial dilutions of the resultant BJAB RT reaction were prepared to generate a relative standard curve to determine Spi-B mRNA levels. Singleplex qRT PCRs were assembled using each RT reaction with 2× TaqMan universal PCR master mix and 20× TaqMan human Spi-B gene assay or 20× TaqMan human β actin or PUM1 endogenous control assay according to the manufacturer's instructions (Applied Biosystems). Relative quantification of Spi-B gene expression was determined using the relative standard curve method described on the Applied Biosystems website (Biosystems, 2008). Spi-B mRNA levels were normalized to input template based on the endogenous control. The BJAB standard is assigned a value of one and the other cell types are reported as values relative to BJAB.
EMSA.
Oligonucleotides with the sequence of the SV40 Spi-B site (5′-CTGAAAGAGGAACTTG-3′), or the JCV Spi-B site L3 (5′-AAAAGGGAAGGTA-3′), L4 (5′-CAAGGGGAAGGGA-3′), L5 (5′-AAAGGGAAGGGA-3′) and L14 (5′-TATAGTGAAACCC-3′) were synthesized with and without 5′ biotinylation by IDT. Mutated versions of the SV40 Spi-B site (5′-CTGAAAGACCAACTTG-3′), and JCV Spi-B L4 (5′-CAAGGCCAAGGGA-3′) and L5 (5′-AAAGCCAAGGGA-3′) were synthesized with 5′ biotinylation. Oligonucleotides with sequences complementary to those listed above were also synthesized with or without 5′ biotinylation as indicated above. The oligonucleotides for the authentic- or mutated-binding sites were annealed to form double-stranded probe at a concentration of 100 ng μl−1. The biotin labelled probes were diluted 1 : 200 in water. Biotin labelled authentic probe or mutant probe was incubated with 5–25 μg nuclear extract from KG-1a cells, BJAB cells or PDA in the presence or absence of a 5-, 100- and 400-fold excess of unlabelled authentic probe. Supershifts were carried out by incubation of the cellular extracts with 2 μl Spi-B antiserum for 30 min on ice before the addition of probe as described previously (Laux et al., 1994). The reactions were incubated at room temperature for 20 min and electrophoresed in a 6 % polyacrylamide–TBE DNA retardation gel (Invitrogen). The complexes were transferred to a positively charged nylon membrane and detected using the LightShift chemiluminescent EMSA kit (Thermo Scientific/Pierce).
Transfection and infection.
Exogenous Spi-B and NF-1X were pre-expressed in human fetal brain-derived progenitor cells or PDN followed by exposure to Mad-4 JCV in a modification of the method described by Messam et al. (2003). Cells were allowed to attach to the wells of PDL-coated six-well dishes at a density of 2–4×105 cells per well for 2 days. Each well of cells was transfected with 1 μg pΔEB-Spi-B DNA or pCHA-NF1X using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). Cells were exposed to medium alone for the mock condition. Approximately 48 h after transfection each well of cells was exposed to 333 haemagglutinaton units (HAU) of Mad-4 JCV or medium alone from the mock condition. Cells were fed with half new medium every 2–3 days. Immunofluorescence was performed on days 5 and 8 post-JCV exposure and T-antigen expression was quantified versus the cell-specific marker expression of nestin for progenitors or β III tubulin for PDN.
Nucleofection.
Archetype, Mad-1, Mad-4, L5 mutant Mad-4, L4 mutant Mad-4 and L5/L4 double-mutant Mad-4 plasmids were introduced into PDAs by nucleofection using the Amaxa basic kit for primary neurons (Amaxa) according to the manufacturer's instructions. Briefly, 1×106 cells were nucleofected with 2 μg DNA using program C13. Cells were allowed to attach for 4 h, followed by replacement of culture medium. Cells were fed with half new medium every 2–3 days. Nucleofection using the pmaxGFP (Amaxa) reporter plasmid was included to determine nucleofection success and efficiency. Immunofluorescence was performed on day 6 post-nucleofection to measure T-antigen expression in GFAP-positive PDAs.
Generation of figures.
Digital Western blot images were detected using fluorescent filters in a FluorChem Q imager (Alpha Innotech). Digital EMSA images were obtained by scanning the exposed film using an hp Scanjet 8250 (Hewlett Packard Development Company). Figures were generated using Canvas II (ACD Systems International Inc.) and Adobe photoshop CS2 (Adobe).
Acknowledgments
We thank Françoise Moreau-Gachelin of the Institut Curie (Paris, France) for generously providing the Spi-B antiserum and human Spi-B plasmid. We thank Ludwig Kappos and Raija Lindberg at the University of Basel (Basel, Switzerland) for their valuable discussions during this work. We thank all of the members of the Laboratory of Molecular Medicine and Neuroscience at the NINDS for all of their hard work, support and invaluable input during this work.
References
- Amemiya, K., Traub, R., Durham, L. & Major, E. O. (1989). Interaction of a nuclear factor-1-like protein with the regulatory region of the human polyomavirus JC virus. J Biol Chem 264, 7025–7032. [PubMed] [Google Scholar]
- Amemiya, K., Traub, R., Durham, L. & Major, E. O. (1992). Adjacent nuclear factor-1 and activator protein binding sites in the enhancer of the neurotropic JC virus. A common characteristic of many brain-specific genes. J Biol Chem 267, 14204–14211. [PubMed] [Google Scholar]
- Andrews, N. C. & Faller, D. V. (1991). A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res 19, 2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki, K., Maeda, H., Wang, J., Kitamura, D. & Watanabe, T. (1988). Purification of a nuclear trans-acting factor involved in the regulated transcription of a human immunoglobulin heavy chain gene. Cell 53, 723–730. [DOI] [PubMed] [Google Scholar]
- Arguello, M., Sgarbanti, M., Hernandez, E., Mamane, Y., Sharma, S., Servant, M., Lin, R. & Hiscott, J. (2003). Disruption of the B-cell specific transcriptional program in HHV-8 associated primary effusion lymphoma cell lines. Oncogene 22, 964–973. [DOI] [PubMed] [Google Scholar]
- Ault, G. S. & Stoner, G. L. (1993). Human polyomavirus JC promoter/enhancer rearrangement patterns from progressive multifocal leukoencephalopathy brain are unique derivatives of a single archetypal structure. J Gen Virol 74, 1499–1507. [DOI] [PubMed] [Google Scholar]
- Bartholdy, B., Du Roure, C., Bordon, A., Emslie, D., Corcoran, L. M. & Matthias, P. (2006). The Ets factor Spi-B is a direct critical target of the coactivator OBF-1. Proc Natl Acad Sci U S A 103, 11665–11670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biosystems, A. (2008). Guide to Performing Relative Quantitation of Gene Expression Using Real-Time Quantitative PCR, pp. 44–51. Technical Note. Foster City. : Applied Biosystems.
- Chang, C. F., Gallia, G. L., Muralidharan, V., Chen, N. N., Zoltick, P., Johnson, E. & Khalili, K. (1996). Evidence that replication of human neurotropic JC virus DNA in glial cells is regulated by the sequence-specific single-stranded DNA-binding protein Pur α. J Virol 70, 4150–4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, N. N. & Khalili, K. (1995). Transcriptional regulation of human JC polyomavirus promoters by cellular proteins YB-1 and Pur α in glial cells. J Virol 69, 5843–5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H., Ray-Gallet, D., Zhang, P., Hetherington, C. J., Gonzalez, D. A., Zhang, D. E., Moreau-Gachelin, F. & Tenen, D. G. (1995a). PU.1 (Spi-1) autoregulates its expression in myeloid cells. Oncogene 11, 1549–1560. [PubMed] [Google Scholar]
- Chen, H. M., Zhang, P., Voso, M. T., Hohaus, S., Gonzalez, D. A., Glass, C. K., Zhang, D. E. & Tenen, D. G. (1995b). Neutrophils and monocytes express high levels of PU.1 (Spi-1) but not Spi-B. Blood 85, 2918–2928. [PubMed] [Google Scholar]
- Chen, N. N., Chang, C. F., Gallia, G. L., Kerr, D. A., Johnson, E. M., Krachmarov, C. P., Barr, S. M., Frisque, R. J., Bollag, B. & Khalili, K. (1995c). Cooperative action of cellular proteins YB-1 and Pur α with the tumor antigen of the human JC polyomavirus determines their interaction with the viral lytic control element. Proc Natl Acad Sci U S A 92, 1087–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl, R., Ramirez-Bergeron, D. L., Rao, S. & Simon, M. C. (2002). Spi-B can functionally replace PU.1 in myeloid but not lymphoid development. EMBO J 21, 2220–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damania, B. & Alwine, J. C. (1996). TAF-like function of SV40 large T antigen. Genes Dev 10, 1369–1381. [DOI] [PubMed] [Google Scholar]
- Daniel, A. M. & Frisque, R. J. (1993). Transcription initiation sites of prototype and variant JC virus early and late messenger RNAs. Virology 194, 97–109. [DOI] [PubMed] [Google Scholar]
- Daniel, A. M., Swenson, J. J., Mayreddy, R. P., Khalili, K. & Frisque, R. J. (1996). Sequences within the early and late promoters of archetype JC virus restrict viral DNA replication and infectivity. Virology 216, 90–101. [DOI] [PubMed] [Google Scholar]
- Dekoninck, A., Calomme, C., Nizet, S., de Launoit, Y., Burny, A., Ghysdael, J. & Van Lint, C. (2003). Identification and characterization of a PU.1/Spi-B binding site in the bovine leukemia virus long terminal repeat. Oncogene 22, 2882–2896. [DOI] [PubMed] [Google Scholar]
- Dorn, A., Fehling, H., Koch, W., le Meur, M., Gerlinger, P., Benoist, C. & Mathis, D. (1988). B-cell control region at the 5′ end of a major histocompatibility complex class II gene: sequences and factors. Mol Cell Biol 8, 3975–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erselius, J. R., Jostes, B., Hatzopoulos, A. K., Mosthaf, L. & Gruss, P. (1990). Cell-type-specific control elements of the lymphotropic papovavirus enhancer. J Virol 64, 1657–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisque, R. J. (1983). Regulatory sequences and virus-cell interactions of JC virus. Prog Clin Biol Res 105, 41–59. [PubMed] [Google Scholar]
- Frisque, R. J., Bream, G. L. & Cannella, M. T. (1984). Human polyomavirus JC virus genome. J Virol 51, 458–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett-Sinha, L. A., Su, G. H., Rao, S., Kabak, S., Hao, Z., Clark, M. R. & Simon, M. C. (1999). PU.1 and Spi-B are required for normal B cell receptor-mediated signal transduction. Immunity 10, 399–408. [DOI] [PubMed] [Google Scholar]
- Garrett-Sinha, L. A., Hou, P., Wang, D., Grabiner, B., Araujo, E., Rao, S., Yun, T. J., Clark, E. A., Simon, M. C. & Clark, M. R. (2005). Spi-1 and Spi-B control the expression of the Grap2 gene in B cells. Gene 353, 134–146. [DOI] [PubMed] [Google Scholar]
- Gerster, T., Matthias, P., Thali, M., Jiricny, J. & Schaffner, W. (1987). Cell type-specificity elements of the immunoglobulin heavy chain gene enhancer. EMBO J 6, 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert, R., Kardas, P., Major, E. O. & Hirsch, H. H. (2010). Rearranged JC virus non-coding control regions found in progressive multifocal leukoencephalopathy increase virus early gene expression and replication rate. J Virol 84, 10448–10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruda, M. C., Zabolotny, J. M., Xiao, J. H., Davidson, I. & Alwine, J. C. (1993). Transcriptional activation by simian virus 40 large T antigen: interactions with multiple components of the transcription complex. Mol Cell Biol 13, 961–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeier, C., Bannister, A. J., Cook, A. & Kouzarides, T. (1993). The activation domain of transcription factor PU.1 binds the retinoblastoma (RB) protein and the transcription factor TFIID in vitro: RB shows sequence similarity to TFIID and TFIIB. Proc Natl Acad Sci U S A 90, 1580–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensold, J. O., Stratton, C. A., Barth, D. & Galson, D. L. (1996). Expression of the transcription factor, Spi-1 (PU.1), in differentiating murine erythroleukemia cells Is regulated post-transcriptionally. Evidence for differential stability of transcription factor mRNAs following inducer exposure. J Biol Chem 271, 3385–3391. [DOI] [PubMed] [Google Scholar]
- Houff, S. A., Major, E. O., Katz, D. A., Kufta, C. V., Sever, J. L., Pittaluga, S., Roberts, J. R., Gitt, J., Saini, N. & Lux, W. (1988). Involvement of JC virus-infected mononuclear cells from the bone marrow and spleen in the pathogenesis of progressive multifocal leukoencephalopathy. N Engl J Med 318, 301–305. [DOI] [PubMed] [Google Scholar]
- Iida, T., Kitamura, T., Guo, J., Taguchi, F., Aso, Y., Nagashima, K. & Yogo, Y. (1993). Origin of JC polyomavirus variants associated with progressive multifocal leukoencephalopathy. Proc Natl Acad Sci U S A 90, 5062–5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, P. N. & Major, E. O. (1999). Viral variant nucleotide sequences help expose leukocytic positioning in the JC virus pathway to the CNS. J Leukoc Biol 65, 428–438. [DOI] [PubMed] [Google Scholar]
- Kenney, S., Natarajan, V. & Salzman, N. P. (1986a). Mapping 5′ termini of JC virus late RNA. J Virol 58, 216–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney, S., Natarajan, V., Selzer, G. & Salzman, N. P. (1986b). Mapping 5′ termini of JC virus early RNAs. J Virol 58, 651–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr, D., Chang, C. F., Chen, N., Gallia, G., Raj, G., Schwartz, B. & Khalili, K. (1994). Transcription of a human neurotropic virus promoter in glial cells: effect of YB-1 on expression of the JC virus late gene. J Virol 68, 7637–7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalili, K., Khoury, G. & Brady, J. (1986). Spacing between simian virus 40 early transcriptional control sequences is important for regulation of early RNA synthesis and gene expression. J Virol 60, 935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Woolridge, S., Biffi, R., Borghi, E., Lassak, A., Ferrante, P., Amini, S., Khalili, K. & Safak, M. (2003). Members of the AP-1 family, c-Jun and c-Fos, functionally interact with JC virus early regulatory protein large T antigen. J Virol 77, 5241–5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs, C. J., McAvoy, M. T. & Kumar, G. (1995). The JC virus minimal core promoter is glial cell specific in vivo. J Virol 69, 2434–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, K. U., Pater, A. & Pater, M. M. (1993). Human JC virus perfect palindromic nuclear factor 1-binding sequences important for glial cell-specific expression in differentiating embryonal carcinoma cells. J Virol 67, 572–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, K. U., Devireddy, L. R., Tang, S. C., Pater, A. & Pater, M. M. (1996). Human JC virus nuclear factor 1 binding motifs and large tumor antigen region required for transactivation of late promoter. J Neurochem 67, 473–481. [DOI] [PubMed] [Google Scholar]
- Laux, G., Dugrillon, F., Eckert, C., Adam, B., Zimber-Strobl, U. & Bornkamm, G. W. (1994). Identification and characterization of an Epstein–Barr virus nuclear antigen 2-responsive cis element in the bidirectional promoter region of latent membrane protein and terminal protein 2 genes. J Virol 68, 6947–6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg, R. L., Achtnichts, L., Hoffmann, F., Kuhle, J. & Kappos, L. (2008). Natalizumab alters transcriptional expression profiles of blood cell subpopulations of multiple sclerosis patients. J Neuroimmunol 194, 153–164. [DOI] [PubMed] [Google Scholar]
- Lynch, K. J. & Frisque, R. J. (1990). Identification of critical elements within the JC virus DNA replication origin. J Virol 64, 5812–5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major, E. O. (2009). Reemergence of PML in natalizumab-treated patients – new cases, same concerns. N Engl J Med 361, 1041–1043. [DOI] [PubMed] [Google Scholar]
- Major, E. O. (2010). Progressive multifocal leukoencephalopathy in patients on immunomodulatory therapies. Annu Rev Med 61, 35–47. [DOI] [PubMed] [Google Scholar]
- Major, E. O., Vacante, D. A., Traub, R. G., London, W. T. & Sever, J. L. (1987). Owl monkey astrocytoma cells in culture spontaneously produce infectious JC virus which demonstrates altered biological properties. J Virol 61, 1435–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, C., Ray-Gallet, D., Tavitian, A. & Moreau-Gachelin, F. (1996). Differential phosphorylations of Spi-B and Spi-1 transcription factors. Oncogene 12, 863–873. [PubMed] [Google Scholar]
- Marshall, L. J. & Major, E. O. (2010). Molecular regulation of JC virus tropism: insights into potential therapeutic targets for progressive multifocal leukoencephalopathy. J Neuroimmune Pharmacol 5, 404–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, J. D., King, D. M., Slauch, J. M. & Frisque, R. J. (1985). Differences in regulatory sequences of naturally occurring JC virus variants. J Virol 53, 306–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzocchetti, A., Wuthrich, C., Tan, C. S., Tompkins, T., Bernal-Cano, F., Bhargava, P., Ropper, A. H. & Koralnik, I. J. (2008). Rearrangement of the JC virus regulatory region sequence in the bone marrow of a patient with rheumatoid arthritis and progressive multifocal leukoencephalopathy. J Neurovirol 14, 455–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda, M., Jona, M., Yasui, K. & Nagashima, K. (1987). Genetic characterization of JC virus Tokyo-1 strain, a variant oncogenic in rodents. Virus Res 7, 159–168. [DOI] [PubMed] [Google Scholar]
- Messam, C. A., Hou, J. & Major, E. O. (2000). Coexpression of nestin in neural and glial cells in the developing human CNS defined by a human-specific anti-nestin antibody. Exp Neurol 161, 585–596. [DOI] [PubMed] [Google Scholar]
- Messam, C. A., Hou, J., Gronostajski, R. M. & Major, E. O. (2003). Lineage pathway of human brain progenitor cells identified by JC virus susceptibility. Ann Neurol 53, 636–646. [DOI] [PubMed] [Google Scholar]
- Monaco, M. C., Sabath, B. F., Durham, L. C. & Major, E. O. (2001). JC virus multiplication in human hematopoietic progenitor cells requires the NF-1 class D transcription factor. J Virol 75, 9687–9695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondek, B., Shepard, A. & Herr, W. (1987). Discrete elements within the SV40 enhancer region display different cell-specific enhancer activities. EMBO J 6, 1017–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petterson, M. & Schaffner, W. (1987). A purine-rich DNA sequence motif present in SV40 and lymphotropic papovavirus binds a lymphoid-specific factor and contributes to enhancer activity in lymphoid cells. Genes Dev 1, 962–972. [DOI] [PubMed] [Google Scholar]
- Pugh, B. F. & Tjian, R. (1991). Transcription from a TATA-less promoter requires a multisubunit TFIID complex. Genes Dev 5, 1935–1945. [DOI] [PubMed] [Google Scholar]
- Rao, S., Garrett-Sinha, L. A., Yoon, J. & Simon, M. C. (1999a). The Ets factors PU.1 and Spi-B regulate the transcription in vivo of P2Y10, a lymphoid restricted heptahelical receptor. J Biol Chem 274, 34245–34252. [DOI] [PubMed] [Google Scholar]
- Rao, S., Matsumura, A., Yoon, J. & Simon, M. C. (1999b). SPI-B activates transcription via a unique proline, serine, and threonine domain and exhibits DNA binding affinity differences from PU.1. J Biol Chem 274, 11115–11124. [DOI] [PubMed] [Google Scholar]
- Ravichandran, V. & Major, E. O. (2008). DNA-binding transcription factor NF-1A negatively regulates JC virus multiplication. J Gen Virol 89, 1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, D., Bosselut, R., Ghysdael, J., Mattei, M. G., Tavitian, A. & Moreau-Gachelin, F. (1992). Characterization of Spi-B, a transcription factor related to the putative oncoprotein Spi-1/PU.1. Mol Cell Biol 12, 4297–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekvig, O. (1997). Polyoma induced autoimmunity to DNA; experimental systems and clinical observations in human SLE. Lupus 6, 325–326. [DOI] [PubMed] [Google Scholar]
- Sabath, B. F. & Major, E. O. (2002). Traffic of JC virus from sites of initial infection to the brain: the path to progressive multifocal leukoencephalopathy. J Infect Dis 186, S180–S186. [DOI] [PubMed] [Google Scholar]
- Schirm, S., Jiricny, J. & Schaffner, W. (1987). The SV40 enhancer can be dissected into multiple segments, each with a different cell type specificity. Genes Dev 1, 65–74. [DOI] [PubMed] [Google Scholar]
- Schmidlin, H., Diehl, S. A., Nagasawa, M., Scheeren, F. A., Schotte, R., Uittenbogaart, C. H., Spits, H. & Blom, B. (2008). Spi-B inhibits human plasma cell differentiation by repressing BLIMP1 and XBP-1 expression. Blood 112, 1804–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer, B. L. & DeKoter, R. P. (2004). Analysis of gene expression and Ig transcription in PU.1/Spi-B-deficient progenitor B cell lines. J Immunol 172, 144–154. [DOI] [PubMed] [Google Scholar]
- Shivakumar, C. V. & Das, G. C. (1994). Biochemical and mutational analysis of the polyomavirus core promoter: involvement of nuclear factor-1 in early promoter function. J Gen Virol 75, 1281–1290. [DOI] [PubMed] [Google Scholar]
- Su, G. H., Ip, H. S., Cobb, B. S., Lu, M. M., Chen, H. M. & Simon, M. C. (1996). The Ets protein Spi-B is expressed exclusively in B cells and T cells during development. J Exp Med 184, 203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, G. H., Chen, H. M., Muthusamy, N., Garrett-Sinha, L. A., Baunoch, D., Tenen, D. G. & Simon, M. C. (1997). Defective B cell receptor-mediated responses in mice lacking the Ets protein, Spi-B. EMBO J 16, 7118–7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner, C., Shinohara, T., Durham, L., Traub, R., Major, E. O. & Amemiya, K. (1996). Expression of multiple classes of the nuclear factor-1 family in the developing human brain: differential expression of two classes of NF-1 genes. J Neurovirol 2, 87–100. [DOI] [PubMed] [Google Scholar]
- Sunden, Y., Semba, S., Suzuki, T., Okada, Y., Orba, Y., Nagashima, K., Umemura, T. & Sawa, H. (2007). DDX1 promotes proliferation of the JC virus through transactivation of its promoter. Microbiol Immunol 51, 339–347. [DOI] [PubMed] [Google Scholar]
- Tamura, T., Inoue, T., Nagata, K. & Mikoshiba, K. (1988). Enhancer of human polyoma JC virus contains nuclear factor I-binding sequences; analysis using mouse brain nuclear extracts. Biochem Biophys Res Commun 157, 419–425. [DOI] [PubMed] [Google Scholar]
- Tan, C. S., Dezube, B. J., Bhargava, P., Autissier, P., Wuthrich, C., Miller, J. & Koralnik, I. J. (2009). Detection of JC virus DNA and proteins in the bone marrow of HIV-positive and HIV-negative patients: implications for viral latency and neurotropic transformation. J Infect Dis 199, 881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacante, D. A., Traub, R. & Major, E. O. (1989). Extension of JC virus host range to monkey cells by insertion of a simian virus 40 enhancer into the JC virus regulatory region. Virology 170, 353–361. [DOI] [PubMed] [Google Scholar]
- Vaz, B., Cinque, P., Pickhardt, M. & Weber, T. (2000). Analysis of the transcriptional control region in progressive multifocal leukoencephalopathy. J Neurovirol 6, 398–409. [DOI] [PubMed] [Google Scholar]
- Wasylyk, B., Hahn, S. L. & Giovane, A. (1993). The Ets family of transcription factors. Eur J Biochem 211, 7–18. [DOI] [PubMed] [Google Scholar]
- Wegner, M., Drolet, D. W. & Rosenfeld, M. G. (1993). Regulation of JC virus by the POU-domain transcription factor Tst-1: implications for progressive multifocal leukoencephalopathy. Proc Natl Acad Sci U S A 90, 4743–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub, S. J., Chow, K. N., Luo, R. X., Zhang, S. H., He, S. & Dean, D. C. (1995). Mechanism of active transcriptional repression by the retinoblastoma protein. Nature 375, 812–815. [DOI] [PubMed] [Google Scholar]
- Yamamoto, H., Kihara-Negishi, F., Yamada, T., Suzuki, M., Nakano, T. & Oikawa, T. (2002). Interaction between the hematopoietic Ets transcription factor Spi-B and the coactivator CREB-binding protein associated with negative cross-talk with c-Myb. Cell Growth Differ 13, 69–75. [PubMed] [Google Scholar]
- Yogo, Y., Kitamura, T., Sugimoto, C., Ueki, T., Aso, Y., Hara, K. & Taguchi, F. (1990). Isolation of a possible archetypal JC virus DNA sequence from nonimmunocompromised individuals. J Virol 64, 3139–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, W., Tuan, J. A. & Comai, L. (1997). SV40 large T antigen binds to the TBP-TAF(I) complex SL1 and coactivates ribosomal RNA transcription. Genes Dev 11, 1605–1617. [DOI] [PubMed] [Google Scholar]
- Zhao, B. & Sample, C. E. (2000). Epstein–Barr virus nuclear antigen 3C activates the latent membrane protein 1 promoter in the presence of Epstein–Barr virus nuclear antigen 2 through sequences encompassing an spi-1/Spi-B binding site. J Virol 74, 5151–5160. [DOI] [PMC free article] [PubMed] [Google Scholar]