Abstract
Low-cost, large-scale production of the baculovirus Autographa californica multiple nucleopolyhedrovirus (AcMNPV) using continuous insect cell culture is seriously hindered by the accumulation of AcMNPV mutants. Specifically, few-polyhedra (FP) mutants, with a reduced yield of occluded virus (polyhedra) and decreased infectivity, usually accumulate upon passaging in cell culture. FP mutations result from transposon insertions in the baculovirus fp25k gene, leading to significantly reduced levels of FP25K protein synthesis. This study evaluated the effects of removing the transposon insertion sites from the wild-type baculovirus fp25k gene; the mutated virus was denoted Ac-FPm. Specifically, this study involved a detailed comparison of wild-type (WT) AcMNPV and Ac-FPm with regard to the proportion of cells having polyhedra, number of polyhedra per cell, the fraction of empty polyhedra, number of occlusion-derived viruses per polyhedron, number of nucleocapsids in the nuclei, FP25K protein synthesis and genetic analysis of the fp25k gene. Removal of TTAA transposon insertion sites from the fp25k gene stabilized FP25K protein synthesis and delayed the appearance of the FP phenotype from passage 5 to passage 10. Electron micrographs revealed that more virus particles were found inside the nuclei of cells infected with Ac-FPm than in the nuclei of cells infected with WT AcMNPV (at passage 10). Abnormalities, however, were observed in envelopment of nucleocapsids and virus particle occlusion within Ac-FPm polyhedra. Thus, the FP phenotype appeared in spite of continued FP25K protein synthesis, suggesting that mechanisms other than fp25k gene disruption can lead to the FP phenotype.
INTRODUCTION
Baculoviruses offer an environmentally friendly approach to controlling insect pests and have been used successfully for the management of various lepidopteran (moth) pests of crops and forests (Moscardi, 1999; Murhammer, 1996; Szewczyk et al., 2009). A potential market for recombinant baculovirus insecticides has already been established (Black et al., 1997; DuPont, 1996; Smith et al., 2000). Baculoviruses can be produced either in insect larvae or by insect cell culture. Production in insect larvae is low cost but labour intensive unless a robotic system is employed (Gard, 1997). Cell culture production in batch bioreactors becomes cheaper than larvae production as the production scale increases (Black et al., 1997; van Lier et al., 1990). Furthermore, it has been estimated that baculoviruses can be produced in a continuous bioreactor system for 50 % of the cost of producing them in batch bioreactors (Rhodes, 1996; Tramper & Vlak, 1986). Unfortunately, baculoviruses are prone to mutant accumulation in continuous cell culture due to repeated passaging. Consistent with this premise, the yield of polyhedra (the form of virus used as the pesticide) and number of viruses per polyhedron decreased significantly by 25 days in a continuous bioreactor system due to few-polyhedra (FP) (Fraser et al., 1983; Harrison & Summers, 1995b) and defective-interfering particle (DIP) mutant accumulation (Kompier et al., 1988; Kool et al., 1991). FP mutations are common in baculoviruses used for the management of lepidopteran pests, including Autographa californica multiple nucleopolyhedrovirus (AcMNPV) (Fraser et al., 1983), Lymantria dispar MNPV (Bischoff & Slavicek, 1997), Galleria mellonella MNPV (GmMNPV) (Fraser & Hink, 1982), Helicoverpa armigera NPV (HaNPV) and Anticarsia gemmatalis MNPV (AgMNPV) (de Rezende et al., 2009). Hence, overcoming such mutations is an important step in enabling continuous large-scale production of baculovirus biopesticides that will make them more cost competitive with chemical pesticides. The work described here focused on investigating strategies to overcome the FP mutation.
Several studies have demonstrated the connection between serial passage of baculovirus in cell culture and the appearance of FP mutants (Cusack & McCarthy, 1989; Fraser & Hink, 1982; Hink & Strauss, 1976; Knudson & Harrap, 1975; MacKinnon et al., 1974; Potter et al., 1976). FP mutants are characterized by fewer cells containing polyhedra, fewer polyhedra per cell and fewer or no viruses per polyhedron (Harrison & Summers, 1995b). Most of these FP mutants result from the incorporation of host cell DNA into a specific region (HindIII-I fragment of AcMNPV) of the baculovirus genome, thereby disrupting FP25K protein synthesis (Beames & Summers, 1988; Carstens, 1987; Fraser et al., 1983). Examples of such insertions are cell-derived transposable elements such as IFP2 (‘piggyBac’; Cary et al., 1989), TFP3 (‘tagalong’; Wang et al., 1989), hitchhiker (Bauser et al., 1996) and TED (Miller & Miller, 1982). There is evidence of host cell insertions in other regions of the AcMNPV genome, e.g. in the 94k and da26 genes, that also lead to the FP phenotype (Friesen & Nissen, 1990; Kumar & Miller, 1987; O'Reilly et al., 1990). FP25K protein synthesis is associated with enhanced biosynthesis and nuclear localization of the polyhedrin protein during the early occlusion phase and acts as a switch from budded virus (BV) production to occlusion-derived virus (ODV) production (Harrison et al., 1996; Jarvis et al., 1992). Furthermore, it has been demonstrated that fp25k gene deletion or an insertion mutation into the fp25k gene leads to the FP phenotype with (i) enhanced BV production (Harrison & Summers, 1995b; Kelly et al., 2008; Wu et al., 2005), (ii) increased synthesis of some BV structural proteins (e.g. GP64, BV E-26 and VP39) (Beniya et al., 1998; Braunagel et al., 1999), (iii) reduced levels of some occluded virus envelope proteins (Harrison et al., 1996), (iv) decreased post-mortem liquefaction of the larval host (Nakanishi et al., 2010), (v) decreased E66 protein synthesis and altered intranuclear transport thereof (Rosas-Acosta et al., 2001), and (vi) production of virions with aberrant morphology (Harrison & Summers, 1995b). Hence, overcoming fp25k gene mutation is a necessary criterion to maintain proper polyhedra and ODV production in continuous cell culture.
Harrison & Summers (1995b) reported that an insertion mutation in the fp25k gene led to the FP phenotype and that normal polyhedron production and the occlusion process could be restored following reinsertion of the wild-type (WT) fp25k gene into the virus. Specifically, the FP phenotype primarily results from transposon insertion into TTAA target sites in the fp25k gene (Beames & Summers, 1990; Fraser et al., 1985). Therefore, this study investigated the effect of modifying the 13 TTAA sites in the fp25k gene, without altering the protein's amino acid sequence, on FP phenotype accumulation.
RESULTS
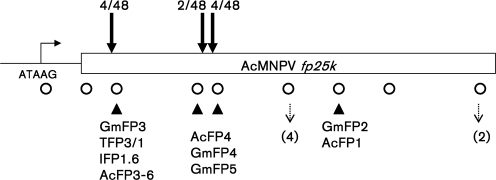
Mutations made in the fp25k gene to remove the TTAA sites (Fig. 1) resulted in a modified AcMNPV, denoted Ac-FPm.
Fig. 1.
TTAA sites mutated in Ac-FPm. Circles represent the 13 TTAA sites in the WT AcMNPV fp25k gene that were changed in Ac-FPm. The names of known mutants involving transposon insertion into TTAA sites are indicated. Closed triangles denote the TTAA sites of known insertions of host DNA/transposons into AcMNPV or GmMNPV mutants (Bischoff & Slavicek, 1997). The sites of multiple TTAA sequences in close proximity are indicated by a dashed arrow, with the number in parentheses denoting the number of TTAA repeats. Sites at which the mutation led to reversion of TTAA during passaging are denoted by solid arrows, with the numbers above the arrows denoting the number of mutations (reversions)/total number of isolates investigated.
Characterization of WT AcMNPV and Ac-FPm virus at passage 1
The initial passage (i.e. passage 1) of WT AcMNPV and Ac-FPm was characterized to provide a basis for assessing changes following passaging in Sf-21 cells. Briefly, the following properties of Sf-21 cells infected with either of these baculoviruses were monitored: (i) cell density and viability, (ii) BV production, (iii) polyhedron production, (iv) FP25K protein synthesis and (v) lethality to insect larvae. Comparing the characteristics of Ac-FPm-infected cells with those of WT AcMNPV-infected cells demonstrated that (i) the time courses of cell density and viability, BV production and FP25K protein production were similar (data not shown) and (ii) there were no significant differences in the distribution of polyhedra per cell (P>0.05, n=3 fields of view). Furthermore, the toxicity of the two baculoviruses against Heliothis virescens (LC50 values) was similar (Table 1). These results indicated that polyhedron production, infectivity, FP25K synthesis and toxicity to insects for WT AcMNPV and Ac-FPm were essentially the same at passage 1.
Table 1.
Dose–mortality response of H. virescens neonates infected with WT AcMNPV and Ac-FPm
P, Passage.
| Virus | Slope±sem | LC50×10−6 (95 % CL)* | Potency ratio (95 % CL)† | Heterogeneity (χ2/n) | g value at 95 % CL‡ |
|---|---|---|---|---|---|
| WT AcMNPV P1 | 1.124±0.151 | 3.171 (1.646–5.194)a | – | 2.316 | 0.207 |
| Ac-FPm P1 | 0.801±0.145 | 3.414 (1.554–6.111)a | 0.841 (0.400–1.711) compared with WT P0 | 1.648 | 0.267 |
| WT AcMNPV P25 | 0.864±0.118 | 11.20 (8.066–16.46)b | 0.279 (0.147–0.487) compared with WT P0 | 0.800 | 0.071 |
| Ac-FPm P25 | 1.242±0.127 | 9.122 (7.195–11.68)b | 0.391 (0.236–0.628) compared with Ac-FPm P0 | 0.550 | 0.040 |
| 1.125 (0.760–1.675) Compared with WT P25 |
*LC50 (polyhedra ml−1) values with 95 % CL were obtained by running the polo probit analysis program. For each treatment, LC50 values with the same letter were not significantly different if the potency ratio 95 % CL included the value 1.0. For each virus, the values with different superscript letters were significantly different at P<0.05.
†The potency ratio is LC50 of the indicated virus/LC50 of another virus. If the potency ratio 95 % CL covered the value 1.0, this indicated that both LC50 values were the same, i.e. no statistical significance; otherwise they were significantly different (Robertson & Preisler, 1992).
‡If g<0.5, then the data fitted the probit model; otherwise, the data did not fit the probit model and the analysis was not valid.
Cells with polyhedra and the FP phenotype
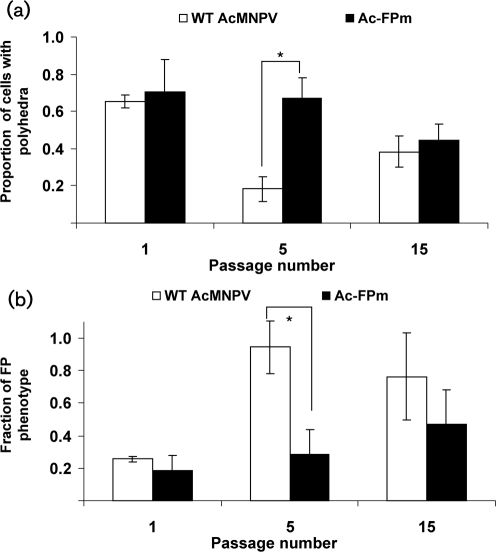
The polyhedral content and FP phenotype (<10 polyhedra per cell) of the infected cells were monitored by light microscopy. For WT AcMNPV-infected cells at 96 h post-infection (p.i.), the percentage of cells with polyhedra in passage 5 was significantly less (P<0.05) than that of WT passage 1 (Fig. 2a). In contrast, for Ac-FPm-infected cells there was no significant difference between passages 1 and 5 (P>0.05). Although a difference was observed between WT AcMNPV and Ac-FPm infection (P<0.05) in the percentage of polyhedra-producing cells at passage 5, no significant difference was observed by passage 15 (Fig. 2a).
Fig. 2.
Comparison of polyhedron production in WT AcMNPV- and Ac-FPm-infected Sf-21 cells after serial passaging at 96 h p.i. (a) Proportion of cells with polyhedra. (b) Fraction of FP phenotype (one to nine polyhedra per cell). The error bars represent the 95 % CL. *, P<0.05 (Student's t-test).
Similarly, a significant increase (P<0.05) in the FP phenotype was noted between passages 1 and 5 of WT AcMNPV-infected cells (Fig. 2b), but no such difference was observed for Ac-FPm-infected cells at 96 h p.i. The FP phenotype for Ac-FPm infection was significantly lower than WT AcMNPV at passage 5, but no significant difference was noted at passage 15 (Fig. 2b).
FP25K protein synthesis
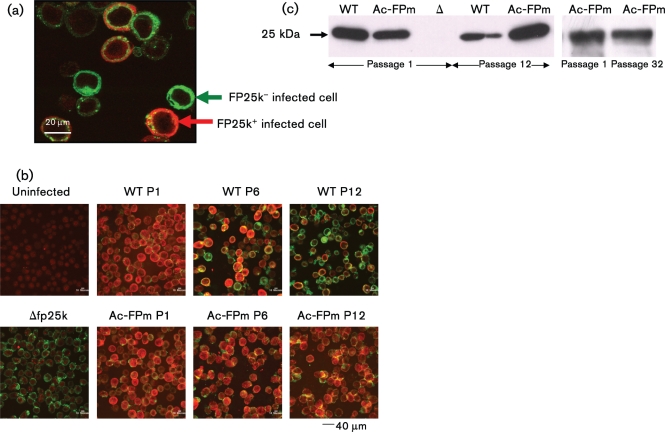
Infected cells isolated at 48 h p.i. and stained with antibodies were examined by immunofluorescence confocal microscopy to identify FP25K and GP64 protein synthesis (GP64 synthesis was used to identify infected cells) (Fig. 3a). The percentage of cells expressing FP25K protein decreased when infected with WT AcMNPV of increasing passage number, i.e. 100, 46.3 and 43.3 % of the infected cell population expressed FP25K protein at passages 1, 6 and 12, respectively. In contrast, no significant change in the percentage of cells containing FP25K protein was observed when infected with Ac-FPm of increasing passage number, i.e. 100, 98.8 and 98.3 % of the infected cell population expressed FP25K protein at passages 1, 6 and 12, respectively (Fig. 3b). These results were confirmed by Western blotting of the proteins from cells infected with the passaged viruses collected at 48 h p.i. (Fig. 3c). Specifically, Western blots of cells infected with passaged WT AcMNPV and Ac-FPm showed a decrease in FP25K protein synthesis in cells infected with WT AcMNPV at passage 12. In contrast, significant FP25K synthesis was maintained in Ac-FPm-infected cells at passage 12 as well as at passage 32.
Fig. 3.
Modification of the fp25k gene by removal of TTAA sites leads to a larger fraction of cells with FP25K synthesis at later passages. (a) Sf-21 cells infected with WT AcMNPV or Ac-FPm at 48 h p.i. were stained for GP64 (green) and FP25K (red) protein to identify the FP25K-positive infected cells. (b) Comparison of the fraction of cells producing FP25K protein infected by WT AcMNPV or Ac-FPm at passages (P) 1, 6 and 12. Uninfected cells (no colour) and cells infected with Δfp25k AcMNPV (green only) were used as controls. The difference between WT AcMNPV- and Ac-FPm-infected cells producing FP25K protein at passage 12 was statistically significant (P<0.05, Student's t-test, n=3 fields of view). (c) Western blot analysis of FP25K protein synthesis at passages 1, 12 and 32 of WT AcMNPV and Ac-FPm viruses at 48 h p.i. Δ, Δfp25k AcMNPV.
Bioassay
Dose–mortality bioassays with neonates of H. virescens demonstrated that the LC50 of both WT AcMNPV and Ac-FPm increased significantly after 25 passages compared with passage 1 (Table 1), indicating a significant reduction in virulence after 25 passages. The LC50 of Ac-FPm from passage 25 was not significantly different from that of WT virus (passage 25) as indicated by the potency ratio (Table 1).
Genetic analysis of the fp25k gene
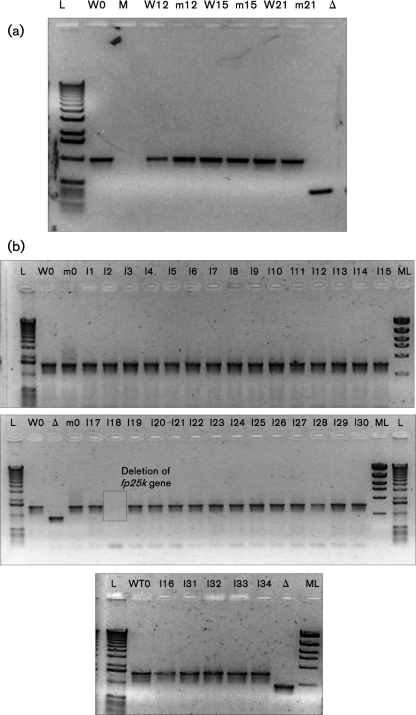
In order to test for transposon insertion during passaging of WT AcMNPV and to evaluate the effect of transposon site removal in Ac-FPm, a detailed sequencing analysis of the fp25k gene was performed. PCR amplification of the fp25k gene from both WT AcMNPV and Ac-FPm viral DNA (of the heterogeneous population) showed no change in size of the PCR product at passages 12, 15 and 21, indicating that no insertions had occurred in either virus at the TTAA sites (Fig. 4a). Genomic variations within these heterogeneous populations were investigated by conducting sequence analyses on individual clonal isolates. Specifically, 34 isolates from passage 12 were obtained following two rounds of plaque purification (Kelly et al., 2007) of the WT AcMNPV and Ac-FPm viruses. PCR amplification of the fp25k gene again demonstrated the absence of insertions in all 34 isolates obtained from WT AcMNPV passage 12 (Fig. 4b). However, evidence of an fp25k gene deletion was found in one of the 34 isolates from WT AcMNPV passage 12 (Fig. 4b). Furthermore, DNA sequencing of the fp25k gene was performed on 16 randomly selected isolates of the WT and Ac-FPm virus clones at passage 12. No evidence of point mutations was found in the WT virus fp25k gene, but 3/16 Ac-FPm clones contained point mutations in the modified fp25k gene (Table 2), some of which involved reversion of the altered TTAA sites back to TTAA (Table 2, Fig. 1). Similar results were obtained from the analysis of DNA from viral isolates at passage 32 (Table 2). No mutations were detected from sequencing the fp25k promoter region (∼400 bp upstream of the fp25k gene start codon) in any of the WT or Ac-FPm virus isolates from passage 12. As the da26 gene is another potential part of the AcMNPV genome that may have insertions or deletions to give the FP phenotype (O'Reilly et al., 1990), further PCR analysis of the da26 gene was performed. No evidence of insertions or deletions was found in the da26 gene in any of the 34 WT and Ac-FPm virus isolates from passage 12 (data not shown).
Fig. 4.
(a) PCR amplification of the fp25k gene region (1 kb) from passaged WT and Ac-FPm virus (mixed population from passages 12, 15 and 21), indicating no insertion in the fp25k gene. (b) PCR amplification of the fp25k gene plus fp25k promoter region (1.54 kb) from 34 isolates of WT AcMNPV (individual plaques) at passage 12 showing no insertion in the fp25k gene; 33/34 of the WT virus isolates retained the complete fp25k gene. L, 1 kb size ladder; ML, molecular mass ladder; M, PCR with no DNA; W at passage 1, WT AcMNPV at passage 1; m0, Ac-FPm0; I, isolate; Δ, Δfp25k AcMNPV.
Table 2.
Point mutations in the Ac-FPm fp25k gene after passaging
P, Passage number, m, virus isolate number.
| Mutants* | No. point mutations | Mutations leading to reversion of TTAA sites | Other mutations |
|---|---|---|---|
| Ac-FPmP12-m2 | 1 | C24T, | |
| Ac-FPmP12-m6 | 3 | C24T, G204A, C237A | |
| Ac-FPmP12-m9 | 2 | C24T, G204A | |
| Ac-FPmP32-m7 | 3 | C24T, C237T | G214A |
| Ac-FPmP32-m20 | 2 | C237T | G214A |
| Ac-FPmP32-m21 | 2 | C237T | G214A |
*Individual virus isolates were plaque purified from passages 12 and 32 of Ac-FPm. Mutations are indicated.
Analysis of polyhedra, ODVs and nucleocapsids outside the polyhedra
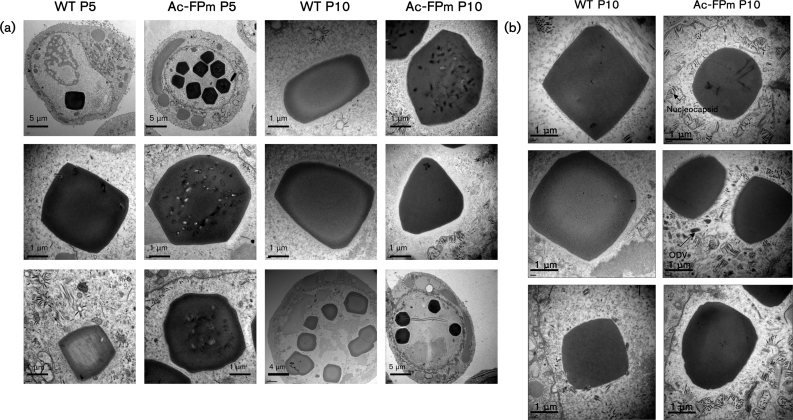
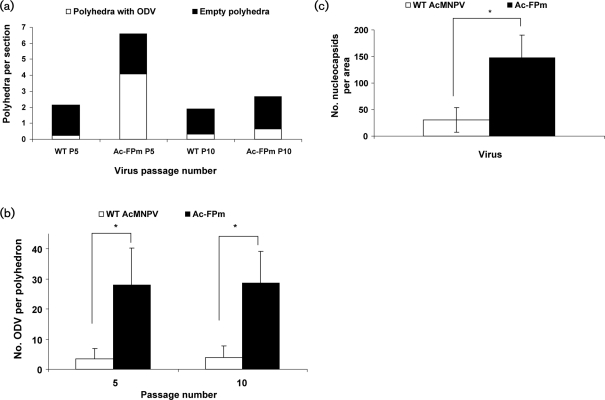
Infected cells were analysed at 72 h p.i. by electron microscopy. The presence of no or few polyhedra per cell in WT AcMNPV-infected cells was detected as early as passage 5 (Fig. 5a). Additionally, a representative fraction of the polyhedra had no ODVs and most of the nucleocapsids outside the polyhedra were not enveloped (Figs 5a and 6a). In contrast, at passage 5, cells having many polyhedra (>10 per cell) and polyhedra having many ODVs were predominant in Ac-FPm-infected cells (Figs 5a and 6a) and the number of ODVs per polyhedron was significantly higher than in WT virus-infected cells (Fig. 6b). At passage 10 of the WT AcMNPV, the virus population was dominated by the FP phenotype, with polyhedra having no ODVs (Figs 5a and 6a). By passage 10, a significant fraction of the Ac-FPm virus population had empty polyhedra (Fig. 6a). Although no significant difference was observed in the number of cells having polyhedra and the number of polyhedra per cell between the two viruses at passage 15 (Fig. 2a, b), the number of ODVs per polyhedron was significantly higher in Ac-FPm-infected cells than WT AcMNPV-infected cells at passage 10 [Fig. 6b, analysis of variance (ANOVA), P<0.05, n=12]. At passage 10, the nucleocapsid density in the nuclei (outside the empty polyhedra) was found to be significantly higher in Ac-FPm-infected cells than in WT AcMNPV-infected cells (Student's t-test, P<0.05, n=15) (Figs 5b and 6c).
Fig. 5.
Electron micrographs of WT AcMNPV- and Ac-FPm-infected Sf-21 cells at passages 5 (P5) and 10 (P10) at 72 h p.i. (a) The FP and MP characteristics showed that formation of the FP phenotype is delayed in Ac-FPm compared with WT AcMNPV. (b) The number of nucleocapsids per area outside the empty polyhedra was greater in Ac-FPm-infected cells than in WT AcMNPV-infected cells at passage 10.
Fig. 6.
Number of polyhedra with ODVs, number of ODVs per polyhedron and number of nucleocapsids per area outside the polyhedra produced by WT AcMNPV- and Ac-FPm-infected Sf-21 cells at passages 5 and 10. (a) The number of polyhedra per cell section and the proportion of polyhedra with ODVs was greater in Ac-FPm-infected cells than in WT AcMNPV-infected cells at passage 5 (n=12 cell sections evaluated for each passage). (b) The number of ODVs per polyhedron was higher in Ac-FPm-infected cells than in WT AcMNPV-infected cells at passages 5 and 10. Results are shown as means±95% CL. *, P<0.05 (Student's t-test, n=10 cross-sections of polyhedra with ODVs for each passage). (c) The nuclei of Ac-FPm-infected cells contained more nucleocapsids and ODVs outside the empty polyhedra than WT AcMNPV-infected cells at passage 10. Results are shown as means±95% CL. *, P<0.05 (Student's t-test, n=15 cross-sections of empty polyhedra for each passage).
DISCUSSION
Effect of virus passaging on FP25K protein synthesis
Although reduced or no FP25K protein synthesis has been correlated with the FP phenotype (Fraser et al., 1983; Beames & Summers, 1988), no previous investigation has evaluated FP25K synthesis in each cell within a cell population infected with passaged virus containing a mixture of FP and MP phenotype. The present immunofluorescence study demonstrated that the fraction of cells with FP25K protein synthesis decreased by more than 50 % from passage 1 to passage 6 in WT AcMNPV-infected cells. In contrast, all of the Ac-FPm-infected cells continued to produce FP25K protein at passage 12 (Fig. 3b). Moreover, a Western blot of proteins from passaged Ac-FPm-infected cells revealed that FP25K synthesis was maintained up to passage 32 (>70 days) (Fig. 3c).
Is transposon insertion the only reason for decreased FP25K protein synthesis?
The evidence suggested that the presence of insertions in the fp25k gene is not the only process through which FP25K protein synthesis can be eliminated. For example, 50 % of WT (passage 12)-infected cells did not express the FP25K protein (Fig. 3b, c) and 33/34 isolates tested by PCR had a normal-length PCR product (the remaining isolate had a deletion), indicating that there were no insertions in the gene. A potential route through which FP25K protein synthesis can be reduced is by altering the synthesis of viral proteins (e.g. E66) that regulate FP25K synthesis (Beniya et al., 1998). Consistent with these results, Lua et al. (2002) and de Rezende et al. (2009) found that the FP phenotype of HaSNPV and AgMNPV, respectively, resulted from passaging in cell culture without any transposon insertions in the fp25k gene. Neither of these investigations, however, monitored FP25K protein synthesis.
Point mutations in the fp25k gene
The point mutations found in passages 12 and 32 in the fp25k gene of Ac-FPm are consistent with results of Lua et al. (2002), who found point mutations in the fp25k gene of passaged HaSNPV. Additionally, point mutations of this type could result from DNA replication errors resulting from DNA polymerase slippage (Bischoff & Slavicek, 1997).
The reversion of altered TTAA sites back to the original TTAA sequence (Table 2 and Fig. 1) suggests that some TTAA sites may have an important role in regulating gene transcription by a cis-acting function, or in regulating mRNA stability, transport and translation. Similar cis-acting functions have been observed with sequences within the AcMNPV genome (but not in the fp25k gene), e.g. a cis-acting element within the 5′ non-coding region of the ie-1 gene (Pullen & Friesen, 1995) and a cis-acting element upstream of the polh promoter (Kumar et al., 2009).
FP phenotype in a heterogeneous infected-cell population
Although many investigations have been performed involving the genetic and electron microscopic analysis of FP clones isolated via plaque purification (Bischoff & Slavicek, 1997; Fraser & Hink, 1982; Harrison & Summers, 1995b; Lua & Reid, 2003; Lua et al., 2002), only a few studies have been conducted to characterize the heterogeneous virus population by electron microscopy (de Rezende et al., 2009; MacKinnon et al., 1974). In this study, the heterogeneous virus population was characterized for passages 5 and 10 with respect to the number of polyhedra-producing cells, the number of polyhedra per cell, the number of ODVs per polyhedron and the number of nucleocapsids outside empty polyhedra (Figs 5 and 6). These results demonstrated that passaging the baculoviruses led to large variations in these parameters compared with the relative homogeneity at passage 1.
Removal of transposon insertion sites delays the emergence of FP characteristics
Comparison of WT AcMNPV- and Ac-FPm-infected cells demonstrated that FP phenotype accumulation was delayed in Ac-FPm-infected cells. An unexpected result of this study was a decrease in the percentage of WT AcMNPV-infected cells having polyhedra from passage 1 to passage 5, followed by an increase from passage 5 to passage 15 (Fig. 2a). One possible reason could be a fluctuation in the number of DIPs (mutant virus in which 40–45 % of the genome is deleted) in the virus population at different times of passage. Cyclic peaks in DIP formation are common during passaging of animal viruses (Dimmock & Marriott, 2006), and DIP genomes could lack the fp25k gene that would lead to the FP phenotype. Consistent with this possibility, gel fractionation of viral genomes from WT-virus infected cells from passage 5 showed that most of the genomes were shorter than those found at passage 1 (data not shown).
The fraction of cells containing polyhedra and the number of polyhedra per cell for Ac-FPm- and AcMNPV-infected cells were comparable by passage 15 (Fig. 2) and the fraction of polyhedra containing viruses were comparable by passage 10 (Fig. 6a). These phenomena occurred in spite of the fact that FP25K protein synthesis in Ac-FPm-infected cells (but not in WT AcMNPV-infected cells) was maintained. This suggested that mutations in genes other than fp25k are involved in FP accumulation. Indeed, mutations in other genes, e.g. da26 and 94k, are known to disrupt occlusion body production (Friesen & Nissen, 1990; O'Reilly et al., 1990). In the present study, it was demonstrated that the da26 gene in WT AcMNPV did not contain insertions at passage 12, but the presence of other types of mutation was not investigated. The FP25K protein may interact with other viral proteins to regulate the viral occlusion process, and mutations in any of the genes involved in this process may lead to the FP phenotype. Also, point mutations in the polyhedrin gene can lead to abnormal polyhedron formation and altered polyhedrin protein localization (Bravo-Patiño & Ibarra, 2000; Katsuma et al., 1999; Lin et al., 2000; Nakazawa et al., 1996).
Whilst the fraction of polyhedra containing ODV in Ac-FPm- and AcMNPV-infected cells was comparable at passage 10, the Ac-FPm-infected cells contained more virus both inside and outside the polyhedra (Figs 5b and 6). These observations support the premise that the FP25K protein is involved in targeting the virions to the nucleus. However, the finding that nucleation of the polyhedrin protein around ODVs was greatly diminished in Ac-FPm-infected cells, even in the presence of significant FP25K protein synthesis, suggests that other proteins are involved in this process.
Conclusion
This investigation demonstrated that removal of TTAA sites from the fp25k gene can maintain FP25K protein synthesis for at least 32 passages in cell culture and can delay, but not stop, FP mutant accumulation. Therefore, further studies need to be conducted to determine the strategy necessary to develop a modified AcMNPV conducive to a continuous process for producing baculovirus biopesticides. This will require a comprehensive understanding of the correlation between FP phenotype and mutations in baculovirus genes. Future work may involve a detailed study of mutation patterns during serial passaging by utilizing microarray analysis (Yamagishi et al., 2003). It would also be interesting to determine the viral proteins that interact with FP25K and are involved in FP phenotype formation. Functional studies of proteins (e.g. E26, E25, ODV-E66, GP64, VP39, 94k and p35) may answer questions concerning the mechanism behind the formation of polyhedral crystals, transport of nucleocapsids and polyhedrin nucleation around ODVs. Finally, viruses with modified TTAA sites in the fp25k gene may lead to a more genetically stable baculovirus expression vector system for recombinant protein production in which many virus passages are required to go from a small virus stock to a large-scale batch bioreactor.
METHODS
Cells and virus.
The Spodoptera frugiperda IPLB-Sf-21AE cell line (Sf-21) (Vaughn et al., 1977) and AcMNPV strain E2 (Ayres et al., 1994) were used. Sf-21 cells were maintained in 125 ml Erlenmeyer flasks (20 ml working volume) on an orbital shaker at 140 r.p.m. and 27 °C in Sf-900 II serum-free medium (Invitrogen). Cell density and viability were monitored with a Vi-Cell (Beckman Coulter) and BV was quantified using an end-point dilution assay as described by O'Reilly et al. (1992). The virus Δfp25K, with the fp25k gene deleted, was used as a control virus (Rosas-Acosta et al., 2001).
Construction of Ac-FPm.
The recombinant baculovirus denoted Ac-FPm was constructed by modifying the TTAA transposon target sites in the AcMNPV fp25k gene. Briefly, plasmid pAcE2HindIII-I containing the HindIII-I fragment of AcMNPV E2 was constructed by ligating the HindIII-I fragment into the HindIII site of pUC18. The coding sequence of fp25k in pAcE2HindIII-I was mutated using template-directed ligation and PCR to alter the 13 potential TTAA transposon target sites without changing the amino acid sequence of the FP25K protein. The 11 TTAA sequences within the coding sequence were altered by changing either the second T or first A without changing the resulting amino acid. Nine different primers were constructed with nucleotide changes incorporated for the TTAA sequences. Two of these primers were used to alter multiple TTAA sequences that occur in close proximity to each other and the other seven primers were used to alter the other seven TTAA sites (Fig. 1). The resulting modified fp25k gene was introduced into the viral genome by co-transfecting Sf-21 cells with the plasmid DNA and AcFPβgal DNA. The AcFPβgal baculovirus contains a β-galactosidase gene fused in frame after nt 373 of the fp25k ORF (Harrison & Summers, 1995b). Following a double homologous recombination event, the baculovirus Ac-FPm was produced. Recombinant baculovirus clones were identified by conducting plaque assays and staining for β-galactosidase activity (the desired clones did not contain this activity). The selected viruses were purified by plaque assay and amplified, and the DNA was extracted to confirm the expected genetic structure by restriction enzyme analysis (O'Reilly et al., 1990). The sequence of the mutated fp25k gene was verified and FP25K protein synthesis was examined by Western blotting. A polyclonal antibody against the FP25K protein was obtained by preparing a fusion protein of FP25K and maltose-binding protein as described by Harrison & Summers (1995a) and then injecting into a rabbit (Elmira Biologicals).
Serial passaging of AcMNPV in vitro.
A serial passaging experiment was designed to simulate the baculovirus passaging that occurs in a continuous bioreactor system. For all passages, a 20 ml working volume of Sf-21 cells (in Sf-900 II SFM medium supplemented with 10 % FBS; Gibco) at a concentration of 0.8×106 cells ml−1 was infected with the corresponding baculovirus. Initially, cells were infected at an m.o.i. of 10 with WT AcMNPV or Ac-FPm. Supernatant containing BV was collected for subsequent passages when the cell viability decreased to 65–70 % (at ∼2–3 days p.i.). This was continued for all passages, with 1.5 ml cell supernatant containing BV being used to infect a fresh cell suspension. All baculovirus stocks were obtained after removing cells by centrifugation at 800 g for 10 min.
Polyhedra quantification.
Sf-21 cells were infected with passaged WT AcMNPV or Ac-FPm at an m.o.i. of 10. Cells (at least 100) harvested at 96 h p.i. were examined by light microscopy for the presence or absence of polyhedra and the number of polyhedra per cell to determine the distribution of the FP phenotype (defined as <10 polyhedra per cell) and MP phenotype (defined as ≥10 polyhedra per cell). Triplicate flasks of infected cells were used for every passage. Cells infected with WT AcMNPV and Ac-FPm were compared using Student's t-test (Shoemaker et al., 1974) with regard to the fraction of cells with polyhedra and the FP phenotype fraction.
Western blot analysis of FP25K synthesis.
At 48 h p.i. for selected passages, infected cell pellets (3×106 cells) were collected and lysed by sonication (30 s). Total cellular protein was quantified by the Bradford method using a BSA standard. Proteins in the cell pellet were separated by 12 % SDS-PAGE (Bio-Rad) at 180 V with each lane loaded with 25 μg protein. The proteins were blotted onto a nitrocellulose membrane (Bio-Rad) using a TransBlot Semi-Dry Transfer Cell (Bio-Rad) and analysed with a rabbit polyclonal antibody against the AcMNPV FP25K protein at a dilution of 1 : 100 000 (Sambrook et al., 1989). Bound rabbit antibody was detected using goat anti-rabbit IgG conjugated with horseradish peroxidase at a 1 : 100 000 dilution (Pierce). A SuperSignal West Pico Chemiluminescent Substrate kit (Pierce) was used to develop the protein bands on the blot and the protein was quantified using ImageJ software version 1.3.
PCR.
The primers fppromoter120up (5′-GCGCTTTACGCTGCTCCGCGGCGGC-3′; forward) and Acfp25k165down (5′-CTCTTACCGTTATAGGGAAGG-3′; reverse) were used to amplify the approximately 1.54 kb fp25k gene region (fp25k gene plus fp25k promoter) by PCR as described by Lua et al. (2002). The PCR fragments were sequenced using the primers fppromoter120up, Acfp25k130up (5′-GGGTCTAATATGAGGTCAAACTC-3′) and Acfp25k165down. Another set of primers, da26200up (5′-CAACAGCTGCCAATGTACCG-3′; forward) and da26200down (5′-CTGAATATAAGCGCTATCAAAGCC-3′; reverse), were used to amplify the da26 gene region to check for transposon insertion (O'Reilly et al., 1990).
Immunofluorescence assay.
Sf-21 cells (0.5×106) were infected at an m.o.i. of 20 and incubated at 27 °C, and an immunofluorescence assay was performed as described by Bjerke & Roller (2006) at 48 h p.i. The primary antibody was a mixture of anti-FP25K (1 : 1000) and anti-GP64 (1 : 2000) (Novagen) and the secondary antibody was a mixture of Alexa Fluor 568 goat anti-rabbit (1 : 1000) and Alexa Fluor 488 goat anti-mouse (1 : 1000) (Invitrogen). The images were photographed with a Bio-Rad MRC-1024 confocal microscope. For each treatment, three fields of view were observed, with each field containing 60–80 cells. FP25K protein synthesis was quantified by calculating the percentage of infected (GP64-positive) cells producing FP25K protein (FP25K positive) per treatment. The percentage of cells infected by WT AcMNPV or Ac-FPm that produced FP25K protein were compared by Student's t-test (Shoemaker et al., 1974) with a 95 % confidence limit (CL) and n=3 (number of fields photographed per treatment).
Bioassay.
Lethal-concentration bioassays were performed using the droplet feeding method (Hughes & Wood, 1981). Polyhedra of WT AcMNPV and Ac-FPm (passages 0 and 25) were assayed against neonate Heliothis virescens larvae as described previously with three replicates of 30 larvae per virus concentration (Harrison & Bonning, 2001). The concentrations used were 1.0×106, 3.0×106, 9.0×106, 18×106 and 36×106 polyhedra ml−1. Larval mortality was scored after mock-infected larvae had pupated. LC50 values were calculated by polo probit analysis and compared by standard lethal concentration ratio comparison (Robertson & Preisler, 1992).
Transmission electron microscopy.
Infected Sf-21 cells from passages 5 and 10 at 72 h p.i. were fixed, embedded in resin and sectioned as described by Harrison & Summers (1995b). Sections (100 nm) were stained with 5 % uranyl acetate and lead citrate. Samples were examined using a JEOL 1230 transmission electron microscope at 120 kV. The images were used to determine the number of cells with polyhedra, the number of ODVs per polyhedra and the number of nucleocapsids outside the empty polyhedra. Statistical analysis of data was performed using ANOVA in matlab with 95 % CL.
Acknowledgments
We are grateful for the technical advice provided by Kathy Walters and Chantal Allamargot from the Central Microscopy Facility at the University of Iowa. We would like to thank Jean Sippy for her assistance in mutant analysis and Sijun Liu for his advice in constructing the Ac-FPm virus. We also thank Aaron Irons for his assistance in evaluating the FP phenotype and genetic analysis of passaged viruses. Finally, we thank Sharon Braunagel and Max Summers for providing the WT AcMNPV and Δfp25K baculovirus used in this research. This research was supported by grants from the Environmental Protection Agency RD-83142101 (D. W. M./B. C. B.), National Science Foundation 0717620 (M. G. F.), National Institutes of Health GM-51611 (M. G. F.) and scholarship support for L. G. provided by the University of Iowa Center for Biocatalysis and Bioprocessing.
References
- Ayres, M. D., Howard, S. C., Kuzio, J., Lopez-Ferber, M. & Possee, R. D. (1994). The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202, 586–605. [DOI] [PubMed] [Google Scholar]
- Bauser, C. A., Elick, T. A. & Fraser, M. J. (1996). Characterization of hitchhiker, a transposon insertion frequently associated with baculovirus FP mutants derived upon passage in the TN-368 cell line. Virology 216, 235–237. [DOI] [PubMed] [Google Scholar]
- Beames, B. & Summers, M. D. (1988). Comparisons of host cell DNA insertions and altered transcription at the site of insertions in few polyhedra baculovirus mutants. Virology 162, 206–220. [DOI] [PubMed] [Google Scholar]
- Beames, B. & Summers, M. D. (1990). Sequence comparison of cellular and viral copies of host cell DNA insertions found in Autographa californica nuclear polyhedrosis virus. Virology 174, 354–363. [DOI] [PubMed] [Google Scholar]
- Beniya, H., Braunagel, S. C. & Summers, M. D. (1998). Autographa californica nuclear polyhedrosis virus: subcellular localization and protein trafficking of BV/ODV-E26 to intracellular membranes and viral envelopes. Virology 240, 64–75. [DOI] [PubMed] [Google Scholar]
- Bischoff, D. S. & Slavicek, J. M. (1997). Phenotypic and genetic analysis of Lymantria dispar nucleopolyhedrosis few polyhedra mutants: mutations in the 25K FP gene may be caused by DNA replication errors. J Virol 71, 1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerke, S. L. & Roller, R. J. (2006). Roles for herpes simplex virus type 1 UL34 and US3 proteins in disrupting the nuclear lamina during herpes simplex virus type 1 egress. Virology 347, 261–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, B. C., Brennan, L. A., Dierks, P. M. & Gard, I. E. (1997). Commercialization of baculoviral insecticides. In The Baculoviruses, pp. 341–388. Edited by Miller, L. K.. New York. : Plenum Press.
- Braunagel, S. C., Burks, J. K., Rosas-Acosta, G., Harrison, R. L., Ma, H. & Summers, M. D. (1999). Mutations within the Autographa californica nucleopolyhedrovirus fp25k gene decrease the accumulation of ODV-E26 and alter its intranuclear transport. J Virol 73, 8559–8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo-Patiño, A. & Ibarra, J. E. (2000). Site-directed mutagenesis of Autographa califonica nucleopolyhedrovirus (AcMNPV) polyhedrin: effect on polyhedron structure. Arch Virol 145, 827–834. [DOI] [PubMed] [Google Scholar]
- Carstens, E. B. (1987). Identification and nucleotide sequence of the regions of Autographa californica nuclear polyhedrosis virus genome carrying insertion elements derived from Spodoptera frugiperda. Virology 161, 8–17. [DOI] [PubMed] [Google Scholar]
- Cary, L. C., Goebel, M., Corsaro, B. G., Wang, H.-G., Rosen, E. & Fraser, M. J. (1989). Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology 172, 156–169. [DOI] [PubMed] [Google Scholar]
- Cusack, T. & McCarthy, W. J. (1989). Effect of serial passage on genetic homogeneity of a plaque variant of Lymantria dispar nuclear polyhedrosis virus (Hamden LDP-67). J Gen Virol 70, 2963–2972. [Google Scholar]
- de Rezende, S. H. M. S., Castro, M. E. B. & Souza, M. L. (2009). Accumulation of few-polyhedra mutants upon serial passage of Anticarsia gemmatalis multiple nucleopolyhedrovirus in cell culture. J Invertebr Pathol 100, 153–159. [DOI] [PubMed] [Google Scholar]
- Dimmock, N. J. & Marriott, A. C. (2006). In vivo antiviral activity: defective interfering virus protects better against virulent influenza virus than avirulent virus. J Gen Virol 87, 1259–1265. [DOI] [PubMed] [Google Scholar]
- DuPont (1996). Notification to conduct small-scale field testing of a genetically altered baculovirus. EPA No. 352-NMP-4.
- Fraser, M. J. & Hink, W. F. (1982). The isolation and characterization of the MP and FP plaque variants of Galleria mellonella nuclear polyhedrosis virus. Virology 117, 366–378. [DOI] [PubMed] [Google Scholar]
- Fraser, M. J., Smith, G. E. & Summers, M. D. (1983). Acquisition of host cell DNA sequences by baculoviruses: relationship between host DNA insertions and FP mutants of Autographa californica and Galleria mellonella nuclear polyhedrosis viruses. J Virol 47, 287–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, M. J., Brusca, J. S., Smith, G. E. & Summers, M. D. (1985). Transposon-mediated mutagenesis of baculoviruses. Virology 145, 356–361. [DOI] [PubMed] [Google Scholar]
- Friesen, P. D. & Nissen, M. S. (1990). Gene organization and transcription of TED, a lepidopteran retrotransposon integrated within the baculovirus genome. Mol Cell Biol 10, 3067–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard, I. E. (1997). Field testing a genetically modified baculovirus. In Microbial Insecticides: Novelty or Necessity? pp. 101–114. BCPC Symposium Proceedings No. 68.
- Harrison, R. L. & Bonning, B. C. (2001). Use of proteases to improve the insecticidal activity of baculoviruses. Biol Control 20, 199–209. [Google Scholar]
- Harrison, R. L. & Summers, M. D. (1995a). Biosynthesis and localization of the Autographa californica nuclear polyhedrosis virus 25K gene product. Virology 208, 279–288. [DOI] [PubMed] [Google Scholar]
- Harrison, R. L. & Summers, M. D. (1995b). Mutations in the Autographa californica multinucleocapsid nuclear polyhedrosis virus 25 kDa protein gene result in reduced virion occlusion, altered intranuclear envelopment and enhance virus production. J Gen Virol 76, 1451–1459. [DOI] [PubMed] [Google Scholar]
- Harrison, R. L., Jarvis, D. L. & Summers, M. D. (1996). The role of AcMNPV 25K gene, “FP25” in baculovirus polh and p10 expression. Virology 226, 34–46. [DOI] [PubMed] [Google Scholar]
- Hink, W. F. & Strauss, E. (1976). Replication and passage of alfalfa looper nuclear polyhedrosis virus plaque variants in cloned cell cultures and larval stages of four host species. J Invertebr Pathol 27, 49–55. [Google Scholar]
- Hughes, P. R. & Wood, H. A. (1981). A synchronous peroral technique for the bioassay of insect viruses. J Invertebr Pathol 37, 154–159. [Google Scholar]
- Jarvis, D. L., Bohlmeyer, D. A. & Gracia, A. (1992). Enhancement of polyhedrin nuclear localization during baculovirus infection. J Virol 66, 6903–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuma, S., Noguchi, Y., Shimada, T., Nagata, M., Kobayashi, M. & Maeda, S. (1999). Molecular characterization of baculovirus Bombyx mori nucleopolyhedrovirus polyhedron mutants. Arch Virol 144, 1275–1285. [DOI] [PubMed] [Google Scholar]
- Kelly, B. J., King, L. A. & Possee, R. D. (2007). Introduction to baculovirus molecular biology. In Baculovirus and Insect Cell Expression Protocols, pp. 25–53. Edited by Murhammer, D. W.. Totowa, New Jersey. : Humana Press.
- Kelly, B. J., Chapple, S. D. J., Allen, C., Pritchard, C., King, L. A. & Possee, R. D. (2008). Extended budded virus formation and induction of apoptosis by an AcMNPV FP-25K/p35 double mutant in Trichoplusia ni cells. Virus Res 133, 157–166. [DOI] [PubMed] [Google Scholar]
- Knudson, D. L. & Harrap, K. A. (1975). Replication of nuclear polyhedrosis virus in a continuous cell culture of Spodoptera frugiperda: microscopy study of the sequence of events of the virus infection. J Virol 17, 254–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kompier, R., Tramper, J. & Vlak, J. M. (1988). A continuous process for the production of baculovirus using insect-cell cultures. Biotechnol Lett 10, 849–854. [Google Scholar]
- Kool, M., Voncken, J. W., van Lier, F. L. J., Tramper, J. & Vlak, J. M. (1991). Detection and analysis of Autographa californica nuclear polyhedrosis virus mutants with defective interfering properties. Virology 183, 739–746. [DOI] [PubMed] [Google Scholar]
- Kumar, S. & Miller, L. K. (1987). Effects of serial passage of Autographa californica nuclear polyhedrosis virus in cell culture. Virus Res 7, 335–349. [DOI] [PubMed] [Google Scholar]
- Kumar, M. S., Ramachandran, A., Hasnain, S. E. & Bashyam, M. D. (2009). Octamer and heat shock elements regulate transcription from the AcMNPV polyhedrin gene promoter. Arch Virol 154, 445–456. [DOI] [PubMed] [Google Scholar]
- Lin, G., Zhong, J. & Wang, X. (2000). Abnormal formation of polyhedra from a single mutation in the polyhedrin gene of Autographa californica multicapsid nucleopolyhedrovirus. J Invertebr Pathol 76, 13–19. [DOI] [PubMed] [Google Scholar]
- Lua, L. H. L. & Reid, S. (2003). Effect of time of harvest of budded virus on the selection of baculovirus FP mutants in cell culture. Biotechnol Prog 19, 238–242. [DOI] [PubMed] [Google Scholar]
- Lua, L. H. L., Pedrini, M. R. S., Reid, S., Robertson, A. & Tribe, D. E. (2002). Phenotypic and genotypic analysis of Helicoverpa armigera nucleopolyhedrovirus serially passaged in cell culture. J Gen Virol 83, 945–955. [DOI] [PubMed] [Google Scholar]
- MacKinnon, E. A., Henderson, J. F., Stoltz, D. B. & Faulkner, P. (1974). Morphogenesis of nuclear polyhedrosis virus under conditions of prolonged passage in vitro. J Ultrastruct Res 49, 419–435. [DOI] [PubMed] [Google Scholar]
- Miller, D. W. & Miller, L. K. (1982). A virus mutant with an insertion of a copia-like transposable element. Nature 299, 562–564. [DOI] [PubMed] [Google Scholar]
- Moscardi, F. (1999). Assessment of the application of baculoviruses for control of Lepidoptera. Annu Rev Entomol 44, 257–289. [DOI] [PubMed] [Google Scholar]
- Murhammer, D. W. (1996). Use of viral insecticides for pest control and production in cell culture. Appl Biochem Biotechnol 59, 199–220. [Google Scholar]
- Nakanishi, T., Goto, C., Kobayashi, M., Kang, W., Suzuki, T., Dohmae, N., Matsumoto, S., Shimada, T. & Katsuma, S. (2010). Comparative studies of lepidopteran baculovirus-specific protein FP25K: development of a novel Bombyx mori nucleopolyhedrovirus-based vector with a modified fp25K gene. J Virol 84, 5191–5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa, H., Kendirgi, F., Belloncik, S., Ito, R., Takagi, S., Minobe, Y., Higo, K., Sumida, M., Matsubara, F. & Mori, H. (1996). Effect of mutations on the intracellular localization of Bombyx mori cytoplasmic polyhedrosis virus polyhedrin. J Gen Virol 77, 147–153. [DOI] [PubMed] [Google Scholar]
- O'Reilly, D. R., Passarelli, A. L., Goldman, I. F. & Miller, L. K. (1990). Characterization of the DA26 gene in a hypervariable region of the Autographa californica nuclear polyhedrosis virus genome. J Gen Virol 71, 1029–1037. [DOI] [PubMed] [Google Scholar]
- O'Reilly, D. R., Miller, L. K. & Luckow, V. A. (1992). Virus methods. In Baculovirus Expression Vectors: a Laboratory Manual, pp. 132–134. New York. : W. H. Freeman.
- Potter, K. N., Faulkner, P. & MacKinnon, E. A. (1976). Strain selection during serial passage of Trichoplusia ni nuclear polyhedrosis virus. J Virol 18, 1040–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullen, S. S. & Friesen, P. D. (1995). Early transcription of the ie-1 transregulator gene of Autographa californica nuclear polyhedrosis virus is regulated by DNA sequences within its 5′ noncoding leader region. J Virol 69, 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes, D. J. (1996). Economics of baculovirus–insect cell production systems. Cytotechnology 20, 291–297. [DOI] [PubMed] [Google Scholar]
- Robertson, J. L. & Preisler, H. K. (1992). Pesticide Bioassays with Arthropods. London. : CRC Press.
- Rosas-Acosta, G., Braunagel, S. C. & Summers, M. D. (2001). Effects of deletion and overexpression of the Autographa californica nuclear polyhedrosis virus fp25k gene on synthesis of two occlusion-derived virus envelope proteins and their transport into virus-induced intranuclear membranes. J Virol 75, 10829–10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989). Detection and analysis of proteins expressed from cloned genes. In Molecular Cloning: A Laboratory Manual, p. 18. Edited by Nolan, C.. New York. : Cold Spring Harbor Laboratory Press.
- Shoemaker, D. P., Garland, C. W. & Steinfeld, J. E. (1974). Experiments in Physical Chemistry, 3rd edn. New York. : McGraw-Hill.
- Smith, C. R., Heinz, K. M., Sansone, C. G. & Flexner, J. L. (2000). Impact of recombinant baculovirus applications on target heliothines and nontarget predators in cotton. Biol Control 19, 201–214. [Google Scholar]
- Szewczyk, B., Rabalski, L., Krol, E., Sihler, W. & Lobo de Souza, M. (2009). Baculovirus biopesticides – a safe alternative to chemical protection of plants. J Biopesticides 2, 209–216. [Google Scholar]
- Tramper, J. & Vlak, J. M. (1986). Some engineering and economic aspects of continuous cultivation of insect cells for the production of baculoviruses. Ann N Y Acad Sci 469, 279–288. [Google Scholar]
- van Lier, F. L. J., van den End, E. J., de Gooijer, C. D., Vlak, J. M. & Tramper, J. (1990). Continuous production of baculovirus in a cascade of insect-cell reactors. Appl Microbiol Biotechnol 33, 43–47. [DOI] [PubMed] [Google Scholar]
- Vaughn, J. L., Goodwin, R. H., Tompkins, G. J. & McCawley, P. (1977). The establishment of two cell lines from the insect Spodoptera frugiperda (Lepidoptera: Noctuidae). In Vitro 13, 213–217. [DOI] [PubMed] [Google Scholar]
- Wang, H. H., Fraser, M. J. & Cary, L. C. (1989). Transposon mutagenesis of baculoviruses: analysis of TFP3 lepidopteran transposon insertions at the FP locus of nuclear polyhedrosis viruses. Gene 81, 97–108. [DOI] [PubMed] [Google Scholar]
- Wu, D., Fei, D., Sun, X., Wang, H., Yuan, L., Vlak, J. M. & Hu, Z. (2005). Functional analysis of FP25K of Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus. J Gen Virol 86, 2439–2444. [DOI] [PubMed] [Google Scholar]
- Yamagishi, J., Isobe, R., Takebuchi, T. & Bando, H. (2003). DNA microarrays of baculovirus genomes: differential expression of viral genes in two susceptible insect cell lines. Arch Virol 148, 587–597. [DOI] [PubMed] [Google Scholar]