Abstract
Nuclear matrix attachment regions (MARs) are regulatory DNA sequences that are important for higher-order chromatin organization, long-range enhancer function, and extension of chromatin modifications. Here we characterize a novel cell type-specific MAR-binding protein, SATB2, which binds to the MARs of the endogenous immunoglobulin μ locus in pre-B cells and enhances gene expression. We found that SATB2 differs from the closely related thymocyte-specific protein SATB1 by modifications of two lysines with the small ubiquitive related modifier (SUMO), which are augmented specifically by the SUMO E3 ligase PIAS1. Mutations of the SUMO conjugation sites of SATB2 enhance its activation potential and association with endogenous MARs in vivo, whereas N-terminal fusions with SUMO1 or SUMO3 decrease SATB2-mediated gene activation. Sumoylation is also involved in targeting SATB2 to the nuclear periphery, raising the possibility that this reversible modification of a MAR-binding protein may contribute to the modulation of subnuclear DNA localization.
Keywords: SUMO, MAR, SATB2, PIAS1, nuclear matrix
Regulation of gene expression is governed in large part by transcription factors that bind to enhancers and promoters. The functions of transcription factors involve both the regulation of chromatin accessibility via the recruitment of histone-modifying enzymes or nucleosome-remodeling complexes, and the stimulation of RNA polymerase via an interaction with the mediator complex (for review, see Zhang and Reinberg 2001; Freiman and Tjian 2003). In addition to enhancers and promoters, nuclear matrix attachment regions (MARs) have been implicated in the regulation of gene expression by altering the organization of eukaryotic chromosomes and augmenting the potential of enhancers to act over large distances (for review, see Scheuermann and Garrard 1999; Bode et al. 2000). MARs have been identified as AT-rich sequences that are associated with the nuclear matrix, a nuclear component that resists salt extraction and digestion with DNase I (Berezney and Coffey 1974; Mirkovitch et al. 1984; Cockerill and Garrard 1986). The nuclear matrix, also referred to as the interchromatin space, contains multiple subnuclear structures that can be visualized microscopically as speckles and that have been proposed to functionally compartmentalize the nucleus (for review, see Schul et al. 1998; Cremer and Cremer 2001; Spector 2001). The association of MARs with the nuclear matrix may serve to structurally define the borders of chromatin domains and participate in the regulation of transcription. In particular, the MARs that flank the enhancers of immunoglobulin heavy- and light-chain genes augment enhancer function in transgenic mice (Xu et al. 1989; Forrester et al. 1994; Lichtenstein et al. 1994). The MARs of the μ and κ loci have also been found to antagonize the effects of DNA methylation, and they were shown to extend an enhancer-induced domain of chromatin accessibility and histone acetylation (Jenuwein et al. 1993; Lichtenstein et al. 1994; Jenuwein et al. 1997; Forrester et al. 1999; Fernandez et al. 2001).
Several proteins have been identified that bind to MAR sequences. In addition to ubiquitous MAR-binding proteins such as SAF-A, Cux/CDP, and MeCP2 (Scheuermann and Chen 1989; Romig et al. 1992; Weitzel et al. 1997), two MAR-binding proteins, SATB1 and Bright, are expressed specifically in T cells and activated B cells, respectively (Dickinson et al. 1992; Herrscher et al. 1995). Bright has been shown to augment immunoglobulin μ gene expression in transfection assays, and it localizes to nuclear matrix-associated promyelocytic leukemia (PML) nuclear bodies (Herrscher et al. 1995; Zong et al. 2000). In contrast, SATB1 has been found to act as a repressor of transcription (Kohwi-Shigematsu et al. 1997; Alvarez et al. 2000). This function of SATB1 correlates with its ability to recruit the Sin3a histone deacetylase and subunits of the ACF nucleosome-mobilizing complex to genes, such as the interleukin receptor 2α (IL2Rα) locus, in a MAR-dependent manner (Yasui et al. 2002). In addition, SATB1 may function as a positive regulator of transcription, because the upregulation of the c-myc gene in stimulated thymocytes is impaired in SATB1-deficient mice (Cai et al. 2003). SATB1 has been proposed to represent a novel type of gene regulator that acts as a three-dimensional protein scaffold upon which MAR sequences are tethered (Cai et al. 2003). However, the regulation of the activities of MAR-binding proteins is still obscure.
Multiple posttranslational mechanisms have been shown to regulate the activity of transcription factors. In addition to phosphorylation, acetylation, and ubiquitination, modification by the covalent conjugation of the small ubiquitin-related modifier SUMO has been found to modulate the activity of transcription factors. SUMO-1 and SUMO2/3 are distantly related to ubiquitin and are also covalently conjugated as 92-97 amino acid polypeptides to internal lysines of substrate proteins (for review, see Melchior 2000; Muller et al. 2001). The enzymatic reactions involved in SUMO modification are similar to those in ubiquitin modification and involve an E1-activating enzyme, consisting of an Aos1/Uba2 heterodimer, the E2-conjugating enzyme Ubc9, and an E3 ligase that promotes the transfer of SUMO from the E2 enzyme to substrate proteins (for review, see Hochstrasser 2001; Jackson 2001). The protein inhibitors of activated STATs (PIAS), the polycomb protein Pc2, and the nuclear pore component RanBP2 have been identified as E3 ligases (Johnson and Gupta 2001; Kahyo et al. 2001; Sachdev et al. 2001; Pichler et al. 2002; Kagey et al. 2003). Mammalian cells contain four members of the PIAS family (PIAS1, 3, x, and y) that share a similar domain structure (Chung et al. 1997; Liu et al. 1998). PIAS proteins have an amino-terminal SAP-domain that mediates the association with the nuclear matrix (Sachdev et al. 2001) and an SP-RING-domain that is important for the sumoylation of substrate proteins (Hochstrasser 2001; Johnson and Gupta 2001; Sachdev et al. 2001).
The effects of sumoylation of proteins are dynamic, as the modification can be reversed by the action of SUMO-specific isopeptitases (Li and Hochstrasser 1999; Best et al. 2002). In addition, the effects of sumoylation are quite diverse. Modifications of proteins with SUMO can alter the association with partner proteins and DNA (Goodson et al. 2001; Hong et al. 2001; Lin et al. 2003), change the subcellular or subnuclear localization (Muller et al. 1998; Sachdev et al. 2001; Kirsh et al. 2002; Ross et al. 2002), or influence protein stability by competing with ubiquitination (Desterro et al. 1998). SUMO modification was found to augment the transcriptional activation potential of some proteins, such as p53 (Gostissa et al. 1999; Rodriguez et al. 1999); however, most often sumoylation and/or association with SUMO E3 ligases antagonizes the activation potential of transcription factors or mediates repression (Ross et al. 2002; Tian et al. 2002; for review, see Gill 2003; Verger et al. 2003). In several cases, SUMO-mediated repression correlates with a targeting of the proteins to PML nuclear bodies (Sachdev et al. 2001; Ross et al. 2002; Schmidt and Muller 2002).
In this study, we identified and characterized a novel cell type-specific MAR-binding protein, SATB2. We found that SATB2 can bind to MAR sequences of the endogenous immunoglobulin μ enhancer region and activate gene transcription. We also show that the transcriptional activation potential and MAR-binding ability of SATB2 can be increased by mutations of two lysines that are modified with SUMO in a PIAS1-dependent manner. In contrast to SUMO-deficient SATB2, which shows a diffuse pattern of nuclear localization, wild-type SATB2 and amino-terminal SUMO-fusions of SATB2 are targeted to the nuclear periphery.
Results
SATB2 is a pre-B cell-enriched MAR-binding protein
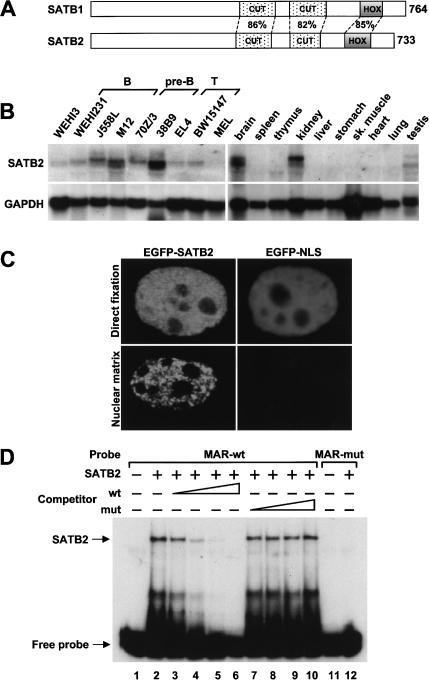
Previous experiments indicated that the nuclear MARs that flank the intragenic μ enhancer on either side markedly augment the expression of a rearranged μ gene in pre-B cells of transgenic mice (Forrester et al. 1994). To identify MAR-binding proteins that are expressed predominantly in early stages of the B-cell lineage, we searched the murine and human databases for sequences that show homologies to known MAR-binding proteins, and we examined their expression in pre-B cells and other cell types. We identified two clones that displayed 67% and 70% overall sequence homology with SATB1 and Bright, respectively. Analysis of the expression patterns of these clones revealed a ubiquitous expression for the Bright homolog and a predominantly pre-B cell-specific pattern for the SATB1 homolog, which we termed SATB2 (Fig. 1; data not shown). Sequence analysis of SATB2 revealed the presence of two CUT domains and a Hox domain, which are highly conserved with the corresponding MAR-binding domains of SATB1 (Fig. 1A; Dickinson et al. 1997; Galande et al. 2001). RNA blot analysis of total RNA from different lymphoid and nonlymphoid cell lines indicated that satb2 transcripts with sizes of ∼6.3 and ∼5.4 kb can be detected most abundantly in cell lines representing pre-B cells and at lower levels in cells representing B and T cells (Fig. 1B, left-hand panels). We also detected satb2 transcripts in primary pre-B cells at levels five- to eightfold lower than the level found in the 38B9 pre-B cell line (data not shown). In a panel of mouse tissues, SATB2 transcripts could be detected predominantly in brain and kidney, although differently sized transcripts were detected at a lower abundance in thymus and testis (Fig. 1B, right-hand panels).
Figure 1.
Cell type-specific expression and nuclear matrix association of SATB2. (A) Schematic representation of the domain structure of SATB1 and SATB2. The amino acid sequence identities in the MAR-binding domain (CUT domains) and the homeodomain (HOX) are indicated in percentages. (B) RNA blot analysis of 20 μg total RNA isolated from various B- and T-lymphoid cell lines, a murine erythroid (MEL) and myeloid (WEHI3) cell line (left-hand panels), and from various mouse tissues (right-hand panels). Abundant expression of ∼6.3- and ∼5.4-kb transcripts that hybridize with a 32P-labeled satb2 DNA probe can be detected in B cells, brain, and kidney (top panels). The quality and quantity of RNA were confirmed by hybridization with a GAPDH probe (bottom panels). (C) Association of SATB2 with the nuclear matrix. 293T cells were transfected with an EGFP-SATB2 or EGFP-NLS gene construct, and after 48 h the fusion proteins were visualized by fluorescence microscopy in directly fixed cells (top panels) or in nuclear matrix preparations (bottom panels). (D) EMSA to detect binding of purified recombinant SATB2 (30 ng) to 32P-labeled wild-type (wt) or mutated (mut) MAR oligonucleotides (lanes 2,12). The specificity of DNA binding by SATB2 was shown by competition with increasing amounts of unlabeled wild-type or mutant MAR oligonucleotides (lanes 3-10).
Previous studies have shown that SATB1 is a component of the nuclear matrix and binds to special AT-rich nucleotide sequences (Dickinson et al. 1992; de Belle et al. 1998). To examine whether SATB2 can associate with the nuclear matrix, we transfected 293T cells with plasmids encoding either an EGFP-SATB2 fusion protein or an EGFP-NLS control protein. We prepared nuclear matrices by DNase I treatment and high salt extraction, and in parallel we fixed the transfected cells directly. Visualization by fluorescence microscopy indicated that EGFP-SATB2, but not EGFP-NLS, was retained in the nuclear matrix preparation (Fig. 1C). We also examined the DNA-binding ability of SATB2 by electrophoretic mobility shift assays (EMSAs) with purified recombinant SATB2 protein. Efficient binding was detected with the wild-type MAR consensus sequence (Dickinson et al. 1992), but not with a mutant oligonucleotide (Fig. 1D). We confirmed the specificity of DNA binding by the addition of excess unlabeled competitor DNA, which impaired DNA binding in a dose-dependent manner. Thus, SATB2 resembles SATB1 in its ability to bind MAR sequences and associate with the nuclear matrix.
SATB2 can augment μ gene transcription and binds to the μ MAR in vivo
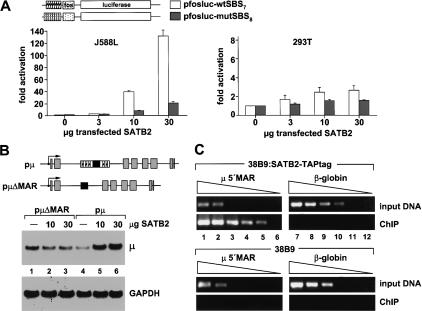
Previous analyses of the functional activity of SATB1 in transfection assays and in knock-out experiments indicated that SATB1 represses transcription of several genes in T cells (Kohwi-Shigematsu et al. 1997; Alvarez et al. 2000). To examine the functional activity of SATB2, we transfected a fos-luciferase reporter construct containing multimerized wild-type or mutated SATB2-binding sites, together with a β-galactosidase control plasmid and increasing amounts of a SATB2 expression plasmid into J558L plasmacytoma cells. SATB2 augmented reporter gene expression up to 150-fold, suggesting that SATB2 can act as an activator of transcription (Fig. 2A, left-hand panel). Significantly reduced stimulation was observed with the reporter construct containing the mutated SATB2-binding sites. Notably, the stimulation of the fos-luciferase reporter with the multimerized wild-type SATB2-binding sites was not observed in transfected 293T cells, suggesting that the transcriptional activation by SATB2 is cell type-specific (Fig. 2A, right-hand panel). Immunoblot analysis to detect epitope-tagged SATB2 indicated that the protein is expressed at higher levels in 293T cells compared to J558L cells (data not shown). Finally, we examined the effects of overexpression of SATB2 on the expression of a cotransfected rearranged wild-type and μΔMAR gene. RNA blot analysis of transfected J558L cells indicated that SATB2 augments the expression of the wild-type μ gene by a factor of approximately five, whereas no significant effect was observed with the μΔMAR gene (Fig. 2B).
Figure 2.
SATB2 augments μ gene expression and binds to endogenous MAR sequences of the μ locus. (A) SATB2 stimulates a luciferase reporter gene containing multimerized SATB2-binding sites in J558L plasmacytoma cells but not in 293T cells. Cells were transfected with 5 μg reporter construct, together with 1 μg β-galactosidase reporter construct for normalization and increasing amounts of SATB2 expression plasmid, as indicated. The levels of luciferase activity, normalized to the activity of the cotransfected β-galactosidase reporter, are expressed as fold-activation relative to the luciferase levels of cells transfected with the reporters alone. All transfections were performed at least three times, and representative experiments with the standard deviations are shown. (B) SATB2 augments the expression of a rearranged μ immunoglobulin gene in a MAR-dependent manner. J558L cells were transfected with 5 μg of wild-type or μΔMAR genes and increasing amounts of SATB2 expression plasmid (10, 30 μg), as indicated. Expression of the μ gene was examined by RNA blot analysis of total RNA (20 μg) with a 32P-labeled VH17.2.25-specific DNA probe (top panel) or GAPDH probe (bottom panel). (C) SATB2 binds to the MAR of the endogenous μ locus. ChIP of extracts from stably transfected 38B9 cells carrying a SATB2-TAPtag expression plasmid, or from control 38B9 cells. Chromatin fragments that had been cross-linked to SATB2-TAPtag were affinity-purified, and the immunoprecipitated DNA was subsequently analyzed by semiquantitative PCR amplification with primers located in the 5′ MAR region of the immunoglobulin μ enhancer (lanes 1-6) or in the β-globin locus (lanes 7-12). The levels of enrichment in the immunoprecipitations were estimated by comparison with the amplification products of DNA isolated from the bulk chromatin extracts (input DNA). Template DNA was used in a linear dilution (3, 1, 0.3, 0.1, 0.03, 0.01 ng) to allow for a semiquantitative determination in the PCR assays.
To examine whether SATB2 binds to the MARs of the endogenous μ gene, we performed chromatin immunoprecipitation (ChIP) experiments. Because of the lack of antibodies that immunoprecipitate SATB2, we generated a pre-B cell line that had been stably transfected with a SATB2-TAPtag gene construct. After a two-step affinity purification of the SATB2-TAPtag protein that had been cross-linked to DNA in vivo, the immunoprecipitated DNA was amplified in serial dilutions by polymerase chain reactions (PCRs) with primers specific for the 5′ MAR region of the intronic μ enhancer (Fig. 2C, lanes 1-6), or with primers specific for β-globin gene sequences as a control (Fig. 2C, lanes 7-12). Significant enrichment (∼100-fold) of μ 5′MAR sequences was detected with the immunoprecipitated DNA from the SATB2-TAPtag-expressing cell line (Fig. 2C, lanes 1-6, top panels), but not with the immunoprecipitated DNA from the parental pre-B cell line (Fig. 2C, lanes 1-6, bottom panels). No enrichment of β-globin sequences was detected (Fig. 2C, lanes 7-12, top panels), indicating that SATB2 is specifically bound to the MAR flanking the intragenic enhancer of the endogenous μ gene.
SATB2 is modified by SUMO conjugation
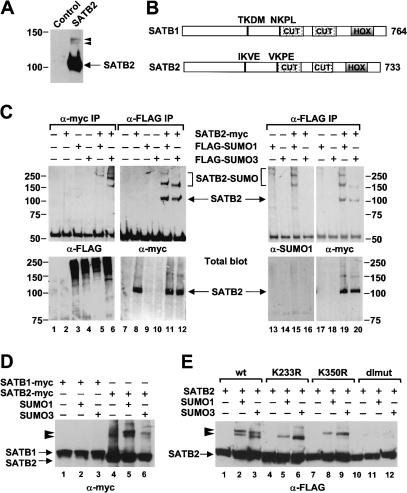
In an immunoblot analysis of lysates from cells transfected with a myc-tagged SATB2 expression plasmid, we detected SATB2 migrating at ∼105 kD and two minor bands migrating at ∼135-140 kD (Fig. 3A). To examine whether the two slower-migrating proteins represent covalent conjugations of SATB2 with the 12-kD SUMO, we transfected the myc-tagged SATB2 expression plasmid, together with a Flag-SUMO1 or Flag-SUMO3 expression plasmid, into 293T cells. Co-immunoprecipitation of proteins with an anti-myc antibody and subsequent immunoblot analysis with an anti-Flag antibody allowed for the detection of the slower-migrating forms of SATB2 (Fig. 3C, lanes 5,6). Likewise, we detected these slower-migrating forms of SATB2 in a reciprocal co-immunoprecipitation with an anti-Flag antibody and an anti-myc immunoblot analysis (Fig. 3C, lanes 11,12). In this experiment, we also detected unmodified SATB2, which could reflect a dimerization of SUMO-modified and unmodified SATB2. Previous experiments showed that SATB1 can dimerize through a PDZ domain that is located upstream of the CUT domains (Galande et al. 2001). To confirm that the slower-migrating forms of SATB2 correspond to a covalent modification with SUMO, we performed a co-immunoprecipitation with the anti-Flag antibody and probed the immunoblot with an anti-SUMO1 antibody (Fig. 3C, lanes 13-16). After stripping, the blot was probed with an anti-myc antibody (Fig. 3C, lanes 17-20). The slower-migrating forms of SATB2, but not the 105-kD form of SATB2, could be detected with the anti-SUMO1 antiserum. Taken together, these data suggest that SATB2 can be modified by conjugation with SUMO1 or SUMO3.
Figure 3.
SATB2 is SUMO-modified in vivo. (A) Immunoblot analysis of total protein extracts from 293T cells transfected transiently with a myc-tagged SATB2 expression plasmid or a control vector. In addition to SATB2, migrating at ∼105 kD, two forms of SATB2 migrating at ∼135 kD and ∼140 kD can be detected with an anti-myc antibody. (B) Schematic representation of the domain structure of SATB1 and SATB2. The two CUT motifs, the homeobox (HOX), and the amino acid sequence of two putative SUMO acceptor sites at positions 233 (IKVE) and 350 (VKPE) of SATB2 are shown. The corresponding sequences in SATB1 do not conform to the consensus SUMO acceptor site ψKXE. (C) Modification of SATB2 by SUMO1 and SUMO3. Myc-tagged SATB2 and Flag-tagged SUMO1 or SUMO3 expression plasmids were transiently transfected into 293T cells. Equivalent amounts (500 μg) of total cellular protein were immunoprecipitated with an anti-myc antibody (lanes 1-6, top panel) or an anti-Flag antibody (lanes 7-20, top panels), and the immunoprecipitated proteins that have been modified with Flag-SUMO proteins were detected with an anti-Flag antibody (lanes 1-6) or an anti-SUMO1 antibody (lanes 13-16). SATB2 and SUMO-modified forms of SATB2 were detected with an anti-myc antibody (lanes 7-12 and 17-20, top panels). Similar expression of SATB2-myc, Flag-SUMO1, and Flag-SUMO3 was confirmed by immunoblot analysis of total cell lysates (bottom panels). (D) SATB2, but not SATB1, can be modified with SUMO proteins. Myc-tagged SATB1 (lanes 1-3) and myc-tagged SATB2 (lanes 4-6) are detected in total cell lysates of transfected cells by an anti-myc immunoblot analysis. SUMO-modified forms of SATB2 are marked by arrowheads. (E) Identification of K233 and K350 of SATB2 as the acceptor sites for sumoylation. Total protein extracts from cells transfected with expression plasmids of Flag-SATB2 (lanes 1-3) or various Flag-tagged SATB2 mutants: K233R (lanes 4-6), K350R (lanes 7-9), or the K233R/K350R double mutant (dlmut, lanes 10-12), alone or together with SUMO1 or SUMO3 expression plasmids, were analyzed by an immunoblot with an anti-Flag antibody. Mutation of either sumoylation site interfered with the appearance of one of the two modified forms of SATB2, whereas the double mutation abrogated SUMO modification. The differences in the migration of the two modified forms of SATB2 are most likely due to the branching position of the sumoylated polypeptides (Hoege et al. 2002).
SUMO acceptor sites have a minimal consensus sequence, ψKXE, in which ψ is a large hydrophobic residue and K is the lysine to which SUMO is added (Rodriguez et al. 2001). Inspection of the amino acid sequence of SATB2 revealed two consensus SUMO acceptor sites at lysines 233 and 350 (Fig. 3B). Interestingly, the corresponding sequences in SATB1 do not conform to the consensus sumoylation sites. Consistent with these sequence differences, we did not detect modifications of myc-tagged SATB1 with either SUMO1 or SUMO3, whereas SATB2 was conjugated with both SUMO proteins (Fig. 3D). To determine whether the putative SUMO conjugation sites in SATB2 are modified in vivo, we mutated the lysines at positions 233 and 350, individually (K233R and K350R) or together (K233R/K350R; dlmut). We examined the effects of these mutations on the sumoylation of SATB2 by cotransfecting wild-type or mutant Flag-SATB2 expression plasmids, together with SUMO1 or SUMO3 plasmids into 293T cells. Immunoblot analysis of total cell lysates indicated that the slower-migrating ∼140-kD band corresponds to a sumoylation at position K233, whereas the faster-migrating band at ∼135 kD corresponds to a SUMO-modification at position K350 (Fig. 3E, lanes 5,6 and 8,9). Consistent with the modification of SATB2 at both lysines, the ∼135-kD and ∼140-kD forms could not be detected in cells expressing the double mutant form of SATB2 (Fig. 3E, lanes 11,12). Thus, the lysines at positions 233 and 350 are required for the SUMO-modification of SATB2.
PIAS1 associates specifically with SATB2 and enhances sumoylation
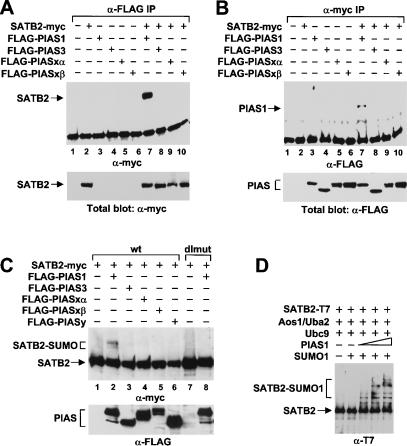
PIAS proteins have been shown to act as SUMO E3 ligases that augment the sumoylation of proteins, and they have been proposed to confer the substrate specificity upon the E2-conjugating enzyme (Johnson and Gupta 2001; Kahyo et al. 2001; Sachdev et al. 2001). To determine whether SATB2 can interact specifically with one of the members of the PIAS family of SUMO E3 ligases, we transfected Flag-tagged gene constructs of PIAS1, PIAS3, PIASxα, and PIASxβ into 293T cells, alone or together with a myc-tagged SATB2 expression plasmid. Co-immunoprecipitation with an anti-myc antibody and subsequent immunoblot analysis with an anti-Flag antibody indicated that PIAS1 was efficiently co-immunoprecipitated (Fig. 4A, lane 7). A weak association was also detected with PIASy but not with the other PIAS proteins (Fig. 4A, lanes 8-10; data not shown). The association between SATB2 and PIAS1 was also detected by the reciprocal co-immunoprecipitation with an anti-myc antibody and an immunoblot analysis with an anti-Flag antibody (Fig. 4B, lane 7). Similar levels of expression of the PIAS proteins and SATB2 were confirmed by a parallel immunoblot analysis of total cell lysates (Fig. 4A,B, lower panels). Thus, SATB2 and PIAS1 can specifically interact in cells expressing both proteins.
Figure 4.
PIAS1 interacts with SATB2 and stimulates SUMO modification of SATB2 in vivo and in vitro. (A,B) PIAS1 interacts with SATB2 in vivo. Myc-tagged SATB2 and Flag-tagged forms of PIAS1, PIAS3, PIASxα, and PIASxβ were transiently expressed in 293T cells. Equivalent amounts of total cellular protein were immunoprecipitated with an anti-Flag antibody (A, lanes 1-10) or an anti-myc antibody (B, lanes 1-10). The co-immunoprecipitated proteins were detected by an immunoblot analysis with an anti-myc antibody (A, lanes 1-10, top panel) or anti-Flag antibody (B, lanes 1-10, top panel). Similar expression of SATB2 and the PIAS proteins was confirmed by immunoblot analysis of total cell lysates (bottom panels). (C) PIAS1 stimulates SUMO conjugation to SATB2 in vivo. Myc-tagged SATB2 or SATB2-dlmut were expressed alone or together with Flag-tagged PIAS proteins in 293T cells. SATB2 and SUMO-modified SATB2 were detected in total cell lysates by immunoblot analysis with an anti-myc antibody. SUMO-modified SATB2 can be detected only in cells expressing PIAS1 (lane 2). (D) PIAS1 stimulates SUMO conjugation to SATB2 in vitro: 300 ng purified His- and T7-double-tagged SATB2 protein was incubated for 30 min with 250 ng purified E1 enzyme (Aos1/Uba2 heterodimer), 250 ng E2 enzyme (Ubc9), and 1 μg SUMO1, alone or with increasing amounts of GST-PIAS1. SATB2 proteins were detected by an anti-T7 immunoblot analysis.
To examine whether PIAS1 can act as a specific SUMO E3 ligase for SATB2, we cotransfected the expression plasmids for wild-type SATB2 or the SATB2-K233R/K350R double mutant (dlmut) together with an expression plasmid for any of the four PIAS family members. SUMO-modified forms of SATB2 were detected by an anti-myc immunoblot analysis specifically in extracts from cells expressing PIAS1 (Fig. 4C, lane 2). As expected, the modified forms of SATB2 were not detected in extracts from cells expressing the SATB2 double mutant (lane 8). Similar levels of expression of the Flag-tagged PIAS proteins were confirmed by a parallel immunoblot analysis with an anti-Flag antibody (Fig. 4C, lower panel). We also examined whether PIAS1 can act as a SUMO E3 ligase for SATB2 in an in vitro reconstituted system, containing purified E1-activating enzyme Aos1/Uba2, E2-conjugating enzyme Ubc9, SUMO1, and ATP. In this experiment, we used recombinant SATB2 protein that had been purified via a His-tag and could be detected by an immunoblot analysis via a T7-tag. Addition of purified GST-PIAS1 augmented the sumoylation of SATB2 in a dose-dependent manner (Fig. 4D). Taken together, these data indicate that PIAS1 acts as a specific E3 ligase for SUMO modification of SATB2 in vivo and in vitro.
Mutations of the sumoylation sites of SATB2 augment its activation potential and binding to MAR sequences in vivo
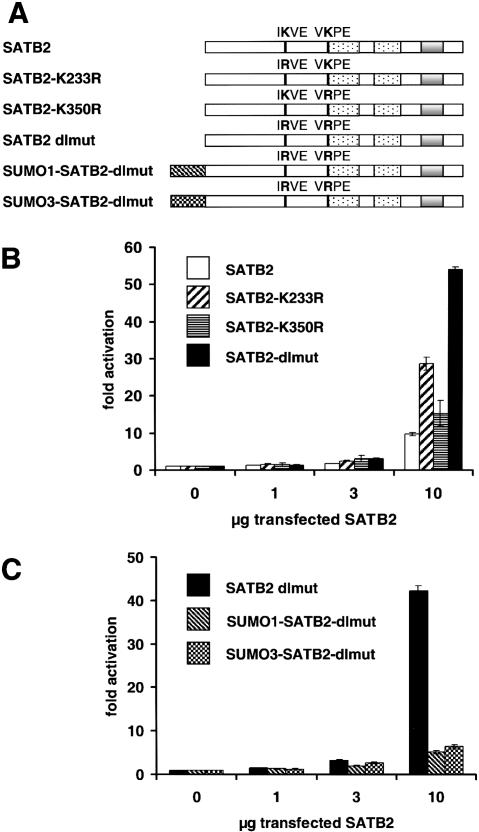
SUMO modification has been found to antagonize the activation potential of several transcription factors (Sachdev et al. 2001; Ross et al. 2002; Sapetschnig et al. 2002; Schmidt and Muller 2002). To examine the effects of sumoylation on the transcriptional activation by SATB2, we transfected J558L cells with expression plasmids encoding wild-type or mutant forms of Flag-tagged SATB2, together with a fos-luciferase reporter construct containing multimerized SATB2-binding sites (Fig. 5A). Wild-type SATB2 augmented reporter gene expression by a factor of up to 10, whereas mutations of both sumoylation sites of SATB2 (dlmut) further increased the level of reporter gene expression by a factor of ∼five (Fig. 5B). Mutation of the sumoylation site at position 233 resulted in a threefold increase of reporter gene expression relative to the stimulation of expression by wild-type SATB2. To examine whether the effects of the mutations of the lysines at positions 233 and 350 are due to a lack of SUMO modification, rather than due to other modifications such as acetylation or methylation, we generated gene constructs in which SUMO1 or SUMO3 is fused to the amino terminus of SATB2-dlmut. Both SUMO1-SATB2-dlmut and SUMO3-SATB2-dlmut were found to activate reporter gene expression at levels five- to sixfold lower than those observed with SATB2-dlmut (Fig. 5C). Immunoblot analysis with an anti-Flag antibody indicated that the various SATB2 proteins are expressed at similar levels (data not shown). Taken together, these data indicate that the sumoylation of SATB2 antagonizes its transcriptional activation potential.
Figure 5.
Sumoylation antagonizes SATB2-mediated transcriptional activation. (A) Schematic representation of wild-type SATB2 and mutant SATB2 proteins containing mutations of the sumoylation sites, individually (K233R, K350R) or in combination (dlmut), and carrying an amino-terminal fusion of SUMO1 (SUMO1-SATB2-dlmut) or SUMO3 (SUMO3-SATB2-dlmut). The amino-terminal fusions of SUMO cannot be hydrolyzed by SUMO isopeptitases. (B) Mutation of both sumoylation sites of SATB2 augments transcriptional activation. J558L cells were transiently transfected by electroporation with 5 μg of a luciferase reporter construct containing multimerized SATB2-binding sites, alone or together with increasing amounts (1, 3, and 10 μg) of expression plasmids encoding wild-type SATB2 or mutated SATB2 proteins. For the normalization of luciferase activities, the activity of a cotransfected β-galactosidase expression plasmid (1 μg) was determined for each sample. The normalized levels of luciferase activity are expressed as fold-activation relative to the level of luciferase activity from cells transfected with the reporter construct alone. (C) Amino-terminal fusions of SUMO1 or SUMO3 antagonize SATB2-mediated transcriptional activation. J558L cells were transiently transfected by electroporation with a luciferase reporter construct, alone or together with increasing amounts of expression plasmids encoding SATB2-dlmut, SUMO1-SATB2-dlmut, or SUMO3-SATB2-dlmut as indicated.
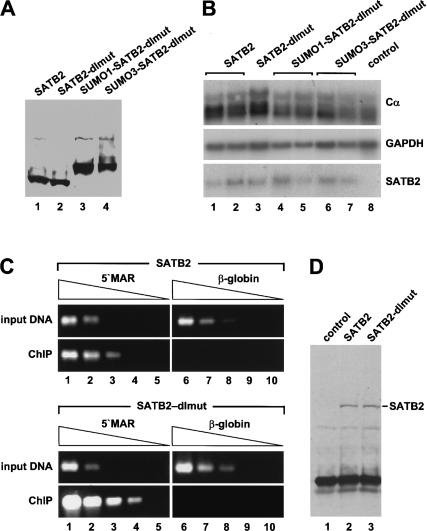
To examine the activation potential of wild-type SATB2 and the mutant SATB2 proteins in a more physiological context, we generated J558L plasmacytoma cell lines that have been stably transfected with Flag-tagged SATB2 constructs, and we then examined the expression of the endogenous immunoglobulin α heavy chain gene (Cα). For this experiment, we chose clones that express the exogenous SATB2 proteins at similar, moderate levels (Fig. 6A). RNA blot analysis indicated that SATB2 and SATB2-dlmut augmented Cα expression, relative to the expression in the parental J558L cells, by a factor of two and three, respectively (Fig. 6B, lanes 1-3,8). In contrast, no activation of Cα expression was observed with the covalent SUMO-SATB2-dlmut fusion proteins (Fig. 6B, lanes 4-7). The lower activation potential of the wild-type SATB2 protein relative to the SUMO-deficient SATB2 double mutant in this and the previous experiment could be accounted for by a SUMO-dependent decrease in the association of SATB2 with chromatin. Toward this end, we examined the binding of wild-type SATB2 and SATB2-dlmut to the μ immunoglobulin MAR sequences in vivo by ChIP experiments with J558L plasmacytoma cells that had been stably transfected with Flag-tagged wild-type and dlmut SATB2 constructs. The results of the ChIP analysis indicated that the SATB2 double mutant associated with the 5′ MAR of the intragenic μ enhancer region at least five times more efficiently than the wild-type SATB2 protein (Fig. 6C). To rule out that the difference in the ChIP efficiency is a result of an altered fractionation of the SATB2 and SATB2-dlmut proteins, we performed an immunoblot analysis of the immunoprecipitated proteins and detected similar amounts of these proteins (Fig. 6D). We also examined the DNA-binding ability of purified recombinant wild-type SATB2, SATB2-dlmut, and SUMO1-SATB2-dlmut proteins in an EMSA and found no significant differences in DNA binding (data not shown). Together, these data suggest that modifications of the sumoylation sites account for the observed differences in DNA binding by wild-type and double mutant SATB2 in vivo.
Figure 6.
Mutations of the sumoylation sites of SATB2 augment the association with MAR sequences of the endogenous immunoglobulin heavy-chain locus. (A) Immunoblot analysis of clones of J558L plasmacytoma cells that have been stably transfected with Flag-tagged wild-type SATB2 and various mutant SATB2 expression plasmids under the control of the EF1α promoter. (B) RNA blot analysis of J558L clones that have been stably transfected with various SATB2 constructs. Five micrograms total RNA was hybridized with probes that detect the endogenous Cα and GAPDH transcripts and the transfected satb2 mRNA. (C) ChIP experiments of J558L cell lines expressing similar amounts of Flag-tagged wild-type SATB2 (top panels) or the sumoylation site-deficient SATB2 double mutant (bottom panels). DNA of chromatin fragments that had been immunoprecipitated with an anti-Flag antibody was subsequently analyzed by semiquantitative PCR, using primers located in the 5′ MAR region of the immunoglobulin μ enhancer (lanes 1-5) or in the β-globin locus (lanes 6-10). Template DNA was used in a fivefold linear dilution series (starting from 5 ng). (D) Anti-Flag immunoblot analysis to detect SATB2 proteins that have been immunoprecipitated under ChIP conditions to control for equal fractionation and recovery of SATB2 and SATB2-dlmut.
SATB2 localizes to the nuclear periphery in a SUMO-dependent manner
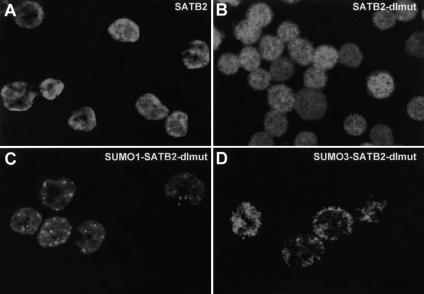
Conjugation of SUMO to target proteins has been shown to influence their subcellular localization. To examine the effects of sumoylation on the subnuclear localization of SATB2, we used the stably transfected J558L cell lines that express wild-type or mutant forms of Flag-tagged SATB2 at similar levels (Fig. 6A). Indirect immunofluorescence analysis with an anti-Flag antibody indicated that wild-type SATB2 is localized predominantly at the nuclear periphery and to some extent in nuclear speckles (Fig. 7A). The SATB2 double mutant was localized more diffusely throughout the nucleus (Fig. 7B). Notably, the covalent fusion of SUMO1 resulted in an accumulation of SATB2 in nuclear speckles, whereas the covalent fusion with SUMO3 resembled the staining of wild-type SATB2 at the nuclear periphery (Fig. 7C,D). This subnuclear distribution of SUMO3-SATB2 is consistent with the observation that SUMO3 is conjugated to SATB2 more efficiently than SUMO1.
Figure 7.
SUMO modification alters the subnuclear localization of SATB2. J558L plasmacytoma cells were stably transfected with plasmids encoding Flag-tagged wild-type and mutant SATB2 proteins, and clones expressing similar amounts of SATB2 protein were chosen for the analysis of the subcellular localization by indirect immunofluorescence (Fig. 6A). (A) Wild-type SATB2 localizes predominantly to the nuclear periphery. (B) SATB2-dlmut is diffusely distributed throughout the nucleus. (C) Covalent attachment of SUMO1 to the amino terminus of SATB2-dlmut localizes the protein into nuclear bodies. (D) Covalent attachment of SUMO3 to the amino terminus of SATB2-dlmut localizes the protein predominantly to the nuclear periphery.
Discussion
SATB2 functions as a transcriptional activator
We have identified SATB2 as a novel cell type-specific MAR-binding protein that is expressed abundantly in pre-B cells, kidney, and brain. The SATB2 protein was shown to bind MAR sequences flanking the enhancer of the endogenous immunoglobulin μ heavy chain (IgH) gene in vivo, and this binding was found to correlate with an increase in the expression of a transfected rearranged μ wild-type gene, but not with a μΔMAR gene lacking the MAR sequences. In addition, the expression of SATB2 and SATB2-dlmut in stably transfected plasmacytoma cells results in a reproducible two- to threefold increase of the high level of expression of the endogenous IgH locus. Finally, SATB2 was found to augment the expression of a reporter gene construct containing multimerized SATB2-binding sites, indicating that SATB2 acts as a bona fide transcriptional activator. Notably, no significant transcriptional activation by SATB2 was observed in nonlymphoid cells (293T, Hela, and NIH3T3) or in the EL4 T cell line (data not shown), suggesting that SATB2 collaborates with a B cell-specific transcriptional co-activator.
In contrast, the closely related thymocyte-specific MAR-binding protein SATB1 has been shown to repress target genes, such as IL-2Rα. In SATB1-knockout thymocytes, which arrest at the CD4+CD8+ double-positive stage of differentiation, IL-2Rα expression is upregulated, and this effect correlates with changes in the acetylation pattern of histone H3 over a large domain (Yasui et al. 2002). Consistent with a repressive effect of SATB1, this protein has been found to associate with the histone deacetylase of the NURD complex and to recruit the chromatin remodeling complexes ACF and ISWI in vitro (Yasui et al. 2002). However, SATB1 was also recently implicated in the positive regulation of genes because the c-myc locus, which contains a MAR region upstream of the transcription start site, is not properly upregulated in PMA-stimulated thymocytes from SATB1-deficient mice (Cai et al. 2003). In this context, SATB1 has been proposed to act as a three-dimensional protein scaffold upon which MAR sequences are tethered. The tethering of MAR sequences is thought to participate in the alteration of chromatin structure over large distances.
The multiple roles of MAR-binding proteins in gene regulation suggest that their function depends on the context of other cis-acting sequences. Although no direct interactions of MAR-binding proteins with transcription factors have been identified to date, three lines of evidence suggest that MAR-binding proteins can functionally collaborate with enhancer/promoter-binding proteins. First, MAR elements that are combined with the μ enhancer generate DNase I-hypersensitive sites and alter chromatin accessibility, whereas MAR elements alone fail to alter the chromatin structure (Jenuwein et al. 1993, 1997). Second, the potentiation of the μ enhancer by the MAR-binding protein Bright has been shown to be dependent on the binding site of the Oct transcription factor in the μ enhancer (Webb et al. 1999). In addition, Bright transactivates only from a subset of binding sites and acts by competing with the repressor Cux/CDP, which recognizes the same nucleotide sequence (Wang et al. 1999; Kaplan et al. 2001). Finally, the position of a MAR element relative to the transcription initiation site can determine whether the MAR element augments or inhibits gene expression (Schubeler et al. 1996). Thus, the context of regulatory sequences may determine whether MAR-binding proteins act as activators or repressors of transcription.
Regulation of SATB2 activity by PIAS1-mediated SUMO modification
The regulation of the activity of MAR-binding proteins is still fairly obscure. No signaling pathways have been linked to MAR-binding proteins, but one known mechanism for the regulation of SATB1 is the irreversible inactivation by proteolytic cleavage in apoptotic T cells during negative selection (Galande et al. 2001). In apoptotic T cells, SATB1 was found to be cleaved by caspase 6 at position 254 (VEMD) between the PDZ domain and the CUT domains, and this proteolysis results in the dissociation of SATB1 from MARs (Galande et al. 2001). Notably, the caspase 6 cleavage site is not conserved in SATB2, and the incubation of recombinant SATB2 with caspase 6 does not result in a proteolytic cleavage in vitro (G. Dobreva, M. Strzelecka, and R. Grosschedl, unpubl.). Instead, we identified SUMO modification as a mechanism that regulates the activity of SATB2. Our analysis of SATB2 indicates that this protein can be modified by SUMO conjugation at two internal lysines. One of these lysines, K233 of SATB2 (IKVE), is located at a similar position as the caspase 6 cleavage site in SATB1, and the other modified lysine, K350 of SATB2, is located immediately upstream of the CUT domains that mediate binding to MAR sequences (Galande et al. 2001). The sumoylation sites of SATB2 are not conserved in SATB1, and we also failed to detect SUMO modification of SATB1 in vivo. Thus, the proteolytic cleavage by caspase 6 and the modification by SUMO conjugation appear to be specific for SATB1 and SATB2, respectively. In contrast to the irreversible inactivation of SATB1 by proteolysis, the possibility of removing SUMO peptides by isopeptitases (Li and Hochstrasser 1999; Best et al. 2002; for review, see Muller et al. 2001) suggest that the regulation of SATB2 activity by sumoylation and desumoylation is dynamic. Two signaling pathways, MAP kinase signaling and CaM kinase signaling, have been found to downregulate sumoylation of proteins (Kirsh et al. 2002; Yang et al. 2003). In addition, sumoylation of proteins is regulated during the cell cycle and cellular stress (Saitoh and Hinchey 2000; Hoege et al. 2002).
Differences in the sumoylation of highly related proteins have also been shown for the Sp1 and Elk families of transcription factors. Sp1, Sp3, and Sp4 recognize the same nucleotide sequences, but only Sp3 can act as both an activator and a repressor of transcription (for review, see Gill 2003). The ability of Sp3 to act as a repressor was shown to depend on the SUMO modification of a lysine in an inhibitory domain that is important for repression of transcription and is not present in Sp1 and Sp4 (Ross et al. 2002; Sapetschnig et al. 2002). Likewise, Elk is the only member of the Elk/SAP subfamily of Ets transcription factors that can be sumoylated (Yang et al. 2003). Thus, SUMO modifications of specific members of protein families may help to diversify the function and/or regulation of highly related proteins.
In our present experiments, we identified PIAS1 as an E3 ligase for SUMO modification of SATB2. Co-expression of SATB2 with PIAS1, but not with other members of the family of PIAS proteins, augmented the sumoylation of SATB2 in vivo and in vitro. The pronounced specificity of PIAS1 as a SUMO E3 ligase for SATB2 contrasts with the lower specificity of PIAS proteins for the sumoylation of various other proteins. For example, p53 and CtBP can be sumoylated in the presence of either PIAS1 or PIASxβ (Schmidt and Muller 2002; Lin et al. 2003). PIAS proteins contain a conserved SAP domain that mediates association with the nuclear matrix and localization to PML nuclear bodies (Sachdev et al. 2001; Ross et al. 2002), whereas other recently identified E3 ligases for SUMO conjugation have distinct subcellular localizations. RanBP2, an E3 ligase for RanGAP, p53, and HDAC4, is localized at the cytoplasmic side of the nuclear pore and has been implicated in the regulation of nucleocytoplasmic shuttling (Kirsh et al. 2002; Pichler et al. 2002). In addition, polycomb group protein-2 (Pc2), which stimulates SUMO modification of CtBP, is a component of Pc nuclear bodies that are involved in the stable repression of genes (Kagey et al. 2003). Therefore, the overlapping substrate specificity and different subcellular localizations of E3 ligases may impart a spatial or temporal regulation of SUMO modification.
SUMO modification of SATB2 reduces chromatin association and transcriptional activation
A functional role for the sumoylation of SATB2 was inferred from the analysis of a mutant protein that cannot be modified with SUMO. First, the SUMO-deficient SATB2 protein was found to associate five times more efficiently with the MAR sequences of the endogenous immunoglobulin μ locus compared to the wild-type SATB2 protein. Second, the SUMO-deficient SATB2 stimulated expression of a cotransfected reporter plasmid carrying multimerized SATB2-binding sites more efficiently than wild-type SATB2. Although lysines can also be modified by ubiquitination, acetylation, or methylation, an amino-terminal fusion of SUMO to the K233R/K350R double mutant of SATB2 results in a decrease of the transcriptional activation potential, suggesting that the effects of the lysine mutations of SATB2 are most likely due to a deficiency in sumoylation. Therefore, SUMO modification appears to contribute to the regulation of the transcriptional activity of SATB2. In this regard, SATB2 differs from LEF1, p53, and STAT1, which are also SUMO-modified by association with PIAS proteins. For these proteins, mutations of the sumoylation sites do not alter the transcriptional activity. In contrast, the sumoylation of SATB2 and Sp3 alter the activity of these proteins, possibly irrespective of the location where they function. Thus, the sumoylation of proteins may generate pools of proteins that are functionally distinct and/or localized to different subnuclear compartments.
Sumoylation changes the subnuclear localization of SATB2
We find a striking correlation of sumoylation of SATB2 and its subnuclear localization. Both SATB2 and the SUMO3-SATB2-dlmut fusion protein are localized predominantly at the nuclear periphery, whereas the sumoylation-deficient SATB2-dlmut protein has a more diffuse nuclear localization. For the determination of protein localization, we used stably transfected plasmacytoma cells that express SATB2 at moderate levels, to avoid artificial targeting of excess proteins to sites of protein storage or degradation. As the SATB2-dlmut and SUMO-SATB2-dlmut proteins do not efficiently associate with PIAS1 (data not shown), the localization of SATB2 appears to be influenced by the added SUMO peptides. The function of another MAR-binding protein, Bright, also correlates with changes in its subnuclear localization. In association with a B cell-specific isoform of the speckle protein 100 (LYSp100B), Bright acts as a transcriptional activator and is localized to specific subnuclear speckles, termed LANDs (Zong et al. 2000). In association with the related Sp100 protein, Bright-mediated activation is repressed by antagonizing tetramerization of Bright and targeting to PML nuclear bodies (Zong et al. 2000).
The differences in the subnuclear localization of wild-type SATB2 and the SUMO-deficient SATB2 double mutant raise the question of whether the sumoylation of this MAR-binding protein may be involved in the changes of the localization of genes during transcriptional activation and/or repression. The immunoglobulin μ gene has been shown to localize at the nuclear periphery in primary T cells that do not express the gene, whereas the μ gene is localized away from the nuclear periphery in pre-B cells in which the gene is activated (Kosak et al. 2002). In addition, the nuclear localization of genes that are silenced by the transcription factor Ikaros is altered in comparison to cells in which Ikaros is not present (Cobb et al. 2000). In this case, the silenced genes colocalize with heterochromatic chromatin in foci that are located near the nuclear periphery. Our experiments indicate that the sumoylation of SATB2 down-regulates its association with MAR sequences and the transcriptional activation of MAR-containing target genes. In addition, sumoylation targets SATB2 to the nuclear periphery. However, we consider it unlikely that these processes are linked and that SUMO modification of SATB2 is involved in localizing silent immunoglobulin genes to the nuclear periphery. Instead, we favor the view that other mechanisms, including epigenetic modifications of chromatin and/or binding to other proteins, may account for the localization of the silent immunoglobulin loci to the nuclear periphery. If SATB2 had a function similar to that of the proposed nuclear scaffold function of SATB1 (Cai et al. 2003), SATB2 could be involved in localizing the immunoglobulin loci to sites of active transcription. According to this scheme, sumoylation of SATB2 could be a mechanism to regulate the tethering during the cell cycle, cellular stress, or extracellular signals that alter the balance between sumoylation and desumoylation.
Materials and methods
Constructs
SATB2 was PCR-amplified from a cDNA pool of Abelson murine leukemia virus (AMuLV)-transformed pre-B cells using the primers 5′-ATGGAGCGGCGGAGCGAGAG and 5′-TTATCTCTGGTCAATTTCGGCAGGTGC, and the products were cloned into pBluescript SK. For generating pcDNA3.1-TAPTag-SATB2, SATB2 was amplified using primers 5′-GCGGATCCATGGAGCGGCGGAGCGAGAG-3′ and 5′-GCGGATCCTTCTCTCTGGTCAATTTCGGCAGGTGC-3′, and after digestion of the PCR product with BamHI, the fragment was cloned into the BamHI site of pcDNA3.1-TAPtag. The SATB2 BamHI fragment was also subcloned in-frame into pEF-Flag or pET21a (Novagen) vectors. The SATB2 point mutants K233R and K350R were generated in the desired expression clone using a QuikChange Site-Directed Mutagenesis Kit (Stratagene). SATB2 K232R/K350R was constructed by the ligation of a BamHI/ScaI fragment derived from pEF-Flag-SATB2K233R and a BamHI/XbaI fragment from pEF-Flag-SATB2K350R into the BamHI/XbaI site of pEF-Flag.
RNA purification and Northern blot analysis
RNA purification was performed using Trizol reagent (Invitrogen). For the detection of satb2 transcripts, an XhoI/Xba fragment from pBs-SATB2 was used. To detect the mouse μ transcript, VH 17.2.25-specific probe was amplified by PCR from the pμ vector (Jenuwein et al. 1993), using primers 5′ GGGATATCCACACCAAACATC 3′ and 5′ AGAGGCCATTCTTACCTG AGG 3′ from pμ. To detect Cα transcripts, a Cα-specific probe was generated using the primers 5′ TGCACAGTTACCCATCCTGA and 5′ AGACGGTCGATGGTCTTCTG. To detect the GAPDH transcripts, a NotI/BamHI fragment from pGAPDH was isolated. Hybridization was performed at 67°C in H-mix (50 mM Na-phosphate buffer, pH 7.0, 1% SDS, 1x Denhardt buffer, 5x SSC, 0.1 mg/mL yeast tRNA).
Cell culture and establishment of stable cell clones
293T cells were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with penicillin-streptomycin-glutamine (PSG) and 10% fetal bovine serum (FBS; Invitrogen Life Technologies). Jurkat, EL4, and J558L cells were cultured in RPMI-1640 medium supplemented with PSG, 50 μM β-mercaptoethanol (Sigma), and 10% FBS. For the generation of stable cell lines, J558L or 38B9 cells were electroporated with 30 μg linearized plasmid DNA, and after 24 h, G418 (GibcoBRL) was added to a final concentration of 2 mg/mL. Single-cell clones were obtained by limiting dilution. Fourteen days after single-cell cloning, G418-resistant clones were expanded in RPMI medium at a final G418 concentration of 200 μg/mL.
Transient transfections and reporter assays
293T cells were transfected with 10 μg plasmid DNA by the calcium phosphate method. EL4, Jurkat, and J558L cell transfections were performed by electroporation with 35 μg of plasmid DNA. The total DNA concentration in each transfection experiment was kept constant by adding vector plasmid DNA. Cells were harvested 24-48 h posttransfection in 200 μL of reporter lysis buffer, and luciferase assays were conducted according to the manufacturer's instructions (Promega). β-galactosidase assays were performed as described (Starr et al. 1996).
Protein expression and purification
His-tagged SATB2 was expressed from pET21a-SATB2 in E. coli strain BL21 after induction with 0.5 mM IPTG. Cells were resuspended in lysis buffer (300 mM NaCl, 5 mM MgCl2, 20 mM sodium phosphate, pH 7.5, 40 mM imidazole, 10% glycerol, 0.1% Triton X-100, 1 mM DTT, and protease inhibitors mix) and sonicated 3 × 30 sec. After centrifugation, the cell lysate was incubated with Ni-NTA beads for 1 h at 4°C, washed with 3×10 mL lysis buffer, and eluted with 300 mM imidazole in lysis buffer. SUMO1, Ubc9, Aos1, and Uba2 were purified as His-tagged proteins from E. coli as described (Sachdev et al. 2001).
Electrophoretic mobility shift assay
The wild-type MAR 5′-TCTTTAATTTCTAATATATTTAGAATTC-3′ or mutant MAR 5′-TCTTTAATTTCTACTGCTTTAGAATTC-3′ oligonucleotides (Dickinson et al. 1992) were radiolabeled with 32P and purified on a 20% native polyacrylamide gel. DNA-binding reactions contained 20 mM HEPES, pH 7.9, 100 mM KCl, 5 mM MgCl2, 0,1 mM EDTA, 8% glycerol, 1 μg poly dI-dC, and 10,000 cpm radiolabeled probe. EMSAs were performed in 0.33x TBE.
In vitro sumoylation assay
SUMO modification reactions were performed in a total volume of 20 μL with 250 ng of Aos1/Uba2, 250 ng Ubc9, 1 μg recombinant SUMO-1, and 300 ng His-epitope-tagged SATB2. The reaction buffer contained 50 mM Tris, pH 7.4, 5 mM MgCl2, and 1 mM ATP. Reactions were incubated at 30°C for 30-45 min and stopped by addition of 2xSDS-PAGE sample buffer.
Immunoprecipitations and immunoblot analysis
For immunoprecipitation and co-immunoprecipitation experiments, cells were harvested in PBS, pelleted, and resuspended in RIPA (10 mM Na-phosphate buffer, pH 7.2, 150 mM NaCl, 1% Na-deoxycholate, 1% Triton X-100, 0.1% SDS, 1 mM DTT, protease inhibitors mix) or co-immunoprecipitation buffer (50 mM Tris-Cl, pH 7.5, 100 mM NaCl, 5 mM EGTA, 0.1% TX-100, 1 mM DTT, protease inhibitors mix). Equivalent amounts of total protein extract—usually 500 μg—were precleared and immunoprecipitated with 1 μg of anti-Flag or anti-myc mAb. For immunoblot analysis, the antibodies were used in following dilutions: anti-T7 mAb (Novagen), 1:5000; anti-Flag M2 mAb (Sigma), 1:5000; anti-myc mAb (Sigma), 1:4000; and anti-SUMO1 (GMP1) mAb (Zymed Laboratories), 1:1000.
Chromatin immunoprecipitations
Proteins were crosslinked to DNA by adding formaldehyde directly in the culture medium to a final concentration of 1% and incubating for 10 min at 37°C. Cells were washed and resuspended in sonication buffer (1% SDS, 10 mM EDTA, 50 mM Tris, pH8.0). DNA was sheared by sonication 15 × 15 sec. Chromatin extracts were diluted 10-fold in immunoprecipitation (IP) buffer (140 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, and protease inhibitors mix). IP using anti-Flag mAb and Protein G-Sepharose or two-step affinity chromatography purification (Rigaut et al. 1999) was performed. The IPs were washed twice with 1 mL of IP buffer, once with 1 mL of IP buffer containing 500 mM NaCl, once with 0.5 mL wash buffer (10 mM Tris, pH 8.0, 250 mM LiCl, 1 mM EDTA, 0.25% sodium deoxycholate), and twice with 0.5 mL of TE buffer. The chromatin bound to the beads was eluted in 300 μL elution buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1% SDS) by heating at 65°C for 15 min. Bound and input chromatin samples were diluted to a final SDS concentration of 0.5%. All samples were incubated overnight at 65°C to reverse the formaldehyde crosslinking. After RNaseA treatment and phenol-chloroform purification of the DNA, semiquantitative PCRs were performed, using primer pairs 5′MAR-CHIP-U (5′ CCTGCAAAAGTCCAGCTTTC 3′) and 5′MAR-CHIP-L (5′ AGAGCCTCACTCCCATTCCT 3′) for the 5′ MAR region in the immunoglobulin intronic enhancer and H7uA and H7uB for the β-globulin locus (Litt et al. 2001).
Nuclear matrix preparation, GFP fluorescence, and indirect immunofluorescence
For indirect immunofluorescence, cells were fixed for 20 min with 3.2% paraformaldehyde in cytoskeletal buffer and subsequently permeabilized for 5 min at room temperature with 0.5% Triton X-100 in cytoskeletal buffer (10 mM PIPES, pH 7.1, 1 mM EGTA, 3 mM MgCl2). Indirect immunofluorescence was conducted essentially as described (Zeng et al. 1997). Nuclear matrix preparations were conducted essentially as described (de Belle et al. 1998).
Note added in proof
The nucleotide sequence has been deposited at the EMBL Nucleotide Sequence Database, Accession Number AJ606045.
Acknowledgments
We thank Drs. Frauke Melchior, Guntram Suske, K. Shuai, and Jorma Palvimo for their generosity in providing plasmids, Heidi Sieber for the purification of the E1 and E2 enzymes, and Boyan Garvalov for his help with the immunohistochemistry. This work was supported by a grant from the German Research Foundation (TR6007).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1153003.
References
- Alvarez J.D., Yasui, D.H., Niida, H., Joh, T., Loh, D.Y., and Kohwi-Shigematsu, T. 2000. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes & Dev. 14: 521-535. [PMC free article] [PubMed] [Google Scholar]
- Berezney R. and Coffey, D.S. 1974. Identification of a nuclear protein matrix. Biochem. Biophys. Res. Commun. 60: 1410-1417. [DOI] [PubMed] [Google Scholar]
- Best J.L., Ganiatsas, S., Agarwal, S., Changou, A., Salomoni, P., Shirihai, O., Meluh, P.B., Pandolfi, P.P., and Zon, L.I. 2002. SUMO-1 protease-1 regulates gene transcription through PML. Mol. Cell 10: 843-855. [DOI] [PubMed] [Google Scholar]
- Bode J., Benham, C., Knopp, A., and Mielke, C. 2000. Transcriptional augmentation: Modulation of gene expression by scaffold/matrix-attached regions (S/MAR elements). Crit. Rev. Eukaryot. Gene Expr. 10: 73-90. [PubMed] [Google Scholar]
- Cai S., Han, H.J., and Kohwi-Shigematsu, T. 2003. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat. Genet. 34: 42-51. [DOI] [PubMed] [Google Scholar]
- Chung C.D., Liao, J., Liu, B., Rao, X., Jay, P., Berta, P., and Shuai, K. 1997. Specific inhibition of Stat3 signal transduction by PIAS3. Science 278: 1803-1805. [DOI] [PubMed] [Google Scholar]
- Cobb B.S., Morales-Alcelay, S., Kleiger, G., Brown, K.E., Fisher, A.G., and Smale, S.T. 2000. Targeting of Ikaros to pericentromeric heterochromatin by direct DNA binding. Genes & Dev. 14: 2146-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerill P.N. and Garrard, W.T. 1986. Chromosomal loop anchorage of the κ immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell 44: 273-282. [DOI] [PubMed] [Google Scholar]
- Cremer T. and Cremer, C. 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2: 292-301. [DOI] [PubMed] [Google Scholar]
- de Belle I., Cai, S., and Kohwi-Shigematsu, T. 1998. The genomic sequences bound to special AT-rich sequence-binding protein 1 (SATB1) in vivo in Jurkat T cells are tightly associated with the nuclear matrix at the bases of the chromatin loops. J. Cell. Biol. 141: 335-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desterro J.M., Rodriguez, M.S., and Hay, R.T. 1998. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell 2: 233-239. [DOI] [PubMed] [Google Scholar]
- Dickinson L.A., Joh, T., Kohwi, Y., and Kohwi-Shigematsu, T. 1992. A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell 70: 631-645. [DOI] [PubMed] [Google Scholar]
- Dickinson L.A., Dickinson, C.D., and Kohwi-Shigematsu, T. 1997. An atypical homeodomain in SATB1 promotes specific recognition of the key structural element in a matrix attachment region. J. Biol. Chem. 272: 11463-11470. [DOI] [PubMed] [Google Scholar]
- Fernandez L.A., Winkler, M., and Grosschedl, R. 2001. Matrix attachment region-dependent function of the immunoglobulin μ enhancer involves histone acetylation at a distance without changes in enhancer occupancy. Mol. Cell Biol. 21: 196-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester W.C., van Genderen, C., Jenuwein, T., and Grosschedl, R. 1994. Dependence of enhancer-mediated transcription of the immunoglobulin μ gene on nuclear matrix attachment regions. Science 265: 1221-1225. [DOI] [PubMed] [Google Scholar]
- Forrester W.C., Fernandez, L.A., and Grosschedl, R. 1999. Nuclear matrix attachment regions antagonize methylation-dependent repression of long-range enhancer-promoter interactions. Genes & Dev. 13: 3003-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiman R.N. and Tjian, R. 2003. Regulating the regulators: Lysine modifications make their mark. Cell 112: 11-17. [DOI] [PubMed] [Google Scholar]
- Galande S., Dickinson, L.A., Mian, I.S., Sikorska, M., and Kohwi-Shigematsu, T. 2001. SATB1 cleavage by caspase 6 disrupts PDZ domain-mediated dimerization, causing detachment from chromatin early in T-cell apoptosis. Mol. Cell Biol. 21: 5591-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G. 2003. Post-translational modification by the small ubiquitin-related modifier SUMO has big effects on transcription factor activity. Curr. Opin. Genet. Dev. 13: 108-113. [DOI] [PubMed] [Google Scholar]
- Goodson M.L., Hong, Y., Rogers, R., Matunis, M.J., Park-Sarge, O.K., and Sarge, K.D. 2001. Sumo-1 modification regulates the DNA binding activity of heat shock transcription factor 2, a promyelocytic leukemia nuclear body associated transcription factor. J. Biol. Chem. 276: 18513-18518. [DOI] [PubMed] [Google Scholar]
- Gostissa M., Hengstermann, A., Fogal, V., Sandy, P., Schwarz, S.E., Scheffner, M., and Del Sal, G. 1999. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 18: 6462-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrscher R.F., Kaplan, M.H., Lelsz, D.L., Das, C., Scheuermann, R., and Tucker, P.W. 1995. The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: AB cell-specific trans-activator that describes a new DNA-binding protein family. Genes & Dev. 9: 3067-3082. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. 2001. SP-RING for SUMO: New functions bloom for a ubiquitin-like protein. Cell 107: 5-8. [DOI] [PubMed] [Google Scholar]
- Hoege C., Pfander, B., Moldovan, G.L., Pyrowolakis, G., and Jentsch, S. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135-141. [DOI] [PubMed] [Google Scholar]
- Hong Y., Rogers, R., Matunis, M.J., Mayhew, C.N., Goodson, M.L., Park-Sarge, O.K., Sarge, K.D., and Goodson, M. 2001. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J. Biol. Chem. 276: 40263-40267. [DOI] [PubMed] [Google Scholar]
- Jackson P.K. 2001. A new RING for SUMO: Wrestling transcriptional responses into nuclear bodies with PIAS family E3 SUMO ligases. Genes & Dev. 15: 3053-3058. [DOI] [PubMed] [Google Scholar]
- Jenuwein T., Forrester, W.C., Qiu, R.Q., and Grosschedl, R. 1993. The immunoglobulin μ enhancer core establishes local factor access in nuclear chromatin independent of transcriptional stimulation. Genes & Dev. 7: 2016-2032. [DOI] [PubMed] [Google Scholar]
- Jenuwein T., Forrester, W.C., Fernandez-Herrero, L.A., Laible, G., Dull, M., and Grosschedl, R. 1997. Extension of chromatin accessibility by nuclear matrix attachment regions. Nature 385: 269-272. [DOI] [PubMed] [Google Scholar]
- Johnson E.S. and Gupta, A.A. 2001. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106: 735-744. [DOI] [PubMed] [Google Scholar]
- Kagey M.H., Melhuish, T.A., and Wotton, D. 2003. The polycomb protein Pc2 is a SUMO E3. Cell 113: 127-137. [DOI] [PubMed] [Google Scholar]
- Kahyo T., Nishida, T., and Yasuda, H. 2001. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell 8: 713-718. [DOI] [PubMed] [Google Scholar]
- Kaplan M.H., Zong, R.T., Herrscher, R.F., Scheuermann, R.H., and Tucker, P.W. 2001. Transcriptional activation by a matrix associating region-binding protein. Contextual requirements for the function of bright. J. Biol. Chem. 276: 21325-21330. [DOI] [PubMed] [Google Scholar]
- Kirsh O., Seeler, J.S., Pichler, A., Gast, A., Muller, S., Miska, E., Mathieu, M., Harel-Bellan, A., Kouzarides, T., Melchior, F., et al. 2002. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J. 21: 2682-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi-Shigematsu T., Maass, K., and Bode, J. 1997. Athymocyte factor SATB1 suppresses transcription of stably integrated matrix-attachment region-linked reporter genes. Biochemistry 36: 12005-12010. [DOI] [PubMed] [Google Scholar]
- Kosak S.T., Skok, J.A., Medina, K.L., Riblet, R., Le Beau, M.M., Fisher, A.G., and Singh, H. 2002. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 296: 158-162. [DOI] [PubMed] [Google Scholar]
- Li S.J. and Hochstrasser, M. 1999. A new protease required for cell-cycle progression in yeast. Nature 398: 246-251. [DOI] [PubMed] [Google Scholar]
- Lichtenstein M., Keini, G., Cedar, H., and Bergman, Y. 1994. B cell-specific demethylation: Anovel role for the intronic κ chain enhancer sequence. Cell 76: 913-923. [DOI] [PubMed] [Google Scholar]
- Lin X., Sun, B., Liang, M., Liang, Y.Y., Gast, A., Hildebrand, J., Brunicardi, F.C., Melchior, F., and Feng, X.H. 2003. Opposed regulation of corepressor CtBP by SUMOylation and PDZ binding. Mol. Cell 11: 1389-1396. [DOI] [PubMed] [Google Scholar]
- Litt M.D., Simpson, M., Gaszner, M., Allis, C.D., and Felsenfeld, G. 2001. Correlation between histone lysine methylation and developmental changes at the chicken β-globin locus. Science 293: 2453-2455. [DOI] [PubMed] [Google Scholar]
- Liu B., Liao, J., Rao, X., Kushner, S.A., Chung, C.D., Chang, D.D., and Shuai, K. 1998. Inhibition of Stat1-mediated gene activation by PIAS1. Proc. Natl. Acad. Sci. 95: 10626-10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F. 2000. SUMO—Nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16: 591-626. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J., Mirault, M.E., and Laemmli, U.K. 1984. Organization of the higher-order chromatin loop: Specific DNA attachment sites on nuclear scaffold. Cell 39: 223-232. [DOI] [PubMed] [Google Scholar]
- Muller S., Matunis, M.J., and Dejean, A. 1998. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 17: 61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S., Hoege, C., Pyrowolakis, G., and Jentsch, S. 2001. SUMO, ubiquitin's mysterious cousin. Nat. Rev. Mol. Cell Biol. 2: 202-210. [DOI] [PubMed] [Google Scholar]
- Pichler A., Gast, A., Seeler, J.S., Dejean, A., and Melchior, F. 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108: 109-120. [DOI] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17: 1030-1032. [DOI] [PubMed] [Google Scholar]
- Rodriguez M.S., Desterro, J.M., Lain, S., Midgley, C.A., Lane, D.P., and Hay, R.T. 1999. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 18: 6455-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M.S., Dargemont, C., and Hay, R.T. 2001. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276: 12654-12659. [DOI] [PubMed] [Google Scholar]
- Romig H., Fackelmayer, F.O., Renz, A., Ramsperger, U., and Richter, A. 1992. Characterization of SAF-A, a novel nuclear DNA binding protein from HeLa cells with high affinity for nuclear matrix/scaffold attachment DNA elements. EMBO J. 11: 3431-3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S., Best, J.L., Zon, L.I., and Gill, G. 2002. SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol. Cell 10: 831-842. [DOI] [PubMed] [Google Scholar]
- Sachdev S., Bruhn, L., Sieber, H., Pichler, A., Melchior, F., and Grosschedl, R. 2001. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes & Dev. 15: 3088-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H. and Hinchey, J. 2000. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275: 6252-6258. [DOI] [PubMed] [Google Scholar]
- Sapetschnig A., Rischitor, G., Braun, H., Doll, A., Schergaut, M., Melchior, F., and Suske, G. 2002. Transcription factor Sp3 is silenced through SUMO modification by PIAS1. EMBO J. 21: 5206-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann R.H. and Chen, U. 1989. A developmental-specific factor binds to suppressor sites flanking the immunoglobulin heavy-chain enhancer. Genes & Dev. 3: 1255-1266. [DOI] [PubMed] [Google Scholar]
- Scheuermann R.H. and Garrard, W.T. 1999. MARs of antigen receptor and co-receptor genes. Crit. Rev. Eukaryot. Gene Expr. 9: 295-310. [DOI] [PubMed] [Google Scholar]
- Schmidt D. and Muller, S. 2002. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl. Acad. Sci. 99: 2872-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D., Mielke, C., Maass, K., and Bode, J. 1996. Scaffold/matrix-attached regions act upon transcription in a context-dependent manner. Biochemistry 35: 11160-11169. [DOI] [PubMed] [Google Scholar]
- Schul W., de Jong, L., and van Driel, R. 1998. Nuclear neighbours: The spatial and functional organization of genes and nuclear domains. J. Cell Biochem. 70: 159-171. [DOI] [PubMed] [Google Scholar]
- Spector D.L. 2001. Nuclear domains. J. Cell Sci. 114: 2891-2893. [DOI] [PubMed] [Google Scholar]
- Starr D.B., Matsui, W., Thomas, J.R., and Yamamoto, K.R. 1996. Intracellular receptors use a common mechanism to interpret signaling information at response elements. Genes & Dev. 10: 1271-1283. [DOI] [PubMed] [Google Scholar]
- Tian S., Poukka, H., Palvimo, J.J., and Janne, O.A. 2002. Small ubiquitin-related modifier-1 (SUMO-1) modification of the glucocorticoid receptor. Biochem. J. 367: 907-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verger A., Perdomo, J., and Crossley, M. 2003. Modification with SUMO. EMBO Rep. 4: 137-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Goldstein, A., Zong, R.T., Lin, D., Neufeld, E.J., Scheuermann, R.H., and Tucker, P.W. 1999. Cux/CDP homeoprotein is a component of NF-muNR and represses the immunoglobulin heavy chain intronic enhancer by antagonizing the bright transcription activator. Mol. Cell Biol. 19: 284-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb C., Zong, R.T., Lin, D., Wang, Z., Kaplan, M., Paulin, Y., Smith, E., Probst, L., Bryant, J., Goldstein, A., et al. 1999. Differential regulation of immunoglobulin gene transcription via nuclear matrix-associated regions. Cold Spring Harb. Symp. Quant. Biol. 64: 109-118. [DOI] [PubMed] [Google Scholar]
- Weitzel J.M., Buhrmester, H., and Stratling, W.H. 1997. Chicken MAR-binding protein ARBP is homologous to rat methyl-CpG-binding protein MeCP2. Mol. Cell Biol. 17: 5656-5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Hammer, R.E., Blasquez, V.C., Jones, S.L., and Garrard, W.T. 1989. Immunoglobulin κ gene expression after stable integration. II. Role of the intronic MAR and enhancer in transgenic mice. J. Biol. Chem. 264: 21190-21195. [PubMed] [Google Scholar]
- Yang S.H., Jaffray, E., Hay, R.T., and Sharrocks, A.D. 2003. Dynamic interplay of the SUMO and ERK pathways in regulating Elk-1 transcriptional activity. Mol. Cell 12: 63-74. [DOI] [PubMed] [Google Scholar]
- Yasui D., Miyano, M., Cai, S., Varga-Weisz, P., and Kohwi-Shigematsu, T. 2002. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature 419: 641-645. [DOI] [PubMed] [Google Scholar]
- Zeng C., Kim, E., Warren, S.L., and Berget, S.M. 1997. Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. EMBO J. 16: 1401-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. and Reinberg, D. 2001. Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes & Dev. 15: 2343-2360. [DOI] [PubMed] [Google Scholar]
- Zong R.T., Das, C., and Tucker, P.W. 2000. Regulation of matrix attachment region-dependent, lymphocyte-restricted transcription through differential localization within promyelocytic leukemia nuclear bodies. EMBO J. 19: 4123-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]