Abstract
Tap has been proposed to play a role in general mRNA export and also functions in expression of RNA with retained introns that contain the MPMV CTE (constitutive transport element). Tap forms a functional heterodimer with NXT/p15. We have previously demonstrated that unspliced intron-containing CTE RNA is efficiently exported to the cytoplasm in mammalian cells. Here we show that Tap and NXT proteins function together to enhance translation of proteins from the exported CTE RNA. Pulse chase experiments show that Tap/NXT significantly increases the rate of protein synthesis. Sucrose gradient analysis demonstrates that Tap and NXT efficiently shift the unspliced RNA into polyribosomal fractions. Furthermore, Tap, but not NXT is detected in polyribosomes. Taken together, our results indicate that Tap and NXT serve a role in translational regulation of RNA after export to the cytoplasm. They further suggest that Tap/NXT may play a role in remodeling of cytoplasmic RNP complexes, providing a link between export pathways and cytoplasmic fate.
Keywords: Unspliced mRNA, mRNA export, translational regulation, Tap/NXT
In higher eukaryotes, the majority of genes produce nascent mRNA transcripts that contain several introns. Export of incompletely spliced RNAs with retained introns from these genes would potentially result in translation of aberrant proteins that could have deleterious effects on the cells. However, cells appear to have developed checkpoints to ensure that only fully spliced mRNAs are exported and expressed (Chang and Sharp 1989; Legrain and Rosbash 1989).
Retroviruses use special mechanisms that allow unspliced and incompletely spliced (that is, intron-containing) viral transcripts to exit the nucleus and escape cellular proofreading mechanisms (Hammarskjöld 1997, 2001). Replication of all of these viruses requires the cytoplasmic expression of intron-containing forms of the initial, genome-length viral RNA transcript, because the unspliced RNA serves both as the mRNA for the viral Gag and GagPol proteins and as the RNA that is packaged into viral particles (Berkowitz et al. 1996). In complex retroviruses such as HIV-1, export of unspliced and incompletely spliced RNA relies on the interaction of the viral Rev protein with a structured RNA sequence present in these RNAs, the Rev responsive element (RRE; Hadzopoulou-Cladaras et al. 1989; Hammarskjold et al. 1989; Malim et al. 1989; Pollard and Malim 1998). The resulting mRNP complex is exported by virtue of the nuclear export signal (NES) in Rev that interacts with the cellular export receptor Crm1 (Fornerod et al. 1997; Neville et al. 1997).
Simple retroviruses do not encode an Rev-like trans-acting protein, and export of their unspliced RNA relies on the interaction of cis-acting RNA elements directly with cellular factors. The first element of this kind was identified in the simian type D retrovirus Mason Pfizer Monkey Virus (MPMV) and was given the name CTE (constitutive transport element; Bray et al. 1994; Ernst et al. 1997a,b; Hammarskjold 2001), because its interaction with cellular factors results in constitutive export of intron-containing RNAs. Although substitution of the RRE by the CTE renders HIV replication Rev-independent (Bray et al. 1994; Zolotukhin et al. 1994), it is clear that the CTE promotes export of intron-containing RNAs by a pathway that is distinct from that used by the RRE-Rev system, because the Crm1 inhibitor Leptomycin B has no effect on CTE-mediated export (Otero et al. 1998; Guzik et al. 2001). Through experiments in the Xenopus oocyte system, it was shown that the CTE pathway uses cellular factors that are also important for cellular mRNA export (Pasquinelli et al. 1997; Saavedra et al. 1997).
Efforts to find a cellular cofactor for CTE-dependent nuclear RNA export led to the identification of Tap as a cellular protein that specifically binds the CTE (Gruter et al. 1998). The Tap protein, first identified as a potential cytoplasmic cofactor for the Tip oncogene encoded by Herpes Virus Saimiri (Yoon et al. 1997), is a 619-amino-acid protein that is a mammalian ortholog of Mex67p, a yeast mRNA export factor (Segref et al. 1997). Tap binds specifically to CTE RNA in vitro and enhances the export of various CTE-containing RNA substrates in Xenopus oocytes (Gruter et al. 1998; Braun et al. 1999; Bachi et al. 2000). In addition, the expression of human Tap increases the expression of proteins from CTE-containing RNA in quail cells (Kang and Cullen 1999). Mex67p has been shown to associate with RNA in vivo, and temperature-sensitive Mex67 mutants display a phenotype of rapid nuclear accumulation of poly(A) RNA at the nonpermissive temperature (Segref et al. 1997). These results, together with the fact that Tap interacts with the nuclear pore complex and shuttles between the nucleus and cytoplasm, have suggested Tap as a major receptor for mRNA export.
NXT1 (p15) has been shown to be an important Tap cofactor (Katahira et al. 1999; Braun et al. 2001; Guzik et al. 2001), and Tap and NXT1 form heterodimers (Fribourg et al. 2001). Nucleoporin binding by Tap, as well as nucleocytoplasmic shuttling, are greatly enhanced by formation of the Tap-NXT1 heterodimer (Levesque et al. 2001; Wiegand et al. 2002). Furthermore, Tap mutants that cannot form Tap-NXT1 heterodimers are unable to support Tap-mediated export of cellular mRNAs and of RNAs where Tap is “tethered” to the RNA using Tap fusion proteins (Guzik et al. 2001; Levesque et al. 2001; Wiegand et al. 2002). A related protein, NXT2, can functionally substitute for NXT1 in the dimer complex (Herold et al. 2000). Although NXT proteins are clearly of importance for Tap function when Tap is recruited to RNA in a non-sequence-specific manner or tethered to RRE RNA through a nonfunctional Rev protein, it has remained unclear whether either of the NXT proteins are essential for Tap function in conjunction with the MPMV CTE.
Many papers published in recent years have demonstrated that cellular intron-containing mRNA receives a protein “mark” at the exon-exon junctions after splicing has been completed. Several proteins are present in this exon junction complex (EJC), which has been proposed to play a crucial role in nonsense-mediated decay (NMD) of RNA containing premature stop codons (Le Hir et al. 2001). Previous studies have also shown that Tap/NXT1 heterodimers can bind to components of the EJC, potentially supporting a role for this complex in export (Kim et al. 2001; Le Hir et al. 2001). Very recently, it has been argued that the EJC may play a role in translational regulation after the RNA reaches the cytoplasm, rather than directly function to promote export. Data from two different laboratories have shown that cellular cDNAs lacking introns are exported from the nucleus, but are less well translated than spliced RNA (Lu and Cullen 2003; Nott et al. 2003). Insertion of removable introns increased the “translational yield,” leading to the hypothesis that the EJC or proteins that associate with this complex enhance cytoplasmic utilization (Le Hir et al. 2003).
In this paper, we present data to show that Tap can act in conjunction with either NXT1 or NXT2 to significantly enhance protein expression from intron-containing RNA containing the MPMV CTE. Specifically, our results indicate that the Tap/NXT complexes enhance polyribosome association and translation of intron-containing RNA that has been exported to the cytoplasm using the CTE. Thus Tap and NXT1 complexes are likely to play an important role in translational regulation beyond their previously proposed function as RNA export receptors.
Results
The combination of Tap and NXT1 dramatically enhances CTE function in 293T cells
We have previously shown that TapRevM10 fusion proteins can efficiently substitute for Rev and act in conjunction with the HIV RRE in 293T and other cells to promote export of intron-containing RNA, providing that the cells are also transfected with a plasmid that expresses NXT1 (Guzik et al. 2001). To analyze whether Tap and/or NXT proteins were also able to enhance expression from constructs containing the MPMV CTE, we used our well-characterized HIV GagPol reporter system (Smith et al. 1990; Bray et al. 1994; Guzik et al. 2001; Coyle et al. 2003). In this system, HIV Gag and GagPol proteins are expressed from an intron-containing RNA that is only exported to the cytoplasm of transfected cells in the presence of a specific export signal in the RNA. Expression of these proteins leads to the production of HIV virus-like particles that are efficiently secreted into the medium of transfected cells. These particles can be quantitated using an enzyme-linked immunosorbent assay (ELISA) that measures p24, the major HIV capsid protein. In the absence of a specific export element, the RNA is not exported from the nucleus and no GagPol expression is observed. To control for transfection efficiency and effects that are not specific to the intron-containing RNA pathway, cells are also transfected with a plasmid expressing secreted alkaline phosphatase (SEAP), which can also be quantitated in the medium. SEAP is expressed from an RNA that contains one intron and is fully spliced before export to the cytoplasm.
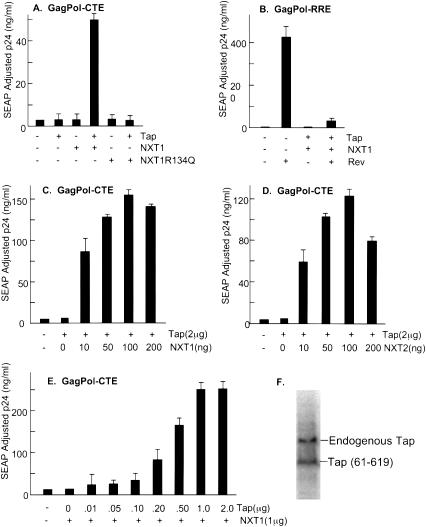
We initially analyzed the effects of transfection of plasmids expressing either Tap or NXT1 on MPMV CTE function in transfected 293T cells using this reporter system. Expression of either of these proteins alone had little effect on the levels of p24 expressed from an HIV GagPol reporter construct containing CTE. In contrast, a very significant increase in the levels of p24 was observed when the NXT1 and Tap plasmids were cotransfected (see Fig. 1A). This effect was specific to the CTE-containing RNA, because SEAP levels did not increase in the presence of Tap and NXT1 (data not shown). The enhancement of p24 was almost completely abrogated when an arginine residue of NXT1 was mutated to glutamine (R134Q). Mutations of this residue in NXT1 have previously been shown to make NXT1 defective for binding to the Tap protein (Fribourg et al. 2001). This suggests that the observed effect of NXT1/Tap cotransfection may reflect a direct interaction between Tap and NXT1.
Figure 1.
Effects of Tap and NXT1 on p24 expression from a GagPolCTE reporter construct in 293T cells. (A) Exogenously added Tap and NXT1 dramatically enhance p24 expression. 293T cells (3 × 106) were transfected with 5 μg of pCMVGagPolCTE and 0.25 μg of pCMVSEAP with or without 2 μg of pCMVTap and 1 μg of pcDNANXT1 or pcDNANXT1R134Q. At 72 h posttransfection, supernatants were collected and analyzed for p24 levels and SEAP activity. The p24 values shown have been normalized for SEAP activity. (B) Rev function is inhibited by expression of exogenous Tap and NXT1. 293T cells (3 × 106) were transfected with 5 μg of pCMVGagPolRRE and 0.25 μg of pCMVSEAP with or without 2 μg of pCMVTap and 1 μg of pcDNANXT1 in the absence or presence of 1 μg of pCMVRev. Supernatants were harvested and analyzed as described in Figure 1A. (C) The effect of increasing amounts of NXT1 on p24 expression from a GagPolCTE reporter in 293T cells. Transfections and analysis are as described in the legend of Figure 1A except that the amount of pcDNANXT1 was varied. (D) The effect of increasing amounts of NXT2 on p24 expression from a GagPolCTE reporter in 293T cells. Transfections and analysis are as described in the legend of Figure 1A except that the amount of pcDNANXT2 was varied. (E) The effect of increasing amounts of Tap on p24 expression from a GagPolCTE reporter in 293T cells. Transfections and analysis are as described in the legend of Figure 1A. (F) Analysis of Tap proteins in transfected 293T cells. Cells (3 × 106) were transfected with 5 μg of GagPolCTE reporter constructs together with 1 μg each of pCMVTap and pcDNANXT1 plasmids, harvested at 65 h posttransfection and analyzed by Western blot. The Western blot was performed on whole-cell lysates using an anti-Tap polyclonal antibody, and 125I-coupled protein A for detection. Exogenous and endogenous Tap proteins are indicated. Tap expressed from the transfected plasmid lacks 60 amino acids at the N-terminal end, compared with the endogenously expressed Tap protein.
To analyze whether the Tap and NXT1 enhancement was specific for the CTE, we performed an experiment in which the pCMVGagPolRRE reporter plasmid was transfected into cells with or without Rev and in the presence or absence of Tap and NXT1. The GagPolRRE plasmid is similar to the GagPolCTE plasmid with the exception that it contains an RRE instead of the CTE and thus requires Rev for p24 expression. The GagPol RNA expressed from this plasmid thus uses the Crm1 pathway for export. As can be seen in Figure 1B, no significant p24 expression was observed with the GagPolRRE plasmid in the absence of Rev, and Rev cotransfection led to production of large amounts of p24, confirming our previously published results (Smith et al. 1990; Bray et al. 1994; Guzik et al. 2001; Coyle et al. 2003). p24 levels did not increase in the presence of Tap and NXT1 alone and cotransfection of Tap and NXT1 with Rev led to decreased levels of p24 compared with the transfection with Rev alone. This experiment has been repeated several times with similar results, showing that the Tap and NXT1 enhancement of p24 expression is specific for the CTE-containing construct and that Tap and NXT1 overexpression inhibits Rev/RRE-mediated p24 expression.
To further analyze the effects of NXT1 and Tap on CTE expression, we performed an experiment in which increasing amounts of either the NXT1 or Tap plasmid were cotransfected with a constant amount of the other plasmid. The results of these experiments demonstrated that a significant enhancement of p24 expression was obtained with as little as 10 ng of the NXT1 plasmid (Fig. 1C). Maximum expression was achieved using 100 ng of plasmid. Essentially identical results were obtained when a plasmid expressing NXT2 (Herold et al. 2000) was used instead of NXT1 (Fig. 1D). In the case of Tap, a dose response was also observed, with maximum enhancement using 1 μg of plasmid (Fig. 1E). As shown in Figure 1F, the levels of “exogenous” Tap expressed in this transfection are similar to those of “endogenous” Tap expressed in these cells. Thus, it is not likely that our results represent effects of high “nonphysiological” levels of Tap/NXT proteins.
Tap/NXT does not significantly increase the nucleo-cytoplasmic export of GagPolCTE RNA in 293T cells
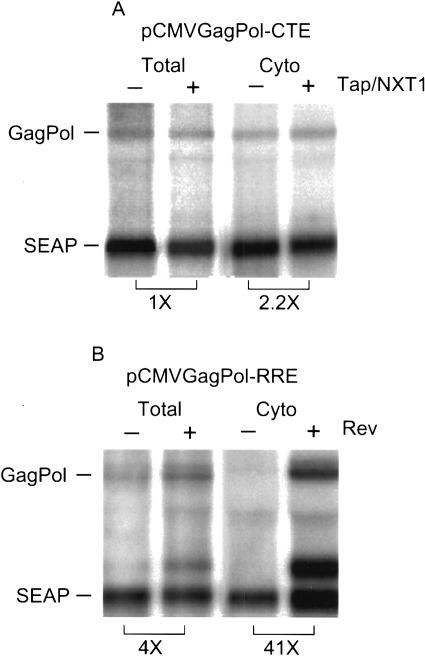
To determine if the Tap and NXT1-mediated enhancement of CTE function occurred at the level of RNA export, Northern blot assays were performed on total and cytoplasmic poly(A)-selected RNAs from transfected cells. The blots were analyzed with a probe specific for the HIV GagPol message, as well as with a probe specific for the control SEAP mRNA. For comparison, a similar analysis was also performed on cells transfected with pCMVGagPolRRE alone or in cotransfections with pCMV-Rev. Previous studies have shown that no GagPolRRE RNA is exported to the cytoplasm unless Rev is provided (Bray et al. 1994; Guzik et al. 2001; Coyle et al. 2003).
As can be seen in Figure 2A, Tap and NXT1 did not increase the levels of GagPolCTE RNA that were detected in total RNA preparations. The CTE-containing RNA was also readily detected in the cytoplasmic fraction even in the absence of transfected Tap and NXT1, and the levels of RNA in the cytoplasm were only slightly increased in the cells cotransfected with Tap and NXT1. This is consistent with our recently published results, showing that the CTE alone mediates efficient export of GagPol RNA in 293T cells (Coyle et al. 2003). In contrast, virtually no GagPolRRE RNA was detected in the cytoplasmic fraction in the absence of Rev, although this RNA was readily detected when total RNA was analyzed (Fig. 2B). This confirms our previous experiments showing that Rev is essential for export of the RRE-containing RNA expressed from the reporter and demonstrates proper fractionation of the cells during the preparation of the cellular extracts.
Figure 2.
Tap and NXT1 do not specifically increase the levels of cytoplasmic CTE RNA in 293T cells. (A) 293T cells (1 × 107) were transfected with 20 μg of the pCMVGagPolCTE reporter plasmid and 5 μg of pCMVSEAP in the presence or absence of 6 μg of pCMVTap, and 3 μg of pcDNANXT1. (B) 293T cells (1 × 107) were transfected with 20 μg of pCMVGagPolRRE and 5 μg of pCMVSEAP in the presence or absence of 5 μg of pCMVRev. Northern blot analyses were performed on cytoplasmic and total poly(A)+ RNA isolated from the 293T cells 65 h after transfection. GagPol- and SEAP-specific mRNAs were detected using specific radiolabeled DNA probes. The blots were analyzed using a Molecular Dynamics PhosphorImager and ImageQuant software. Brackets show the fold difference between the indicated lanes in the levels of the GagPol RNA bands after adjustment for variations in SEAP RNA levels.
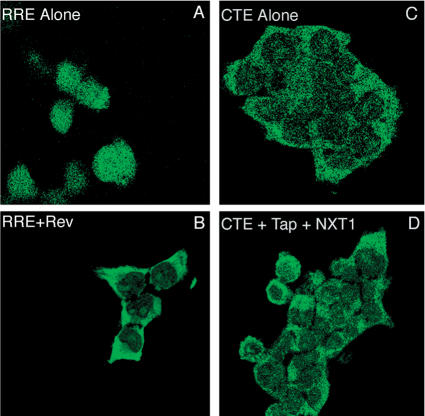
To further analyze the localization of GagPolCTE RNA in the absence or presence of Tap and NXT1, we performed an RNA in situ hybridization experiment using transfected 293T cells. Cells expressing GagPolRRE RNA with or without Rev coexpression were used as controls in this experiment. The results of this experiment are shown in Figure 3. As expected, cells transfected with the GagPolRRE plasmid alone showed only nuclear fluorescence (Fig. 3, upper left panel). In contrast, the fluorescence was almost exclusively cytoplasmic in the presence of Rev (Fig. 3, lower left panel). This is consistent with previous studies and the Northern blot results presented above and confirms that Rev is essential for the cytoplasmic appearance of GagPolRRE RNA. In cells transfected with pCMVGagPolCTE alone, a signal was observed mainly in the cytoplasm, and the localization did not visibly change in the presence of Tap and NXT1 (Fig. 3, right panels). In fact, the localization and intensity of the signal observed in the cells expressing GagPolCTE RNA both in the absence and in the presence of Tap and NXT1 were very similar to those observed with GagPolRRE in the presence of Rev. Taken together with the Northern blot results, this suggested that Tap and NXT1 may function to specifically increase the utilization of CTE-containing RNA in the cytoplasm, as was previously shown for Sam68 (Coyle et al. 2003).
Figure 3.
In situ RNA hybridization of RNA produced from pCMVGagPolCTE and pCMVGagPolRRE. (A,B) Rev coexpression causes relocalization of GagPolRRE RNA to the cytoplasm. (C,D) Tap and NXT1 coexpression does not visibly change the cytoplasmic localization of GagPolCTE RNA. 293T cells were transfected with the indicated constructs as described in Figure 1A,B. At 48 h after transfection, cells were fixed and hybridized to an anti-sense digoxigenin (DIG)-labeled DNA oligonucleotide probe complementary to a Gag sequence. Hybridization was detected with an anti-DIG antibody conjugated to FITC. Hybridization signals were visualized by LSM5 PASCAL confocal microscope (Zeiss).
Tap and NXT1 increase the rate of synthesis of proteins translated from GagPolCTE RNA
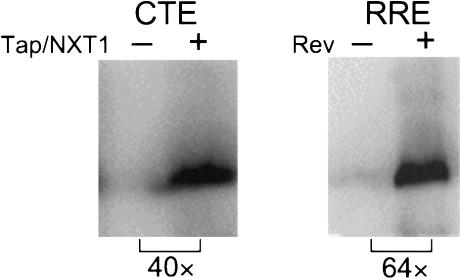
Because our results showed that Tap and NXT1 increased p24 accumulation without a corresponding increase in RNA levels, we next decided to analyze whether this was a direct result of an increased rate of protein synthesis. GagPolCTE and GagPolRRE constructs normally give rise to polyproteins that are cleaved by the HIV protease and bud from transfected cells as virus-like particles. Because of this, it is difficult to directly visualize the primary translation product in transfected cells. This precludes a straightforward measurement of the rate of protein synthesis by examining the accumulating protein directly. To overcome this problem, we therefore constructed two new plasmids, pCMVGagPolCTE(myr- pro-) and CMVGagPolRRE(myr- pro-). These plasmids have point mutations in the HIV protease active site that make the protease inactive (pro-) and in the N terminus of Gag that destroy the myristoylation signal (myr-). Thus only nonmyristoylated Pr55Gag and Pr160Gag-Pol precursors are expected to be produced from these vectors (Smith et al. 1993). Because myristoylation is required for virus particle budding, these proteins would be expected to remain inside the transfected cell in an uncleaved form. This was verified by showing that no p24 appeared in the medium supernatant (data not shown) and that only uncleaved Pr55Gag proteins accumulated in the cytoplasm of transfected cells (see below).
In initial experiments, we transfected 293T cells with these modified constructs ±Rev or Tap and NXT1. Pr55Gag accumulation was measured by Western blot analysis 65 h posttransfection (Fig. 4). As can be seen in this figure, Tap and NXT1 increased protein accumulation translated from the CTE construct ∼40-fold, whereas Rev increased expression of Pr55Gag from the RRE construct 64-fold.
Figure 4.
Analysis of Pr55Gag in 293T cells. 293T cells (3 × 106) were transfected with 5 μg of pCMVGagPolRRE(myr- pro-) or 5 μg of pCMVGagPolCTE(myr- pro-) reporter constructs, with or without 1 μg of pCMVRev or 2 μg of pCMVTap and 1 μg of pcDNANXT1 as indicated. Cells were harvested at 65 h and analyzed by Western blot. The Western blot was performed on whole-cell lysates of transfected cells with an anti-p24 monoclonal antibody, and 125I-coupled protein A for detection. Brackets show the fold difference between the indicated lanes in the levels of the Pr55Gag protein bands.
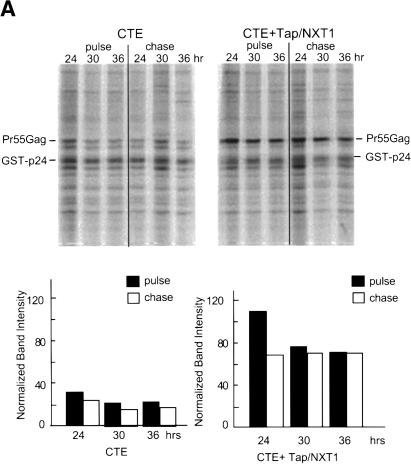
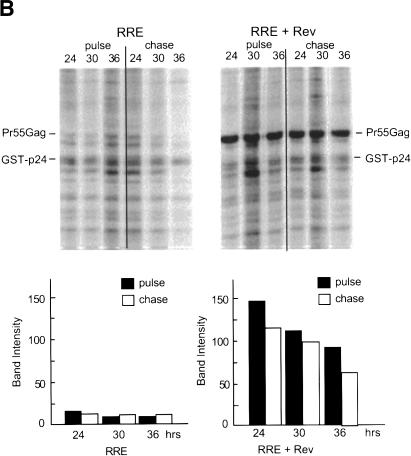
To directly analyze if Tap/NXT1 affected the rate of protein synthesis and/or stability of the synthesized proteins, we performed pulse-chase experiments on cells transfected with the modified (myr- pro-) CTE construct. As before, we compared this with protein synthesis from the RRE ± Rev. At 24, 30, and 36 h posttransfection, cells were pulsed with 35S-Translabel (methionine and cysteine) for 15 min. The cells were then washed twice with PBS and harvested (pulse), or re-fed with growth medium containing excess cold amino acids, and harvested 10 h later (chase). Cell lysates were subjected to immunoprecipitation using a mouse anti-Gag(p24) monoclonal antibody and analyzed for Pr55Gag by SDS-PAGE and autoradiography. To compensate for potential differences in immunoprecipitation efficiency between samples, in vitro purified GST-p24 protein was added to the cell lysates before the immunoprecipitation. A second immunoprecipitation was also performed on the supernatants after the initial immunoprecipitation to judge the efficiency of the first precipitation. Greater than 95% of each protein was pulled down in the first precipitation (data not shown).
The results of this pulse chase experiment are shown in Figure 5. Only a very small amount of Pr55Gag was observed in the cells transfected with the GagPolCTE plasmid alone, irrespective of whether cells were labeled at 24, 30, or 36 h after transfection (Fig. 5A). The amount of labeled protein did not decrease significantly after a 10-h chase, indicating that the proteins produced were stable during this period. A significant increase in the amount of Pr55Gag produced was observed in the presence of Tap and NXT1. Again, the protein produced was stable during a 10-h chase period. In the case of the GagPolRRE plasmid (Fig. 5B), as expected, little Pr55Gag was seen in the absence of Rev, whereas large amounts of stable Pr55Gag were observed in the presence of this protein. These results demonstrate that expression of Tap and NXT1 increases the rate of protein synthesis from the GagPolCTE RNA in transfected 293T cells. However, protein stability is not majorly affected by the expression of these proteins, although the calculated half-lives from the data shown in Figure 5 indicate that the protein made in the absence of Tap/NXT1 is somewhat less stable than Pr55Gag expressed in the presence of these proteins (24 h vs. 36 h). This clearly cannot account for the differences in the rate of synthesis we observed in the 15-min pulse, and we do not even know if it is significant because of the difficulties in accurately measuring the low amounts of proteins that are synthesized in the absence of Tap/NXT1.
Figure 5.
Pulse-chase analysis of Pr55Gag protein. (A) Protein expressed from pCMVGagPolCTE(myr- pro-) in transfected 293T cells at various times posttransfection. 293T cells (3 × 106) were transfected with 5 μg of pCMVGagPolCTE(myr- pro-) with or without 2 μg of pCMVTap and 1 μg of pcDNANXT1. (B) Protein expressed from pCMVGagPolRRE (myr- pro-) in transfected 293T cells at various times posttransfection. 293T cells (3 × 106) were transfected with 5 μg of pCMVGagPolRRE (myr- pro-) with or without 1 μg of pCMVRev. Cells were pulsed with 35S Trans-label (methionine/cysteine) for 15 min at the indicated times after transfection and chased for 10 h. (Upper panels) Lysates were made, labeled GST-p24 was added as a recovery control, and immunoprecipitation was performed using an anti-p24 monoclonal antibody (183-H12-5C). (Lower panels) The precipitates were analyzed on a SDS polyacrylamide gel using a PhosphorImager. The locations of the immunoprecipitated Pr55Gag and the control GST-p24 protein are indicated. The amount of radioactivity in each Pr55Gag protein band is shown for each time point both before and after the chase period. The data have been normalized using the intensity of the recovery control protein GST-p24 in each sample. The analysis was performed using ImageQuant software. Band intensity units represent the radioactivity in each band derived from the pixel intensity on the PhosphorImager screen.
Tap and NXT1 promote polyribosome association of CTE-containing RNA
To directly determine the fraction of GagPolCTE RNA that was free in the cytoplasm or associated with polyribosomes, we performed a polyribosome analysis on cells transfected with GagPolCTE ± Tap and NXT1. As before, cells transfected with GagPolRRE ± Rev served as controls. To perform the polyribosome analysis, Triton X-100-treated cell lysates were centrifuged on sucrose gradients that were fractionated. RNA was isolated from each of the fractions and analyzed by semiquantitative RT-PCR using primers specific for the GagPol mRNA (Fig. 6). To verify that the RT-PCR reactions were indeed semiquantitative, we performed a similar analysis on 10-fold diluted RNA samples. This gave signals that were 5-10-fold lower (data not shown).
Figure 6.
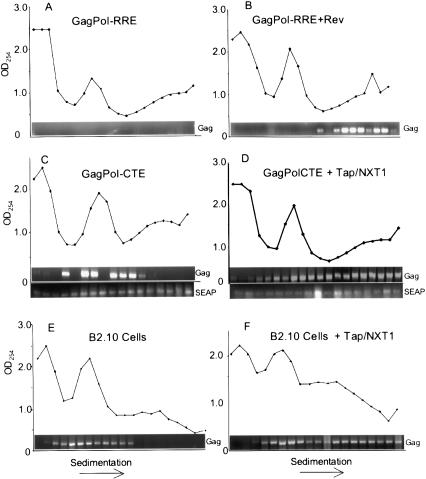
Polyribosome profile analysis of GagPol mRNA. (A-D) Polyribosome profiles in transfected 293T cells. 293T cells (1 × 107) were transfected with the indicated constructs as described in Figure 2, A and B. (E,F) Polyribosome profiles in B2.10 cells stably expressing GagPolCTE RNA. B2.10 cells (1 × 107) that were transfected with 6 μg of pCMVTap and 3 μg of pcDNANXT1 or not transfected were analyzed. Forty-eight hours after transfection, cells were harvested and subjected to sucrose gradients centrifugation as described in Materials and Methods. The gradients were fractionated, and the O.D.254 of each fraction was measured. RNA was then prepared from each fraction by phenol extraction and assayed for GagPol or SEAP mRNA by semiquantitative RT-PCR. The absorbance profile of each gradient, as well as the RT-PCR product derived from each fraction, is shown.
In cells transfected with GagPolRRE, no specific RNA signal was detected in any part of the gradient (Fig. 6A). This is consistent with the fact that no RRE RNA reaches the cytoplasm in the absence of Rev. In contrast, a strong RNA signal was obtained in the polyribosomal fractions in cells cotransfected with Rev (Fig. 6B). Little or no RNA was detected in the lighter fractions of the gradient.
In cells transfected with GagPolCTE alone, specific RNA was detected in several of the lighter gradient fractions, but little RNA was associated with polyribosomes (Fig. 6C). Some of the cytoplasmic RNA sedimented in the “monosome” region of the gradient, whereas additional RNA was detected in lighter fractions. When cotransfected with Tap and NXT1, a substantial portion of the RNA was shifted to the polyribosomal part of the gradient (Fig. 6D). However, some of the RNA remained in the nonpolyribosomal part of the gradient, probably reflecting the fact that not all cells are cotransfected with both Tap and NXT1. In contrast to the CTE RNA, the spliced SEAP mRNA expressed from the control plasmid was present in the polyribosomal fractions both in the absence and presence of cotransfected Tap/NXT1.
These results demonstrate that, in the absence of exogenously added Tap and NXT1, GagPolCTE RNA resides in stable complexes in the cytoplasm of 293T cells that are not associated with polyribosomes. Coexpression of Tap and NXT1 causes the movement of this RNA into polyribosomes. This is consistent with all of the p24 and Pr55Gag expression results presented above. The data show that Tap and NXT1 promote polyribosome association and protein translation of GagPolCTE RNA, in addition to the role in RNA export previously proposed for these proteins.
All of the above experiments were performed on cells that were transiently transfected with GagPolCTE RNA. To analyze whether Tap and NXT1 would have similar effects on a CTE-containing RNA that was expressed from integrated DNA, we established 293T cell lines using pCMVGagPolCTE and a plasmid expressing hygromycin. Several independent cell lines were established and shown to produce only very low amounts of p24. To analyze the effects of Tap and NXT1, we chose one of these cell lines (B2.10) for further experiments. Cotransfection of Tap- and NXT1-expressing plasmids led to a very significant increase in p24 expression from this cell line (data not shown). A polyribosome analysis of transfected cells demonstrated that whereas the GagPolCTE RNA was present almost exclusively in the nonpolyribosomal fractions in cells lacking exogenously added Tap and NXT1, a significant amount of RNA shifted to the polyribosomal fractions after transfection of plasmids expressing these proteins (Fig. 6E,F). These results confirmed that Tap and NXT1 promote polyribosome association and translation of proteins also in cell lines stably expressing intron-containing RNA.
Tap, but not NXT1, associates with polyribosomes in transfected 293T cells
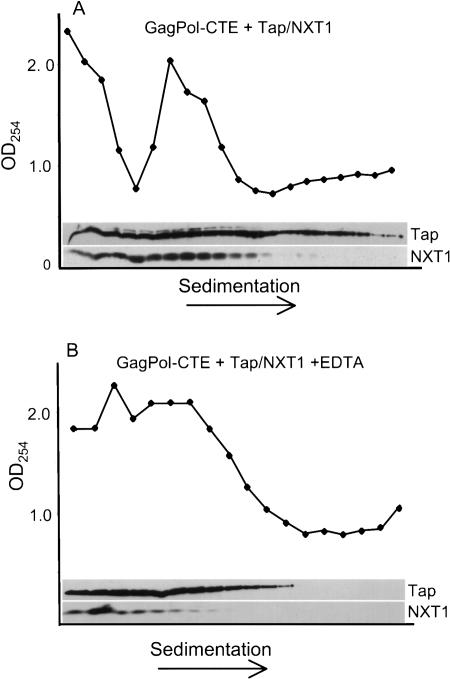
Finally, to determine whether either Tap or the NXT1 protein was associated with polyribosomes, we subjected the fractions from the polyribosome gradient to immunoprecipitation using Flag antibodies coupled to agarose beads. These antibodies immunoprecipitate both Tap and NXT1, because both of the expressed proteins contain N-terminal Flag epitopes. This was confirmed in immunoprecipitation experiments on lysates from transfected cells, which showed that both Tap and NXT 1 were efficiently immunoprecipitated using Flag beads (data not shown). When gradient fractions were analyzed, both Tap and NXT1 proteins were immunoprecipitated from the lighter portions of the gradient (Fig. 7A). In contrast, only Tap was detected in the polyribosome fractions. When EDTA was added to the lysate to dissociate the polyribosomes (Fig. 7B), Tap was no longer present in the denser part of the gradient and was found primarily in the same fractions where the GagPolCTE mRNP complex was detected in the absence of Tap/NXT1 (Fig. 6C). This is consistent with the fact that the EDTA was used at a concentration of 15 mM, which has been shown to cause the dissociation of ribosomes from mRNAs without disrupting the majority of nonribosomal RNA-protein complexes (Calzone et al. 1982; Johannes and Sarnow 1998). These results suggest that Tap stays associated with the CTE-containing RNA in polysomes or binds to ribosomal components, whereas NXT1 is dissociated from the translation complex prior to, or at the start of translation.
Figure 7.
Tap, but not NXT1, is present in the polyribosome fractions containing GagPolCTE RNA. (A) 293T cells (1 × 107) were transfected with 15 μg of pCMVGagPolCTE, 6 μg of pcDNAFLAGTap, and 3 μg of pcDNANXT1 and subjected to polyribosome analysis as described in Materials and Methods. The gradients were fractionated and the O.D.254 of each fraction was measured. Proteins from each fraction were immunoprecipitated using an anti-FLAG antibody (Sigma), and the precipitates were resolved by SDS-PAGE and analyzed by Western blotting using the same anti-FLAG antibody. The bands for Tap and NXT1 proteins are indicated. (B) The same as in A, except that 15 mM EDTA was added to the lysate before gradient analysis. This has been shown to cause the dissociation of ribosomes from mRNAs without disrupting the majority of nonribosomal RNA-protein complexes (Calzone et al. 1982; Johannes and Sarnow 1998).
Discussion
The results presented in this paper indicate that Tap and NXT1 or NXT2 play a role in promoting efficient translation of unspliced RNA that contains the MPMV CTE, in addition to the previously proposed roles for these proteins in mRNA export. They also establish a role for NXT in CTE function.
Although Tap and NXT predominantly localize to the nucleus at “steady state,” previous studies have shown that they are shuttling proteins (Bear et al. 1999; Black et al. 1999; Kang and Cullen 1999). The experiments presented here show that both the Tap and NXT1 proteins can be readily detected in the cytoplasm of transfected cells. However, only Tap is associated with polyribosomes. This suggests that NXT1 leaves the RNA concomitant with translation initiation, whereas Tap remains on the RNA or associates with ribosomal components. The association of Tap/NXT-RNA complexes with the translation initiation machinery may lead to a remodeling of the mRNP complex, resulting in the release of NXT. Recent experiments using intron-containing and intronless cellular cDNA constructs have indicated that the EJC may serve to promote translation (Le Hir et al. 2003; Lu and Cullen 2003; Nott et al. 2003), in addition to its previously proposed roles in RNA export and nonsense-mediated decay (NMD; Maquat and Carmichael 2001). Because there is evidence that the Tap/NXT heterodimer associates with EJC complexes in the nucleus and accompanies the RNA to the cytoplasm (Kim et al. 2001; Le Hir et al. 2001), it seems possible that the ability of the EJC to promote translation could be another manifestation of Tap/NXT function at the cytoplasmic level. In this context, it is of interest to note that it has previously been shown that Y14, a component of the EJC complex, remains associated with mRNAs in the cytoplasm until they are translated (Dostie and Dreyfuss 2002), and translation is required to remove Y14 from the mRNA complex. In view of these studies, we propose that the ability of the CTE to directly recruit the Tap/NXT heterodimer could serve as a “mark” in lieu of components of the EJC to promote efficient translation of the unspliced RNA.
Another recent study using 293T cells showed that Tap and NXT1 or NXT2 increased CAT expression from unspliced RNA containing the CAT reporter ORF in the HIV env intron (Braun et al. 2001). Although the effects of overexpression of Tap and NXT seen in this study were attributed entirely to increased RNA export, this conclusion was based mainly on CAT expression levels. Thus, it seems plausible that at least some of the observed enhancement could reflect effects on translation similar to the ones seen with the GagPol reporters that were used in this study.
Tap is a member of a family of evolutionarily conserved genes, the NXF (Nuclear Export Factor) family (Herold et al. 2000). As indicated by the name, the initial belief was that all of these proteins would serve more or less related roles in RNA export. It was therefore surprising, when it was shown that at least one of the NXF proteins (NXF5) localizes exclusively to the cytoplasm. This protein is expressed in neuronal cells, and mutations in this gene have been linked to a hereditary form of male mental retardation (Jun et al. 2001; Frints et al. 2003). The current hypothesis is that this protein plays a role in neuronal mRNA localization or translation or both. Thus a cytoplasmic role for NXF proteins is not unprecedented, and it will be of clear interest to see whether other proteins in this family also function in cellular processes other than export.
The fact that moderate overexpression of Tap and NXT results in dramatic effects on protein expression suggests that 293T cells are deficient in Tap and NXT function. This is surprising, because Tap and the NXT proteins have been proposed to play major roles in export of most, if not all mRNAs, and the CTE-containing RNA is efficiently exported from the nucleus in 293T cells. If the endogenous Tap/NXT proteins are sufficient to export the RNA to the cytoplasm, it is not intuitive why more should be needed for efficient translation. One possibility is that the essential export function of Tap/NXT is served by other export receptor proteins in these cells. Another possibility is that Tap is altered in 293T cells, so that its export function remains intact, but its translation enhancement function is defective. Experiments are presently under way to determine if the CTE RNA and other mRNAs are still efficiently exported to the cytoplasm in 293T cells, when the levels of Tap and NXT are further reduced using transdominant negative proteins and siRNA strategies.
In this study, GagPolCTE RNA was shown to be inefficiently translated in 293T cells in the absence of additional Tap/NXT1 expression. We have previously shown that expression of the cellular Sam68 protein also leads to increased cytoplasmic utilization of the CTE-containing RNA in 293T cells (Coyle et al. 2003). In addition, we showed that Sam68 and Tap/NXT1 were able to synergistically activate expression from the GagPolCTE RNA in quail cells. A similar synergistic effect of Sam68 and Tap/NXT1 has also been observed in 293T cells (data not shown), suggesting the possibility that these factors act in concert to promote translation. In contrast to the results with GagPolCTE, RRE-containing RNA is efficiently translated in 293T cells in the presence of the Rev protein. This suggests that Rev serves roles in the cytoplasm, in addition to the well-established role in RNA export. The fact that protein expression from the RRE construct decreases in the presence of Tap/NXT1 might reflect a competition for limiting cytoplasmic cellular factors involved in translational regulation.
Even in the absence of transfected Tap/NXT or Sam68, the exported GagPolCTE RNA is present in stable cytoplasmic mRNP complexes in 293T cells. It will be of clear interest to determine the components of this complex. Also, because RNAs that are exported to the cytoplasm without appropriate EJC complexes are believed to be subject to degradation by the cytoplasmic NMD machinery, it seems possible that the presence of a CTE enables the RNA to avoid this fate.
Previous studies have shown that another family of proteins, the ELAV or HuR proteins, appear to bind to elements in the 3′-UTR of certain classes of mRNAs in the nucleus, ultimately promoting their translation in the cytoplasm (Antic and Keene 1998; Fan and Steitz 1998; Antic et al. 1999; Brennan et al. 2000). This has been proposed to be part of a mammalian posttranscriptional operon mechanism, in which nuclear mRNA-binding proteins regulate mRNAs as subpopulations during cell growth and development (Keene and Tenenbaum 2002). Our results seem to provide further evidence for the regulation of RNAs in this manner.
Materials and methods
Plasmids
To facilitate identification, all of the plasmids used in this study were indexed as numbers in the form pHRXXXX. The subgenomic HIV-1 reporter constructs pCMVGagPolRRE (pHR354) and pCMVGagPolCTE (pHR1361; Srinivasakumar et al. 1997); pCMVTap (pHR2128), a plasmid that expresses the Tap protein (Guzik et al. 2001); pcDNANXT1 (pHR2283; Black et al. 1999), a plasmid that expresses a FLAG-tagged NXT1 protein; pCMVRev (pHR30; Smith et al. 1993), a plasmid that expresses the HIV Rev protein; and pCMVSEAP (pHR1831), a plasmid that expresses secreted alkaline phosphatase (SEAP; Cullen and Malim 1992), have been described previously. pcDNANXT2 (pHR2606) was constructed by inserting the NcoI fragment from pGEX-p15a (a kind gift from Elisa Izaurralde, EMBL) into the BamHI site of pcDNA-FLAG. pcDNAFLAGTap (pHR2352) and pcDNANXT1R134Q (pHR2600) were kindly provided by Lyne Levesque and Bryce Paschal (University of Virginia). pCMVGagPolRRE(myr- pro-) (pHR2899) and pCMVGagPolCTE(myr- pro-) (pHR2900), containing point mutations in the myristoylation signal and pro region of the HIV gagpol gene, were made by SOE PCR (Horton et al. 1990; Horton 1995). To make the myristoylation signal mutation (myr-), a G-to-T change was made in the second codon of Gag, resulting in a glycine-to-valine substitution in the second amino acid of MA(p17). To make the protease mutation (pro-), an A was changed to a G in a codon that resulted in an aspartate-to-glycine mutation in the active site of the protease at amino acid 25.
Cell lines and transient transfections
293T/17 cells (Pear et al. 1993) were maintained in Iscove's Modified Dulbecco's medium supplemented with 10% bovine calf serum. 293T/17 cells were transfected by using a calcium phosphate transfection protocol (Graham and van der Eb 1973). B2.10, a 293T-derived cell line stable expressing GagPolCTE RNA, has been described previously (Coyle et al. 2003). Transfections were performed with the amounts of plasmids stated in the figure legends.
p24 ELISA and SEAP quantitation
Supernatants from transfected cells were collected at 65 to 72 h posttransfection and subjected to a short spin in a microcentrifuge to remove residual cells and debris. p24 expression levels were determined by an ELISA protocol using a p24 monoclonal antibody (183-H12-5C) and pooled human anti-HIV immunoglobulin G (Wehrly and Chesebro 1997). The p24 antibody was obtained from the AIDS Research and Reference Reagent Program and was contributed by Bruce Chesebro (National Institute of Allergy and Infectious Diseases-Rocky Mountain Laboratories). SEAP activity in the supernatants was measured with the Phospha-Light Chemiluminescent Reporter Kit (Tropix).
Western blot analysis and antibodies
The Western blot analysis was performed essentially as previously described (Hammarskjöld et al. 1986; Guzik et al. 2001). For detection of HIV Gag proteins, blots were probed with the mouse anti-p24 monoclonal antibody (183-H12-5C). For detection of FLAGTap and FLAGNXT1, blots were probed with anti-FLAG M2 antibody (Sigma). To detect endogenous Tap, blots were probed with a polyclonal Tap antibody raised in rabbits against a GST-Tap(61-619) fusion protein produced from the vector pGEX2T-Tap (pHR2123). Blots were normally developed using a horseradish peroxidase-coupled second antibody and the ECL system (Amersham Biosciences). For quantitation, after washing to remove unbound first antibody, blots were probed with a second species-specific antibody and 10 μCi/ml of 125I-conjugated protein A. Quantitation was performed with a Molecular Dynamics PhosphorImager and ImageQuant analysis software.
Purification of pGEXp24 fusion protein
To prepare glutathione S-transferase (GST)-p24, Escherichia coli DH5α cells were transformed with the plasmid pGEXp24 (pHR163) and cultured in 2 mL of M9 minimal medium containing 18 amino acids lacking methionine and cysteine at 37°C. The overnight culture was transferred to 50 mL of medium, and incubation continued until the O.D. at 600 nm reached 0.6 ∼ 0.8. At this time, 1 mM isopropylthiogalactopyranoside (IPTG) was added, and incubation was continued for an additional 4 h. Cells were then labeled with 5 μCi/mL 35S Translabel (methionine and cysteine) for 1 h. Cells were harvested and disrupted by sonication in lysis buffer (50 mM Tris-HCl at pH 8.0, 1% Triton X-100, and 1× protease inhibitor cocktail; Sigma). Cleared supernatant was incubated with glutathione-agarose beads, and the bound GST fusion protein was eluted with freshly prepared elution buffer (100 mM Tris-HCl at pH 8.0 and 10 mM glutathione), containing the protease inhibitor cocktail (Sigma).
35S labeling and immunoprecipitations
Transfected 293T cells were washed twice with methionine- and cysteine-free Dulbecco's modified Eagle's medium (GIBCO/Invitrogen) and then incubated for 15 min at 37°C in labeling medium containing 200 μCi 35S Trans-label (ICN) and 10% dialyzed bovine calf serum. Cells were lysed by adding 1 mL of lysis buffer (150 mM NaCl, 10 mM Tris-HCl at pH 7.4, 1% NP-40, 1% deoxycholate, 1.5% sodium dodecyl sulfate). Lysates were cleared by centrifugation, and protein concentration was determined by colorimetric analysis (Bio-Rad). 35S-labeled GST-p24 protein was added as an internal control. Pr55Gag was immunoprecipitated from 200 μg of total cell lysates with 10 μg of mouse anti-p24 monoclonal antibody (183-H12-5C) followed by 30 μL of 50% protein G-Sepharose beads (Amersham Biosciences). The protein-bead mixture was incubated for 1.5 h at 4°C. Samples were washed two times with 500 μL of lysis buffer and one time with 500 μL of wash buffer (10 mM Tris-HCl at pH 8.0, 100 mM NaCl, 1 mM EDTA at pH 8.0) with centrifugation at 500g for 3 min. The beads were then resuspended in 1× SDS sample buffer, boiled for 5 min, and centrifuged briefly. Eluted proteins were resolved by electrophoresis on a 15% SDS-polyacrylamide gel. Quantitation was performed using a Molecular Dynamics PhosphorImager on the dried gel and ImageQuant analysis software.
Polyribosome analysis
Transfected cells (2 × 107) were exposed to 50 μg/mL cycloheximide for 5 min at 48 h posttransfection, harvested by trypsinization, washed twice with cold PBS, and resuspended in 0.5 mL of cold RSB buffer (10 mM Tris-HCl at pH 7.4, 10 mM NaCl, 3 mM MgCl2). The trypsin and PBS also contained 50 μg/mL cycloheximide. After the addition of an equal volume of RSB containing 1% Triton X-100, 1% deoxycholate, and 2% Tween 20, the suspension was incubated for 10 min on ice and centrifuged for 10 min at 10,000g. In some experiments, EDTA was added to a final concentration of 15 mM in order to disrupt the polysomes (Calzone et al. 1982; Johannes and Sarnow 1998). The supernatant lysates were loaded onto gradients of 10% to 50% (w/v) sucrose in 10 mM Tris-HCl (pH 7.6), 75 mM KCl, and 3 mM MgCl2. After centrifugation in an SW41 rotor for 2 h at 36,000 rpm and 4°C, the gradient was collected from the top into 20 fractions and the O.D.254 was determined for each fraction. Sodium dodecyl sulfate (SDS) was added to a final concentration of 0.5%, and each fraction was extracted with phenol and then twice with phenol-chloroform. The nucleic acids in the aqueous phase (the bottom phase at high sucrose concentrations) were precipitated with ethanol and resuspended in water. After DNAse I (NEB) treatment, they were assayed by the reverse transcriptase-polymerase chain reaction method (RTPCR). The primers for RT-PCR of the GagPol mRNA were top strand primer, 5′-AGACCGGTTCTATAAAACTCTAAGAGCCG-3′, and bottom strand primer, 5′-GGCCAGATCTTCCCTAAAAAAATTAGCCTGTCTCTCAG-3′.
The primers used for RT-PCR of the SEAP mRNA were top strand primer, 5′-TTCCAGACCATTGGCTTGAGT-3′, and bottom strand primer, 5′-TTGCCGCGTGTCGTGTT-3′.
RNA was used to synthesize cDNA using these primers and AMV-RT (Roche) for 45 min at 48°C. The cDNAs were amplified using PCR for 25 cycles. Each cycle consisted of 30 sec of denaturation at 94°C and 120 sec of annealing/extension at 60°C. At the end of the PCR cycles, incubation was continued for 7 min at 62°C. PCR products were analyzed on a 1% agarose gel. To ensure that these assays were semiquantitative, 10-fold dilutions of the samples were also amplified and verified to give signals that were ∼10-fold lower.
RNA fractionation and Northern blot analysis
The methods used for nuclear and cytoplasmic RNA extraction, poly(A) mRNA selection, and Northern blot analysis were previously described (Hammarskjöld et al. 1986, 1994). 293T cells were harvested at 65 h posttransfection. A SacI-Bgl II (nucleotides 682-2093) fragment of the HIV-1 BH10 clone and BamHI fragment of the human SEAP cDNA (nucleotides 213-1698) were labeled with [α-32P]dCTP by using the T7 Quickprime Kit (Pharmacia). Northern blots were quantitated with a Molecular Dynamics PhosphorImager and ImageQuant analysis software.
RNA in situ hybridization
293T cells were cotransfected with 5 μg of either pCMVGagPolRRE or CTE reporter plasmids with or without 1 μg of pCMVRev (pHR30) or 2 μg of pCMVTap (pHR2128) and 1 μg of pcDNANXT1 (pHR2283). Twenty-four hours posttransfection, cells were seeded onto 0.1% gelatin-coated coverslips and incubated for another 24 h. Cells were then fixed with 4% paraformaldehyde-PBS, and in situ hybridization was carried out as previously described (Coyle et al. 2003).
Acknowledgments
We thank Eliza Izaurralde (EMBL, Heidelberg), Lyne Levesque (University of Virginia), and Bryce Paschal (University of Virginia) for gifts of reagents used in this study; and Bruce Chesebro for help with the establishment of the p24 ELISA and his gift to the AIDS Reagent Repository of p24 monoclonal antibodies. Joy Morgenegg provided expert tissue culture assistance. This work was supported by National Institutes of Health grants AI34721 and AI054335 to M.-L.H. and AI47008 to D.R. Salary support for M.L.H. and D.R was provided by the Charles H. Ross Jr. and Myles H. Thaler Endowments at the University of Virginia.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1155703.
References
- Antic D. and Keene, J.D. 1998. Messenger ribonucleoprotein complexes containing human ELAV proteins: Interactions with cytoskeleton and translational apparatus. J. Cell Sci. 111 (Pt 2): 183-197. [DOI] [PubMed] [Google Scholar]
- Antic D., Lu, N., and Keene, J.D. 1999. ELAV tumor antigen, Hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes & Dev. 13: 449-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachi A., Braun, I.C., Rodrigues, J.P., Pante, N., Ribbeck, K., von Kobbe, C., Kutay, U., Wilm, M., Gorlich, D., Carmo-Fonseca, M., et al. 2000. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA 6: 136-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear J., Tan, W., Zolotukhin, A.S., Tabernero, C., Hudson, E.A., and Felber, B.K. 1999. Identification of novel import and export signals of human TAP, the protein that binds to the constitutive transport element of the type D retrovirus mRNAs. Mol. Cell. Biol. 19: 6306-6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowitz R., Fisher, J., and Goff, S.P. 1996. RNA packaging. In Morphogenesis and maturation of retroviruses (ed. H.-G. Krausslich), pp. 177-218. Springer, Berlin.
- Black B.E., Levesque, L., Holaska, J.M., Wood, T.C., and Paschal, B.M. 1999. Identification of an NTF2-related factor that binds Ran-GTP and regulates nuclear protein export. Mol. Cell. Biol. 19: 8616-8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun I.C., Rohrbach, E., Schmitt, C., and Izaurralde, E. 1999. TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J. 18: 1953-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun I.C., Herold, A., Rode, M., Conti, E., and Izaurralde, E. 2001. Overexpression of TAP/p15 heterodimers bypasses nuclear retention and stimulates nuclear mRNA export. J. Biol. Chem. 276: 20536-20543. [DOI] [PubMed] [Google Scholar]
- Bray M., Prasad, S., Dubay, J.W., Hunter, E., Jeang, K.T., Rekosh, D., and Hammarskjöld, M.L. 1994. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc. Natl. Acad. Sci. 91: 1256-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C.M., Gallouzi, I.E., and Steitz, J.A. 2000. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell Biol. 151: 1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzone F.J., Angerer, R.C., and Gorovsky, M.A. 1982. Regulation of protein synthesis in Tetrahymena: Isolation and characterization of polysomes by gel filtration and precipitation at pH 5.3. Nucleic Acids Res. 10: 2145-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D.D. and Sharp, P.A. 1989. Regulation by HIV Rev depends upon recognition of splice sites. Cell 59: 789-795. [DOI] [PubMed] [Google Scholar]
- Coyle J.H., Guzik, B.W., Bor, Y.C., Jin, L., Eisner-Smerage, L., Taylor, S.J., Rekosh, D., and Hammarskjöld, M.L. 2003. Sam68 enhances the cytoplasmic utilization of intron-containing RNA and is functionally regulated by the nuclear kinase Sik/BRK. Mol. Cell. Biol. 23: 92-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B.R. and Malim, M.H. 1992. Secreted placental alkaline phosphatase as a eukaryotic reporter gene. Methods Enzymol. 216: 362-368. [DOI] [PubMed] [Google Scholar]
- Dostie J. and Dreyfuss, G. 2002. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr. Biol. 12: 1060-1067. [DOI] [PubMed] [Google Scholar]
- Ernst R.K., Bray, M., Rekosh, D., and Hammarskjöld, M.-L. 1997a. A structured retroviral RNA element that mediates nucleocytoplasmic export of intron-containing RNA. Mol. Cell. Biol. 17: 135-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1997b. Secondary structure and mutational analysis of the Mason-Pfizer monkey virus RNA constitutive transport element. RNA 3: 210-222. [PMC free article] [PubMed] [Google Scholar]
- Fan X.C. and Steitz, J.A. 1998. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 17: 3448-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M., Ohno, M., Yoshida, M., and Mattaj, I.W. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90: 1051-1060. [DOI] [PubMed] [Google Scholar]
- Fribourg S., Braun, I.C., Izaurralde, E., and Conti, E. 2001. Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Mol. Cell 8: 645-656. [DOI] [PubMed] [Google Scholar]
- Frints S.G., Jun, L., Fryns, J.P., Devriendt, K., Teulingkx, R., Berghe, L.V., Vos, B.D., Borghgraef, M., Chelly, J., Portes, V.D., et al. 2003. Inv(X)(p21.1;q22.1) in a man with mental retardation, short stature, general muscle wasting, and facial dysmorphism: Clinical study and mutation analysis of the NXF5 Gene. Am. J. Med. Genet. 119A: 367-374. [DOI] [PubMed] [Google Scholar]
- Graham F.L. and van der Eb, A.J. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52: 456-467. [DOI] [PubMed] [Google Scholar]
- Gruter P., Tabernero, C., von, K.C., Schmitt, C., Saavedra, C., Bachi, A., Wilm, M., Felber, B.K., and Izaurralde, E. 1998. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell 1: 649-659. [DOI] [PubMed] [Google Scholar]
- Guzik B.W., Levesque, L., Prasad, S., Bor, Y.C., Black, B.E., Paschal, B.M., Rekosh, D., and Hammarskjöld, M.L. 2001. NXT1 (p15) is a crucial cellular cofactor in TAP-dependent export of intron-containing RNA in mammalian cells. Mol. Cell. Biol. 21: 2545-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadzopoulou-Cladaras M., Felber, B.K., Cladaras, C., Athanassopoulos, A., Tse, A., and Pavlakis, G.N. 1989. The rev (trs/art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a cis-acting sequence in the env region. J. Virol. 63: 1265-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarskjöld M.L. 1997. Regulation of retroviral RNA export. Semin. Cell Dev. Biol. 8: 83-90. [DOI] [PubMed] [Google Scholar]
- ____. 2001. Constitutive transport element-mediated nuclear export. Curr. Top. Microbiol. Immunol. 259: 77-93. [DOI] [PubMed] [Google Scholar]
- Hammarskjöld M.L., Wang, S.-C., and Klein, G. 1986. High-level expression of the Epstein-Barr virus EBNA1 protein in CV1 cells and human lymphoid cells using a SV40 late replacement vector. Gene 43: 41-50. [DOI] [PubMed] [Google Scholar]
- Hammarskjöld M.L., Heimer, J., Hammarskjöld, B., Sangwan, I., Albert, L., and Rekosh, D. 1989. Regulation of human immunodeficiency virus env expression by the rev gene product. J. Virol. 63: 1959-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarskjöld M.L., Li, H., Rekosh, D., and Prasad, S. 1994. Human immunodeficiency virus env expression becomes Rev-independent if the env region is not defined as an intron. J. Virol. 68: 951-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold A., Suyama, M., Rodrigues, J.P., Braun, I.C., Kutay, U., Carmo-Fonseca, M., Bork, P., and Izaurralde, E. 2000. TAP (NXF1) belongs to a multigene family of putative RNA export factors with a conserved modular architecture. Mol. Cell. Biol. 20: 8996-9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton R.M. 1995. PCR-mediated recombination and muta-genesis. SOEing together tailor-made genes. Mol. Biotechnol. 3: 93-99. [DOI] [PubMed] [Google Scholar]
- Horton R.M., Cai, Z.L., Ho, S.N., and Pease, L.R. 1990. Gene splicing by overlap extension: Tailor-made genes using the polymerase chain reaction. Biotechniques 8: 528-535. [PubMed] [Google Scholar]
- Johannes G. and Sarnow, P. 1998. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA 4: 1500-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun L., Frints, S., Duhamel, H., Herold, A., Abad-Rodrigues, J., Dotti, C., Izaurralde, E., Marynen, P., and Froyen, G. 2001. NXF5, a novel member of the nuclear RNA export factor family, is lost in a male patient with a syndromic form of mental retardation. Curr. Biol. 11: 1381-1391. [DOI] [PubMed] [Google Scholar]
- Kang Y. and Cullen, B.R. 1999. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes & Dev. 13: 1126-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J., Strasser, K., Podtelejnikov, A., Mann, M., Jung, J.U., and Hurt, E. 1999. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 18: 2593-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene J.D. and Tenenbaum, S.A. 2002. Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell 9: 1161-1167. [DOI] [PubMed] [Google Scholar]
- Kim V.N., Yong, J., Kataoka, N., Abel, L., Diem, M.D., and Dreyfuss, G. 2001. The Y14 protein communicates to the cytoplasm the position of exon-exon junctions. EMBO J. 20: 2062-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Gatfield, D., Izaurralde, E., and Moore, M.J. 2001. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 20: 4987-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Nott, A., and Moore, M.J. 2003. How introns influence and enhance eukaryotic gene expression. Trends Biochem. Sci. 28: 215-220. [DOI] [PubMed] [Google Scholar]
- Legrain P. and Rosbash, M. 1989. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell 57: 573-583. [DOI] [PubMed] [Google Scholar]
- Levesque L., Guzik, B., Guan, T., Coyle, J., Black, B.E., Rekosh, D., Hammarskjöld, M.L., and Paschal, B.M. 2001. RNA export mediated by Tap involves NXT1-dependent interactions with the nuclear pore complex. J. Biol. Chem. 276: 44953-44962. [DOI] [PubMed] [Google Scholar]
- Lu S. and Cullen, B.R. 2003. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA 9: 618-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim M.H., Hauber, J., Le, S.V., Maizel, J.V., and Cullen, B.R. 1989. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338: 254-257. [DOI] [PubMed] [Google Scholar]
- Maquat L.E. and Carmichael, G.G. 2001. Quality control of mRNA function. Cell 104: 173-176. [DOI] [PubMed] [Google Scholar]
- Neville M., Stutz, F., Lee, L., Davis, L.I., and Rosbash, M. 1997. The importin-β family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr. Biol. 7: 767-775. [DOI] [PubMed] [Google Scholar]
- Nott A., Meislin, S.H., and Moore, M.J. 2003. A quantitative analysis of intron effects on mammalian gene expression. RNA 9: 607-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero G.C., Harris, M.E., Donello, J.E., and Hope, T.J. 1998. Leptomycin B inhibits equine infectious anemia virus Rev and feline immunodeficiency virus rev function but not the function of the hepatitis B virus posttranscriptional regulatory element. J. Virol. 72: 7593-7597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli A.E., Ernst, R.K., Lund, E., Grimm, C., Zapp, M.L., Rekosh, D., Hammarskjöld, M.L., and Dahlberg, J.E. 1997. The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses a cellular mRNA export pathway. EMBO J. 16: 7500-7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear W.S., Nolan, G.P., Scott, M.L., and Baltimore, D. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. 90: 8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard V.W. and Malim, M.H. 1998. The HIV-1 Rev protein. Annu. Rev. Microbiol. 52: 491-532. [DOI] [PubMed] [Google Scholar]
- Saavedra C., Felber, B., and Izaurralde, E. 1997. The simian retrovirus-1 constitutive transport element, unlike the HIV-1 RRE, uses factors required for cellular mRNA export. Curr. Biol. 7: 619-628. [DOI] [PubMed] [Google Scholar]
- Segref A., Sharma, K., Doye, V., Hellwig, A., Huber, J., Luhrmann, R., and Hurt, E. 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 16: 3256-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.J., Cho, M.I., Hammarskjöld, M.L., and Rekosh, D. 1990. Human immunodeficiency virus type 1 Pr55gag and Pr160gag-pol expressed from a simian virus 40 late replacement vector are efficiently processed and assembled into viruslike particles. J. Virol. 64: 2743-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.J., Srinivasakumar, N., Hammarskjöld, M.L., and Rekosh, D. 1993. Requirements for incorporation of Pr160gag-pol from human immunodeficiency virus type 1 into virus-like particles. J. Virol. 67: 2266-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasakumar N., Chazal, N., Helga, M.C., Prasad, S., Hammarskjöld, M.L., and Rekosh, D. 1997. The effect of viral regulatory protein expression on gene delivery by human immunodeficiency virus type 1 vectors produced in stable packaging cell lines. J. Virol. 71: 5841-5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrly K. and Chesebro, B. 1997. p24 antigen capture assay for quantification of human immunodeficiency virus using readily available inexpensive reagents. Methods 12: 288-293. [DOI] [PubMed] [Google Scholar]
- Wiegand H.L., Coburn, G.A., Zeng, Y., Kang, Y., Bogerd, H.P., and Cullen, B.R. 2002. Formation of Tap/NXT1 heterodimers activates Tap-dependent nuclear mRNA export by enhancing recruitment to nuclear pore complexes. Mol. Cell. Biol. 22: 245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon D.W., Lee, H., Seol, W., DeMaria, M., Rosenzweig, M., and Jung, J.U. 1997. Tap: A novel cellular protein that interacts with Tip of herpesvirus saimiri and induces lymphocyte aggregation. Immunity 6: 571-82. [DOI] [PubMed] [Google Scholar]
- Zolotukhin A.S., Valentin, A., Pavlakis, G.N., and Felber, B.K. 1994. Continuous propagation of RRE- and Rev-RRE- human immunodeficiency virus type 1 molecular clones containing a cis-acting element of simian retrovirus type 1 in human peripheral blood lymphocytes. J. Virol. 68: 7944-7952. [DOI] [PMC free article] [PubMed] [Google Scholar]