Abstract
Vietnam is one of the countries most affected by highly pathogenic H5N1 influenza A viruses. To evaluate the potential pathogenicity in mammals of H5N1 viruses isolated from humans in Vietnam, we determined the sequences of all eight genes of 22 human isolates collected between 2003 and 2008 and compared their virulence in mice. The isolates were classified into clade 1 and clade 2.3.4 and differed in pathogenicity for mice. Whilst lysine at position 627 of PB2 (PB2-627K) is a critical virulence determinant for clade 2.3.4 viruses, asparagine at position 701 of PB2 and other unknown virulence determinants appear to be involved in the high pathogenicity of clade 1 viruses, warranting further studies to determine the factors responsible for the high virulence of H5N1 viruses in mammals.
Since late 2003, highly pathogenic H5N1 avian influenza viruses have spread among poultry and wild birds in Asia, Africa and Europe (WHO, 2010b). These highly pathogenic H5N1 viruses have caused not only outbreaks in birds, but also several hundred human infections. As of 8 June 2010, >495 humans around the world have been infected and approximately 60 % of these infections have been fatal (WHO, 2010b). Upon acquisition of transmissibility among humans, H5N1 viruses will cause a severe pandemic.
Vietnam is one of the countries most affected by highly pathogenic H5N1 viruses. Since 2003, H5N1 virus outbreaks in poultry have been reported in more than 45 of the 64 Vietnamese provinces (OIE, 2010). Nationwide vaccination programmes, which began in 2005, may have contributed, in part, to the reduction in outbreaks among poultry in 2006. However, H5N1 viruses re-emerged and outbreaks in poultry have again been reported since 2007. Since late 2003, when the first human infection was reported, Vietnam has seen many H5N1 virus patients; 119 cases have been reported to date. The fatality rate remains high: 45 % (93 patients with 42 fatal cases) for 2003–2005 and 65 % (26 patients with 17 fatal cases) for 2007–2010 (WHO, 2010b).
H5N1 viruses isolated from patients vary in pathogenicity, as measured in a mouse model; some replicate systemically with lethal outcomes, whereas others do not (Gao et al., 1999; Maines et al., 2005). Using this animal model, determinants of virulence in mammals for H5N1 viruses have been identified. High haemagglutinin (HA) cleavability conferred by the presence of a series of basic amino acids at the cleavage site (Hatta et al., 2001) is a critical determinant for virus systemic infection and high lethality not only in birds, but also in mice. The amino acid at position 627 of the PB2 protein (PB2-627), which is a host-range determinant (Subbarao et al., 1993; Naffakh et al., 2000), is a virulence determinant in mice (Hatta et al., 2001). In addition, PB2-627K is important for virus replication in the upper respiratory tract of mice, suggesting that the amino acid residue at position 627 of the PB2 protein could facilitate person-to-person transmission of H5N1 viruses (Hatta et al., 2007). Asparagine at position 701 of the PB2 protein (PB2-701N) is also a genetic marker of high virulence for H5N1 viruses in mammals, conferring efficient replication in mammalian cells (Gabriel et al., 2005; Li et al., 2005). In addition, four amino acid residues at the C terminus of the NS1 protein and serine at position 66 of the PB1-F2 protein (PB1-F2-66S) also contribute to high pathogenicity of H5N1 viruses in mice (Conenello et al., 2007; Jackson et al., 2008).
In this study, to examine the pathogenicity in mammals of H5N1 viruses isolated from humans in northern Vietnam between 2003 and 2008, we infected mice with these H5N1 viruses and compared their virulence. We then analysed the genome sequences of these viruses to identify potential determinants of virulence in mammals.
Madin–Darby canine kidney (MDCK) cells and an MDCK cell line overexpressing the human β-galactoside α-2,6-sialyltransferase I gene (MDCK-ST6GalI) (Hatakeyama et al., 2005) were maintained in minimal essential medium (MEM) containing 5 % newborn calf serum at 37 °C in 5 % CO2. Nasal swabs, pharyngeal swabs and tracheal aspirates were collected from avian H5N1 influenza virus-infected patients in northern Vietnam and were sent to the NIHE in Vietnam (Dinh et al., 2006). For H5N1 virus isolation, clinical specimens were inoculated onto MDCK cells in MEM containing 0.3 % BSA and incubated at 37 °C. MDCK-ST6GalI cells were used only for UT31203A virus, as it could not be isolated in MDCK cells. For UT31244II and UT31244III viruses, 10-day-old embryonated chicken eggs at 35 °C were used, as they were could not be isolated in MDCK or MDCK-ST6GalI cells. Stock viruses were propagated in MDCK cells at 37 °C, except for UT31244II and UT31244III, which were propagated in eggs at 35 °C and stored at −80 °C.
Viral RNAs were extracted with ISOGEN (Nippon Gene) or a viral RNA mini kit (Qiagen) according to the manufacturers’ instructions. Extracted RNAs were reverse-transcribed with SuperScript III reverse transcriptase (Invitrogen) and an oligonucleotide complementary to the 12 nt sequence at the 3′ end of the viral RNA and amplified by PCR with Pfu-ultra (Stratagene) or Phusion (Finnzymes) high-fidelity DNA polymerase and primers specific for each segment of the H5N1 influenza viruses. Primer sequences are available upon request. The PCR products were cloned into the pCR-Blunt II-TOPO vector (Invitrogen). At least three clones for each sample were sequenced by using a BigDye Terminator version 3.1 Cycle Sequencing kit on an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems). The GenBank accession numbers for the nucleotide sequences obtained in this study are HM114446–HM114621.
Phylogenetic analysis of the sequence data was performed with clustal w software, which relies on neighbour-joining methods to generate phylogenetic trees. Estimates of the phylogenies were calculated by performing 100 neighbour-joining bootstrap replicates.
To determine the 50 % mouse lethal dose (MLD50), groups (n=4 per group) of 6-week-old female BALB/c mice (Japan SLC) were anaesthetized with sevoflurane and infected intranasally with 50 μl of serial 10-fold dilutions of viruses, thereby creating doses ranging from 100 to 105 p.f.u. Mice were monitored daily for clinical signs of infection for 14 days post-infection. MLD50 values were calculated by using the method of Reed & Muench (1938).
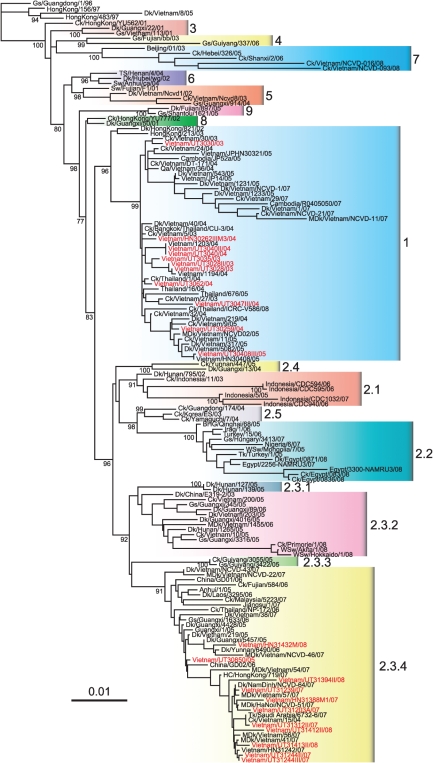
We sequenced the entire genomes of 22 H5N1 influenza viruses isolated from patients between December 2003 and February 2008 in northern Vietnam. The dates and locations of the virus isolations are summarized in Table 1 and Supplementary Fig. S1 (available in JGV Online). To understand the evolution of H5N1 influenza viruses in Vietnam, we performed phylogenetic analysis of the HA genes of these 22 strains in addition to other available sequences of H5N1 viruses isolated in Vietnam. According to the recent nomenclature system for highly pathogenic H5N1 viral HA genes, the viruses isolated from poultry in Vietnam were classified into seven different subclades: clades 0, 1, 2.3.2, 2.3.4, 3, 5 and 7, as reported previously (Wan et al., 2008; WHO, 2008, 2010a). The human H5N1 isolates studied here belonged to only two clades: the viruses isolated between 2003 and early 2005 were clade 1 and those isolated since late 2005 were clade 2.3.4 (Fig. 1).
Table 1.
Human H5N1 viruses analysed in this study
| Virus strain | Abbreviation | Date of collection | Province of collection* | Clinical outcome | Collection site of specimen | Passage history† |
|---|---|---|---|---|---|---|
| A/Vietnam/UT3028/2003‡ | UT3028 | Dec 2003 | Ha Nam | Died | Trachea | C2 |
| A/Vietnam/UT3028II/2003‡ | UT3028II | Dec 2003 | Ha Nam | Died | Trachea | C2 |
| A/Vietnam/UT3030/2003 | UT3030 | Dec 2003 | Nam Dinh | Died | Trachea | C2 |
| A/Vietnam/UT3035/2003 | UT3035 | Dec 2003 | Bac Giang | Recovered | Nose | C2 |
| A/Vietnam/UT3040/2004§ | UT3040 | Jan 2004 | Bac Ninh | Died | Pharynx | C2 |
| A/Vietnam/UT3040II/2004§ | UT3040II | Jan 2004 | Bac Ninh | Died | Trachea | C2 |
| A/Vietnam/UT3047III/2004 | UT3047III | Jan 2004 | Thai Binh | Died | Pharynx | C2 |
| A/Vietnam/UT3062/2004 | UT3062 | Jan 2004 | Bac Giang | Died | Pharynx | C2 |
| A/Vietnam/UT30259/2004 | UT30259 | Jul 2004 | Ha Tay | Died | Trachea | C2 |
| A/Vietnam/HN30262IIIM3/2004 | HN30262IIIM3 | Aug 2004 | Ha Tay | Died | Trachea | C4 |
| A/Vietnam/UT30408III/2005 | UT30408III | Feb 2005 | Thai Binh | Recovered | Pharynx | C2 |
| A/Vietnam/UT30850/2005 | UT30850 | Oct 2005 | Ha Noi | Died | Trachea | C2 |
| A/Vietnam/UT31203A/2007 | UT31203A | May 2007 | Vinh Phuc | Recovered | Pharynx | M1C1 |
| A/Vietnam/UT31239/2007 | UT31239 | Jun 2007 | Thanh Hoa | Recovered | Nose | C2 |
| A/Vietnam/UT31244II/2007|| | UT31244II | Jun 2007 | Ha Nam | Died | Pharynx | E2 |
| A/Vietnam/UT31244III/2007|| | UT31244III | Jun 2007 | Ha Nam | Died | Pharynx | E2 |
| A/Vietnam/UT31312II/2007 | UT31312II | Jul 2007 | Ha Tay | Died | Trachea | C2 |
| A/Vietnam/HN31388M1/2007 | HN31388M1 | Dec 2007 | Son La | Died | Trachea | C2 |
| A/Vietnam/UT31394II/2008 | UT31394II | Jan 2008 | Tuyen Quang | Died | Trachea | C2 |
| A/Vietnam/UT31412II/2008 | UT31412II | Feb 2008 | Hai Duong | Died | Trachea | C2 |
| A/Vietnam/UT31413II/2008 | UT31413II | Feb 2008 | Ninh Binh | Died | Trachea | C2 |
| A/Vietnam/HN31432M/2008 | HN31432M | Feb 2008 | Phu Tho | Died | Pharynx | C2 |
*See location map (Supplementary Fig. S1).
†C, MDCK cells; M, MDCK-ST6GalI cells; E, eggs. The number indicates the number of passages.
‡A/Vietnam/UT3028/2003 and A/Vietnam/UT3028II/2003 were isolated from the same individual, but A/Vietnam/UT3028II/2003 was isolated 1 day later than A/Vietnam/UT3028/2003.
§A/Vietnam/UT3040/2004 and A/Vietnam/UT3040II/2004 were isolated from the same individual, but A/Vietnam/UT3040II/2004 was isolated 1 day later than A/Vietnam/UT3040/2004.
||A/Vietnam/UT31244II/2007 and A/Vietnam/UT31244III/2007 were isolated from the same individual, but A/Vietnam/UT31244III/2007 was isolated 10 days later than A/Vietnam/UT31244II/2007.
Fig. 1.
Phylogenetic relationships among the HA genes of H5N1 viruses isolated from patients in Vietnam. Numbers at branch nodes indicate neighbour-joining bootstrap values. Analysis was based on nt 77–1672 of the HA gene. The HA gene tree was rooted to A/goose/Guangdong/1/96. Viruses analysed in this study are shown in red. Bar, 0.01 nucleotide substitutions per site. Abbreviations: BHG, bar-headed goose; Ck, chicken; Dk, duck; Gs, goose; HC, house crow; MDk, Muscovy duck; Qa, quail; Tk, turkey; TS, tree sparrow; WSw, whooper swan.
Next, we investigated the phylogenetic relationships of the other viral genes. With the exception of the neuraminidase (NA) gene, the phylogenetic trees of seven gene segments showed similar evolutionary relationships to that of the HA gene (Supplementary Fig. S2, available in JGV Online). For the NA gene, although the phylogenetic relationship was similar to that of the HA gene, the clade 2.3.4 viruses were separated into two sublineages. These results indicate that the 22 human isolates analysed here belonged to two genetic groups: 11 viruses were in clade 1, whereas nine viruses were in clade 2.3.4, with UT30850 and HN31432M being slightly different from the others in their NA genes.
To evaluate the pathogenicity of these human isolates in mammals, BALB/c mice were infected intranasally with these viruses and MLD50 values were determined. As shown in Table 2, the human isolates differed in virulence, ranging in their MLD50 from >105 p.f.u. for UT31413II to 0.46 p.f.u. for UT31239. Both virulent and avirulent viruses were found in clade 1 and clade 2.3.4, and did not correlate with clinical outcomes (Tables 1 and 2).
Table 2.
Virulence in BALB/c mice and molecular characterization of H5N1 viruses
| Virus strain | Clade classification | Amino acid | MLD50 [log10 (p.f.u.)] | Virulence in mice* | ||
|---|---|---|---|---|---|---|
| PB2 | C terminus of NS1 | |||||
| 627 | 701 | |||||
| UT3028 | 1 | K | D | E-S-E-V | 2.3 | Low |
| UT3028II | 1 | K | D | E-S-E-V | 2.5 | Low |
| UT3030 | 1 | E | N | E-S-E-V | 2.5 | Low |
| UT3035 | 1 | E | D | E-S-E-V | 2.5 | Low |
| UT3040 | 1 | K | D | 10 aa deletion | 0.3 | High |
| UT3040II | 1 | K | D | 10 aa deletion | 3.3 | Low |
| UT3047III | 1 | E | N | E-S-E-V | 3.5 | Low |
| UT3062 | 1 | K | D | E-S-E-V | 0.4 | High |
| UT30259 | 1 | K | D | E-S-E-V | 1.3 | High |
| HN30262IIIM3 | 1 | E | N | E-S-E-I | 1.4 | High |
| UT30408III | 1 | K | D | E-S-E-V | 3.5 | Low |
| UT30850 | 2.3.4′† | K | D | E-S-E-V | 1.0 | High |
| UT31203A | 2.3.4 | K | D | G-S-E-V | 0.2 | High |
| UT31239 | 2.3.4 | K | D | G-S-E-V | −0.3 (0.46 p.f.u.) | High |
| UT31244II‡ | 2.3.4 | E | D | G-S-E-V | 4.7 | Low |
| UT31244III‡ | 2.3.4 | E | D | G-S-E-V | 3.3 | Low |
| UT31312II | 2.3.4 | E | D | G-S-E-V | 4.0 | Low |
| HN31388M1 | 2.3.4 | E7/K1§ | D | G-S-E-V | 2.5 | Low |
| UT31394II | 2.3.4 | K | D | G-S-E-V | 0.3 | High |
| UT31412II | 2.3.4 | K | D | G-S-E-V | 0.6 | High |
| UT31413II | 2.3.4 | E | D | G-S-E-V | >5.0 | Low |
| HN31432M | 2.3.4′† | E | D | 10 aa deletion | 2.9 | Low |
*Viruses with an MLD50 <102 p.f.u. were considered to be of high virulence in this study.
†Clade 2.3.4′ indicates that these isolates differ slightly from the others in the NA gene.
‡UT31244II and UT31244III lack 11 aa at the C terminus of the PB1-F2 protein.
§Number of clones possessing K or E of a total of eight clones.
We then correlated the results of the mouse pathogenicity data with the sequence information (Table 2). All isolates examined here contained polybasic amino acids at the cleavage site of the HA protein, which are necessary for the high virulence of H5N1 viruses in mice (Hatta et al., 2007). No isolate possessed glutamic acid at position 92 of the NS1 protein (NS1-92E), which is associated with high virulence of these viruses in pigs (Seo et al., 2002). Similarly, no isolates possessed PB1-F2-66S, which is another marker for high virulence of H5N1 viruses in mice. The PB1-F2 protein of isolates UT31244II and UT31244III possessed an 11 aa deletion at the C terminus.
Although all of the isolates in clade 1 except for UT3035 possessed either PB2-627K or PB2-701N, which are responsible for high pathogenicity in mice, some viruses (UT3028, UT3028II, UT3030, UT3040II, UT3047III and UT30408III) showed relatively low virulence for mice. UT3035, which has neither PB2-627K nor PB2-701N, had low virulence. These results indicate that although PB2-627K and PB2-701N contribute to high pathogenicity in mice, they are not sufficient to confer high virulence in mice. On the other hand, all of the clade 2.3.4 isolates that possessed PB2-627K showed high virulence in mice, whereas those that possessed PB2-627E (avian type) were of low virulence.
Four amino acid residues (E-S-E-V) at the C terminus of NS1 are associated with virulence of H5N1 viruses in mice (Jackson et al., 2008). Among the clade 1 viruses tested, some (UT3028, UT3028II, UT3030, UT3035, UT3047III and UT30408III) were avirulent even though they possessed this motif. Whilst most of the clade 2.3.4 viruses in this study lacked this sequence motif, some were virulent, indicating that, like PB2-627K and PB2-701N, there may be some other sequence motif that can substitute for this NS1 sequence motif. These results suggest that, in addition to the HA cleavage-site sequence, PB2-627K, PB2-701N, the C terminus of NS1 and PB1-F2-66S, there are other virulence determinants that have yet to be discovered.
In this study, we phylogenetically investigated 22 H5N1 viruses isolated from humans in northern Vietnam and determined their pathogenicity in mice. All of the Vietnamese human isolates examined in this study belonged to either clade 1 or clade 2.3.4. Both of these lineages contain human H5N1 viruses isolated in other Asian countries besides Vietnam. Although none of these human isolates from northern Vietnam belonged to clades 2.3.2, 3, 5 or 7, a limited number of human isolates from China in 2009 were assigned to clades 2.3.2 and 7 (WHO, 2010a).
We also found differences in pathogenicity for mice between two strains that were isolated from the same individual but on different days and are genetically very closely related to each other: UT3040 showed high virulence, whereas UT3040II was of low virulence, even though both strains possessed the human-type PB2-627K. These two strains differ in their amino acid sequences by only three residues (one each in PB1, PA and NP; Supplementary Table S1, available in JGV Online). The amino acid residues found in PB1 and NP of UT3040II are specific for this virus among the 22 human isolates analysed here, implying that these amino acids may contribute to its attenuated phenotype in mice. Because glutamic acid at position 142 of the PA of UT3040 was also found in UT3028II and HN31432M, which were of low virulence, it remains unclear whether this amino acid residue in PA contributes to pathogenicity. To determine whether these mutations in the polymerase complex affect polymerase activity, the polymerase activity of UT3040 and UT3040II was assessed by use of a plasmid-based minigenome assay essentially as described by Ozawa et al. (2007) (Supplementary Fig. S3, available in JGV Online). Briefly, human embryonic kidney 293 cells were co-transfected with plasmids for the expression of the viral polymerase complex proteins (i.e. PB2, PB1, PA and NP) and a firefly luciferase-encoding influenza viral minigenome together with pGL4.74[hRluc/TK] (Promega), which expresses Renilla luciferase and served as an internal control. Firefly luciferase activity was measured by using a dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions. The viral polymerase complex from UT3040 exhibited significantly higher activity than that from UT3040II, suggesting that the difference in virulence between UT3040 and UT3040II originates from the difference in their polymerase activities. Reverse-genetics studies will help to determine the amino acid residues responsible for the difference in virulence between these viruses.
Supplementary Material
Acknowledgments
We thank Susan Watson for editing the manuscript. This work was supported by ERATO (Japan Science and Technology Agency), a Grant-in-Aid for Specially Promoted Research, a contract research fund for the Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases from the Ministry of Education, Culture, Sports, Science, and Technology, grants-in-aid from the Ministry of Health, Labor, and Welfare of Japan, National Institute of Allergy and Infectious Disease Public Health Service research grants and the Center for Research on Influenza Pathogenesis (CRIP) funded by the National Institute of Allergy and Infectious Diseases (contract HHSN266200700010C).
Footnotes
The GenBank/EMBL/DDBJ accession numbers for the sequences obtained in this study are HM114446–HM114621.
Three supplementary figures and a supplementary table are available with the online version of this paper.
References
- Conenello, G. M., Zamarin, D., Perrone, L. A., Tumpey, T. & Palese, P. (2007). A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog 3, 1414–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh, P. N., Long, H. T., Tien, N. T., Hien, N. T., Mai, L. T. Q., Phong, L. H., Tuan, L. V., Tan, H. V., Nguyen, N. B. & other authors (2006). Risk factors for human infection with avian influenza A H5N1, Vietnam, 2004. Emerg Infect Dis 12, 1841–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel, G., Dauber, B., Wolff, T., Planz, O., Klenk, H. D. & Stech, J. (2005). The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc Natl Acad Sci U S A 102, 18590–18595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, P., Watanabe, S., Ito, T., Goto, H., Wells, K., McGregor, M., Cooley, A. J. & Kawaoka, Y. (1999). Biological heterogeneity, including systemic replication in mice, of H5N1 influenza A virus isolates from humans in Hong Kong. J Virol 73, 3184–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama, S., Sakai-Tagawa, Y., Kiso, M., Goto, H., Kawakami, C., Mitamura, K., Sugaya, N., Suzuki, Y. & Kawaoka, Y. (2005). Enhanced expression of an α2,6-linked sialic acid on MDCK cells improves isolation of human influenza viruses and evaluation of their sensitivity to a neuraminidase inhibitor. J Clin Microbiol 43, 4139–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta, M., Gao, P., Halfmann, P. & Kawaoka, Y. (2001). Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293, 1840–1842. [DOI] [PubMed] [Google Scholar]
- Hatta, M., Hatta, Y., Kim, J. H., Watanabe, S., Shinya, K., Nguyen, T., Lien, P. S., Le, Q. M. & Kawaoka, Y. (2007). Growth of H5N1 influenza A viruses in the upper respiratory tracts of mice. PLoS Pathog 3, 1374–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, D., Hossain, M. J., Hickman, D., Perez, D. R. & Lamb, R. A. (2008). A new influenza virus virulence determinant: the NS1 protein four C-terminal residues modulate pathogenicity. Proc Natl Acad Sci U S A 105, 4381–4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Chen, H., Jiao, P., Deng, G., Tian, G., Li, Y., Hoffmann, E., Webster, R. G., Matsuoka, Y. & Yu, K. (2005). Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J Virol 79, 12058–12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines, T. R., Lu, X. H., Erb, S. M., Edwards, L., Guarner, J., Greer, P. W., Nguyen, D. C., Szretter, K. J., Chen, L. M. & other authors (2005). Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. J Virol 79, 11788–11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naffakh, N., Massin, P., Escriou, N., Crescenzo-Chaigne, B. & van der Werf, S. (2000). Genetic analysis of the compatibility between polymerase proteins from human and avian strains of influenza A viruses. J Gen Virol 81, 1283–1291. [DOI] [PubMed] [Google Scholar]
- OIE (2010). Update on highly pathogenic avian influenza in animals (type H5 and H7). Accessed 1 March 2010. http://www.oie.int/downld/AVIAN%20INFLUENZA/A_AI-Asia.htm.
- Ozawa, M., Fujii, K., Muramoto, Y., Yamada, S., Yamayoshi, S., Takada, A., Goto, H., Horimoto, T. & Kawaoka, Y. (2007). Contributions of two nuclear localization signals of influenza A virus nucleoprotein to viral replication. J Virol 81, 30–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, L. J. & Muench, H. (1938). A simple method of estimating fifty per cent endpoints. Am J Hyg 27, 493–497. [Google Scholar]
- Seo, S. H., Hoffmann, E. & Webster, R. G. (2002). Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat Med 8, 950–954. [DOI] [PubMed] [Google Scholar]
- Subbarao, E. K., London, W. & Murphy, B. R. (1993). A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J Virol 67, 1761–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, X. F., Nguyen, T., Davis, C. T., Smith, C. B., Zhao, Z. M., Carrel, M., Inui, K., Do, H. T., Mai, D. T. & other authors (2008). Evolution of highly pathogenic H5N1 avian influenza viruses in Vietnam between 2001 and 2007. PLoS One 3, e3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2008). Towards a unified nomenclature system for highly pathogenic avian influenza virus (H5N1). Emerg Infect Dis 14, e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2010a). Antigenic and genetic characteristics of influenza A(H5N1) and influenza A(H9N2) viruses and candidate vaccine viruses developed for potential use in human vaccines. Accessed 1 March 2010. http://www.who.int/csr/disease/avian_influenza/guidelines/h5n1virus/en/index.html. [PubMed]
- WHO (2010b). Cumulative number of confirmed human cases of avian influenza A/(H5N1) reported to WHO. Accessed 11 June 2010. http://www.who.int/csr/disease/avian_influenza/country/cases_table_2010_06_08/en/index.html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.