Abstract
Pancreatic ductal adenocarcinoma ranks among the most lethal of human malignancies. Here, we assess the cooperative interactions of two signature mutations in mice engineered to sustain pancreas-specific Cre-mediated activation of a mutant Kras allele (KrasG12D) and deletion of a conditional Ink4a/Arf tumor suppressor allele. The phenotypic impact of KrasG12D alone was limited primarily to the development of focal premalignant ductal lesions, termed pancreatic intraepithelial neoplasias (PanINs), whereas the sole inactivation of Ink4a/Arf failed to produce any neoplastic lesions in the pancreas. In combination, KrasG12D expression and Ink4a/Arf deficiency resulted in an earlier appearance of PanIN lesions and these neoplasms progressed rapidly to highly invasive and metastatic cancers, resulting in death in all cases by 11 weeks. The evolution of these tumors bears striking resemblance to the human disease, possessing a proliferative stromal component and ductal lesions with a propensity to advance to a poorly differentiated state. These findings in the mouse provide experimental support for the widely accepted model of human pancreatic adenocarcinoma in which activated KRAS serves to initiate PanIN lesions, and the INK4A/ARF tumor suppressors function to constrain the malignant conversion of these PanIN lesions into lethal ductal adenocarcinoma. This faithful mouse model may permit the systematic analysis of genetic lesions implicated in the human disease and serve as a platform for the identification of early disease markers and for the efficient testing of novel therapies.
Keywords: Pancreatic cancer, Ink4a/Arf, PanIN, metastasis, mouse model
Pancreatic ductal adenocarcinoma has a median survival of 6 months and a 5-year survival of <5%, making it one of the most lethal human cancers (Warshaw and Fernandez-del Castillo 1992). This poor prognosis relates to the uniformly advanced disease stage at the time of diagnosis and to its profound resistance to existing therapies. A number of key challenges must be addressed to permit improvements in patient outcome, including the need to understand more definitively the cellular origins of this disease, to elucidate the biological interactions of the tumor cell and stromal components, to determine the role of specific genetic lesions and their signaling surrogates in the initiation and progression of the tumor, and to uncover the basis for the intense therapeutic resistance of these cancers (Kern et al. 2001).
This malignancy is thought to arise from the pancreatic ducts on the basis of its histological and immunohistochemical relationship to this cell type (Solcia et al. 1995). Consistent with a ductal origin, premalignant lesions, known as pancreatic intraepithelial neoplasms (PanINs)—which are thought to arise from the smaller pancreatic ducts—are found in close physical contiguity with advanced malignant tumors (Cubilla and Fitzgerald 1976; Hruban et al. 2001). PanINs appear to progress toward increasingly atypical histological stages and display the accumulation of clonal genetic changes, suggesting that they are precursors of ductal adenocarcinoma (Moskaluk et al. 1997; Yamano et al. 2000; Luttges et al. 2001; Klein et al. 2002). The cell-of-origin question is complicated by the developmental plasticity of the pancreas that enables transdifferentiation between cell lineages (Sharma et al. 1999; Meszoely et al. 2001; Bardeesy and DePinho 2002). Acinar cells have been shown to undergo metaplastic conversion to duct-like cells, both in culture and under a variety of stresses in vivo (Jhappan et al. 1990; Sandgren et al. 1990; Hall and Lemoine 1992; Rooman et al. 2000). The development of pancreatic tumors with ductal features following a process of acinarductal metaplasia—in transgenic mice expressing TGF-α in the acini—has suggested a progenitor role for acinar cells in this malignancy (Meszoely et al. 2001; Wagner et al. 2001). Other experimental studies have suggested that islets cells or a putative pancreatic stem cell population may also give rise to pancreatic adenocarcinomas (Yoshida and Hanahan 1994; Pour et al. 2003). Finally, it remains possible that pancreatic adenocarcinoma arises from any one of these differentiated cell types or from tissue stem cells and, rather, that specific genetic lesions dictate the phenotypic endpoint of the tumor regardless of the originating cellular compartment. This paradigm has been previously suggested in malignant glioma (Holland et al. 1998; Bachoo et al. 2002).
Activating KRAS mutations, present in virtually all pancreatic adenocarcinomas, occur with increasing frequency in progressively later stage PanINs (Klimstra and Longnecker 1994; Moskaluk et al. 1997; Rozenblum et al. 1997). The early onset of KRAS mutations suggests a role in tumor initiation. In support of this notion, transgenic mice expressing an activated Kras allele under the control of the acinar-specific elastase-1 promoter develop premalignant ductal lesions (Grippo et al. 2003). Loss of function of the G1 cyclin-dependent kinase inhibitor, INK4A, also appears to be a near universal event in human pancreatic adenocarcinoma, and its pathogenetic relevance is underscored by the increased susceptibility in kindreds harboring germline INK4A mutations (Goldstein et al. 1995; Whelan et al. 1995; Rozenblum et al. 1997). Moreover, in sporadic tumors, the frequent inactivation of the INK4A locus by homozygous deletion has raised the possibility that the physically linked ARF tumor suppressor, encoded in an alternate reading frame and distinct first exon in the INK4A/ARF locus (Quelle et al. 1995), may also play a role in human pancreatic adenocarcinoma. Although ARF is a known activator of the p53 pathway (Kamijo et al. 1998; Pomerantz et al. 1998; Stott et al. 1998; Zhang et al. 1998) and its loss clearly attenuates the function of that critical tumor suppressor, more recent studies have demonstrated that ARF has p53-independent functions—including the repression of ribosomal RNA synthesis, the inhibition of NF-κB activity, and the targeting of E2F for degradation—suggesting that a number of other pathways are impaired by INK4A/ARF loss (Martelli et al. 2001; Rocha et al. 2003; Sugimoto et al. 2003). INK4A/ARF mutations occur in moderately advanced PanIN lesions (at a later stage than do KRAS mutations), an observation consistent with a role for INK4A/ARF in constraining malignant progression (Moskaluk et al. 1997; Wilentz et al. 1998). Finally, mutations in SMAD4/DPC4 and p53—encountered in >50% of tumors—are late events, implying a role in full malignant progression, including acquisition of an invasive phenotype (DiGiuseppe et al. 1994; Wilentz et al. 2000; Luttges et al. 2001).
The extensive genetic and molecular data in human tumors provide a strong framework in which to model how a given genetic lesion—or combinations of mutations—governs specific tumor biological features of the disease during its evolution. In this regard, genetically engineered mouse models have proven useful in the systematic dissection of these and related issues for a number of cancer types (Van Dyke and Jacks 2002). In the case of modeling cancer of the exocrine pancreas, transgenic mouse lines targeting a series of oncogenes to the acinar cell compartment have produced acinar carcinomas, mixed acinar-ductal tumors, or cystic tumors (Ornitz et al. 1987; Quaife et al. 1987; Sandgren et al. 1991; Glasner et al. 1992; Bardeesy et al. 2002a; Grippo et al. 2003). An important advance was provided by the observation of pancreatic tumors with ductal features in a subset of Elastase-TGFα; p53+/- mice (Wagner et al. 2001). These tumors were associated with widespread acinar-ductal metaplasia, a phenomenon whose relevance to human adenocarcinoma remains an area of active investigation. A notable recent report has described the impact of activated Kras expression directed to the differentiated pancreatic ductal epithelium by the cytokeratin 19 (Ck-19) promoter (Brembeck et al. 2003). These transgenic mice did not develop overt neoplastic lesions of the ducts, suggesting that cooperating mutations are needed for tumor initiation or that the differentiated ducts are insensitive to the transforming potential of the activated Kras allele. With regard to the latter possibility, there is a notable lack of tumors in transgenic mice expressing activated Hras in the β-cells of the pancreatic islet (Efrat et al. 1990). Mice with constitutive deletion of both or either component of the Ink4a/Arf locus do not develop spontaneous pancreatic adenocarcinoma; however, the rapid onset of lymphomas and sarcomas in these mice may preclude the study of more latent effects of deletions of this locus on pancreatic neoplasia (Serrano et al. 1996; Kamijo et al. 1997; Krimpenfort et al. 2001; Sharpless et al. 2001). These previous studies have provided significant insights into disease pathogenesis and underscore the need for continued efforts directed toward the construction of multiallelic mouse models that recapitulate the genesis and progression of the human disease.
Results
Generation of mouse strains with pancreas-specific KrasG12D expression and Ink4a/Arf deletion
To model the unique and cooperative interactions of two signature genetic lesions encountered in human pancreatic adenocarcinoma, we characterized mouse strains harboring a latent KrasG12D knock-in allele (LSL-Kras) pioneered by Jackson et al. (2001) and/or a conditional Ink4a/Arf knockout allele in the germline. As described previously (Jackson et al. 2001), the LSL-KrasG12D allele is expressed at endogenous levels after Cre-mediated excision of a transcriptional stopper element and may aide in the directed expression of KrasG12D in the appropriate cell of origin for a given cancer type. The conditional Ink4a/Arf allele (Ink4a/Arflox) was engineered to sustain Cre-mediated excision of exons 2 and 3, thereby eliminating both p16INK4A and p19ARF proteins (Supplemental Fig. 1). The performance of this allele was assessed genetically and functionally: (1) crosses of Ink4a/Arflox/lox mice to the EIIA-Cre constitutive deleter strain produced offspring with the expected deletion product; (2) Ink4a/Arflox/lox mouse embryonic fibroblasts (MEFs) showed normal levels of p16INK4A and p19ARF and underwent passage-induced senescence with similar kinetics to wild-type cells, whereas infection of these cells with retroviruses encoding Cre recombinase caused extinction of expression of both p16INK4A and p19ARF and resulted in immortal cell growth; and (3) Ink4a/Arflox/lox mice showed similar tumor incidences and life spans to wild-type mice (see Supplemental material). These data indicate that the Ink4a/Arflox allele retains wild-type function and can be rendered null for both Ink4a and Arf by Cre recombinase activity. To express the KrasG12D allele and to delete both copies of the conditional Ink4a/Arf allele specifically in the pancreas, we used the Pdx1-Cre transgene, shown previously to delete efficiently loxP-containing alleles in all pancreatic lineages (endocrine, exocrine, and duct cells; Gu et al. 2002).
KrasG12D provokes premalignant ductal lesions
Histopathologic and genetic analyses of human specimens have generated a staged progression model for premalignant and malignant pancreatic ductal lesions and their corresponding mutational profile (Fig. 1A). To examine the role of KrasG12D on the initiation and progression of pancreatic neoplasia, a cohort of Pdx1-Cre; LSL-KrasG12D mice was generated and assessed for pancreatic pathology. Compound Pdx1-Cre and LSL-KrasG12D strains were born at the expected Mendelian ratios and without evidence of clinical compromise (data not shown). Up to an age of 30 weeks, Pdx1-Cre; LSL-KrasG12D mice (n = 15) showed no outward signs of ill health. Correspondingly, serum glucose, amylase, lipase, albumin, and total bilirubin measurements in these mice (n = 5) were within normal limits—an observation suggesting normal pancreatic structure and physiology. Furthermore, histologically normal islets, acini, and ducts were clearly evident in the pancreata of these mice at all stages analyzed (Fig. 1B). Serial histological surveys (9, 12, 18, and 26 weeks), however, revealed pancreatic ductal lesions strongly reminiscent of human PanINs. In younger mice, primarily small PanIN-1 lesions—consisting of elongated mucinous ductal cells—were detected (Fig 1C). By 12 weeks, larger and more proliferative ductal lesions were noted (Fig. 1D), and these changes became more pronounced with age. At 26 weeks, extensive regions of the pancreatic parenchyma had been replaced by PanINs, surrounded by a pronounced fibrous stroma (Fig. 1E). Although PanINs increased in number and size with age, no invasive tumors were seen up to 30 weeks of age in any of the 15 mice analyzed. Although there was no evidence of neoplasia in the acinar cell compartment, focal reactive metaplastic changes were observed—likely relating to regional ductal obstruction by PanINs. Similarly, some pancreatic islets appeared moderately enlarged yet showed no evidence of neoplasia. The modest impact of KrasG12D expression on these compartments is concordant with the normal weight gains and serum chemistries noted above. Mice harboring either the LSL-KrasG12D or the Pdx1-Cre alleles alone showed no pancreatic abnormalities. These results are in line with an extensive independent study by Tuveson and colleagues of mice harboring LSL-KrasG12D and either a different Pdx1-Cre allele or a p48-Cre allele (Kawaguchi et al. 2002; S.R. Hingorani, E.F. Petricoin, A. Maitra, V. Rajapaske, C. King, M.A. Jacobetz, B.A. Hitt, Y. Kawaguchi, L.A. Liotta, H.C. Crawford, et al., in prep.). In this study both compound strains displayed progressive premalignant lesions with ductal histology—an observation consistent with an initiating role for activated KRAS in pancreatic ductal adenocarcinoma.
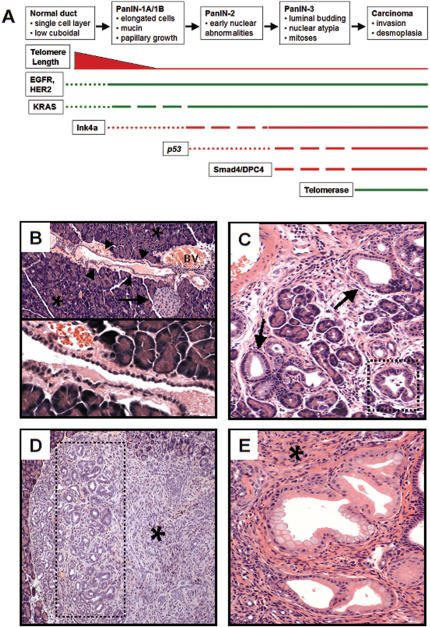
Figure 1.
Preinvasive ductal lesions arising in Pdx1-Cre; LSL-KrasG12D mice. (A) Genetic progression model of human pancreatic adenocarcinoma. The cellular phenotypes of the increasing grades of ductal neoplastic lesions are indicated. Previous studies have cataloged the presence of genetic alterations at specific disease stages, as depicted in the temporal sequence. The thickness of the line corresponds to the frequency of a lesion. Loss-of-function events are depicted in red, whereas gain-of-function lesions are shown in green. (B, top) Hematoxylin and eosin (H&E) stain showing a normal islet (arrow) and duct (arrowheads) in the background of normal acinar tissue (asterisks) in a 12-week-old Pdx1-Cre; LSL-KrasG12D mouse. An adjacent blood vessel (BV) is also indicated. (Bottom) Higher-power view of the single-layer cuboidal ductal epithelium. (C) PanIN-1 lesions detected in a 9-week-old Pdx1-Cre; LSL-KrasG12D mouse (H&E staining). Note PanIN lesions with mucinous columnar epithelium (arrows) and papillary architecture (dashed box). (D) Focus of ductal proliferation (dashed box) with prominent stromal response (asterisk) in a 12-week-old Pdx1-Cre; LSL-KrasG12D mouse (H&E staining). (E) Extensive PanIN lesions with a classical picture of intimately associated fibrotic stroma (asterisk) in the pancreas of a Pdx1-Cre; LSL-KrasG12D mouse at 26 weeks of age (H&E staining).
Ink4a/Arf loss causes malignant progression in the pancreas
The Ink4a/Arf locus has been proposed to restrain the oncogenic potential of activated Ras genes, a concept in line with the high degree of coincident mutations of these genes in human cancers and the role of Ink4a/Arf loss in facilitating the oncogenicity of activated Hras in vitro and in vivo (Serrano et al. 1996, 1997; Chin et al. 1997; Kamijo et al. 1997; Rozenblum et al. 1997; Ruas and Peters 1998). The lack of progression of the PanIN lesions to advanced cancer in Pdx1-Cre; LSL-KrasG12D mice prompted an assessment of the combined impact of KrasG12D expression and Ink4a/Arf deletion in the pancreas. Southern blots of whole pancreas DNA from Pdx1-Cre; Ink4a/Arflox/+ mice revealed efficient deletion of the Ink4a/Arflox allele (Fig. 2A). Up to an age of 7 weeks, mice of each genotype were clinically normal; however, between 7 and 11 weeks of age, all Pdx1-Cre; LSL-KrasG12D; Ink4a/Arflox/lox animals (n = 26) became moribund (Fig. 2B, survival curve), commonly presenting with weight loss, ascites, jaundice (Fig. 2C), and a palpable abdominal mass (Table 1). Autopsies revealed the presence of solid pancreatic tumors ranging in diameter from 4 to 20 mm (Fig 2C; Table 1). Grossly, these tumors were firm with irregular and ill-defined margins, frequently adhering to adjacent organs and to the retroperitoneum. In some cases, more than one distinct tumor nodule was apparent, suggesting that these neoplasms may be multifocal in nature. The tumors were highly invasive, frequently involving the duodenum and/or spleen and occasionally obstructing the common bile duct (Table 1). Liver and lung metastases were not grossly evident. No tumors were observed in control cohorts, including the Pdx1-Cre; LSL-KrasG12D; Ink4a/Arflox/+ and Pdx1-Cre; Ink4a/Arflox/lox animals, up to an age of 21 weeks. The absence of pancreatic cancers in mice lacking Ink4a/Arf in the pancreas, together with the powerful synergy of this lesion in promoting the advancement of KrasG12D-induced PanINs, points to a role of Ink4a/Arf in constraining the malignant progression of early-stage ductal neoplasms rather than in regulating the initiating phases of tumorigenesis.
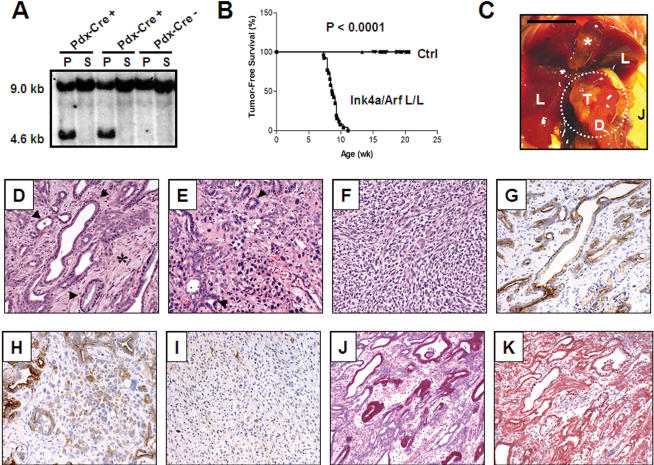
Figure 2.
Ink4a/Arf deficiency promotes progression to invasive pancreatic adenocarcinoma. (A) Complete excision of the Ink4a/Arf locus in the pancreas with Pdx1-Cre. PstI Southern blot on pancreas (P) or spleen (S) DNA from Ink4a/Arflox/+ mice that harbor or lack the Pdx1-Cre transgene. The wild-type or lox allele migrates at 9.0 kb, and the recombined Ink4a/Arf-null allele corresponds to the 4.6-kb band. (B) Kaplan-Meier pancreatic tumor-free survival curve for Pdx1-Cre; LSL-KrasG12D; Ink4a/Arflox/lox mice (denoted Ink/Arf L/L; n = 26 mice) and control cohorts (denoted Ctrl; all combinations of Pdx1-Cre, LSL-KrasG12D, and Ink4a/Arflox/lox alleles, n = 186 mice). Clinically, mice presented in a moribund state and were killed for autopsy. (C) Gross photograph of a pancreatic adenocarcinoma obstructing the common bile duct and causing dilation of the gall bladder (asterisk). Note that jaundice is readily apparent in the abdominal skin (J). T indicates tumor; D, duodenum; and L, liver. Bar, 0.6 cm. Dashed circle denotes the tumor. (D) Well-differentiated ductal adenocarcinoma observed in a Pdx1-Cre; LSL-KrasG12D; Ink4a/Arflox/lox animal at 7.9 weeks of age. Glandular tumor cells (arrowheads) are surrounded by abundant stroma (asterisk). (E) Poorly differentiated adenocarcinoma arising in the same mouse as that from D. Irregular, ill-formed formed glands (arrowheads) are present with mixture of highly mitotic, atypical tumor cells. (F) Region of tumor with sarcomatoid features from the same mouse as in D and E. (G-I) Regions of well-differentiated (G), poorly differentiated (H), and sarcomatoid (I) tumor stained for the ductal marker, cytokeratin-19. (J) PAS+D stain for apical mucins (maroon) in well-differentiated tumor cells. (K) Trichrome stain for collagen (blue) reveals the fibrotic nature of the tumor stroma.
Table 1.
Data from clinically compromised Pdx1-Cre LSL-KRAS cInk4a/Arflox/lox animals
| ID | Age (wk) | Tumor size (cm) | Location | Weight loss | Biliary obstruct | Jaundice | Bloody ascites | Invasion/metastasis (m) |
|---|---|---|---|---|---|---|---|---|
| 30 | 7.9 | 1 | Head | Y | N | N | Y | D, C, S, A |
| 31 | 7.9 | 1.4 | Head | N | Y | Y | Y | D. S |
| 32 | 7.9 | 2 | Head | Y | Y | N | N | D, PN, L, S, RN (m) |
| 33 | 7.9 | 1.4 | Tail | Y | N | N | Y | SP, RP, DP |
| 35 | 7.4 | 0.5 | Body | Y | N | N | N | V |
| 43 | 8.4 | >2 | Entire pancreas | Y | N | N | Y | D, S |
| 44 | 7.3 | 2 | Entire pancreas | N | N | N | N | V |
| 45 | 8.6 | 1 | Head | Y | N | N | Y | SP, L, E, S, D |
| 46 | 8.9 | 0.6 | Head | Y | Y | Y | N | D |
| 52 | 8.1 | 1 | Head | N | N | N | N | — |
| 58 | 8.7 | 0.5 | Head | Y | N | N | N | V, PN |
| 59 | 9.3 | 1.5 | Entire pancreas | Y | Y | Y | N | RP, RN |
| 60 | 9.9 | 0.9 | Head | N | Y | Y | Y | D, PN, V, L (m), RP, A, DP |
| 61 | 9.3 | N/A | Head | Y | Y | Y | N/A | PN, L (m), D |
| 62 | 10.4 | 1.5 | Head | Y | Y | N | Y | PN, S, D, C |
| 63 | 9.3 | N/A | Entire pancreas | Y | Y | N | Y | GB, L (m), V, PN, S, D |
| 64 | 9.1 | 1.5 | Entire pancreas | Y | Y | N | Y | S, C, PN, SP |
| 65 | 9.7 | 0.4 | Head | Y | Y | Y | N | PN (m), D |
| 76 | 11.1 | 0.75 | Head | N | N | N | N | SP, D |
| 81 | 8.6 | 1.5 | Head | Y | N | N | Y | V, LN |
| 90 | 8.4 | 2 | Entire pancreas | N | N | N | N | PN, D |
| 91 | 8.3 | 1 | Head | N | N | N | N | D, RN |
| 94 | 8.3 | N/A | Inferior pancreas | Y | NA | NA | N/A | LN, D |
| 97 | 9.4 | 1 | Tail | N | N | N | N | LN |
Tumor size is given as the approximate diameter of largest distinct tumor nodule when multiple foci were present. Tumors were often large and invasive, making size approximation difficult.
Location indicates relation of the pancreatic tumor to the adjacent organs, with head, body, and tail used in reference to the human anatomy. Inferior pancreas refers to the location of mouse pancreatic tissue affixed to the lower intestines, for which there is no analogous human pancreatic tissue.
Weight loss was judged by signs of physical wasting and decreased weight compared to that of littermates.
Jaundice and bloody ascites were clinical observations made on autopsy.
Invasion/metastasis are noted by location as follows: duodenum (D), stomach (S), colon (C), peripancreatic lymph node (PN), renal lymph node (RN), adrenal gland (A), liver (L), spleen (SP), retroperitoneum (RP), diaphragm (DP), venous (V), esophagus (E), unspecified lymph node (LN), and gall bladder (GB). Metastases are indicated by (m) after the indicated location.
Murine pancreatic tumors recapitulate the pathologic features of human pancreatic adenocarcinoma
Microscopic examination of the tumors revealed a well-formed glandular morphology with resemblance to well-differentiated and moderately differentiated human pancreatic ductal adenocarcinomas (Fig. 2D). Human pancreatic adenocarcinomas predominantly express ductal markers and lack expression of exocrine and endocrine markers (Solcia et al. 1995). Consistent with a ductal phenotype, the glandular components of the murine tumors were positive for Ck-19 immunohistochemistry (Fig. 2G,H), reacted with DBA lectin (data not shown), and showed mucin content by periodic acid-Schiff plus diastase (PAS+D; Fig. 2J) and alcian blue staining (data not shown). Furthermore, the tumors showed stromal collagen deposition, as evidenced by positive trichrome staining (Fig. 2K). None of the tumors showed reactivity for amylase and insulin, markers for acinar and β-cell differentiation, respectively (Supplemental Fig. 2). The histological and immunohistochemical profiles of these experimental tumors bear striking resemblance to human pancreatic ductal adenocarcinoma.
In addition to well-differentiated and moderately differentiated ductal adenocarcinoma, regions of undifferentiated/anaplastic (sarcomatoid) tumor (Fig. 2E,F) were observed in most cases. These anaplastic regions show weak or patchy reactivity with anti-Ck-19 antibodies and the DBA lectin (Fig. 2H,I; data not shown). These tumors exhibited high mitotic activity, severe nuclear atypia, and extensive cellular pleomorphism. In some tumors, several grades of differentiation ranging from well-differentiated to anaplastic carcinoma were noted. The epithelial nature of these anaplastic regions was confirmed by electron microscopy, which revealed the presence of intermembranous junctions and microvilli (data not shown).
Invasion and metastasis of murine pancreatic adenocarcinomas
Local invasion and metastatic spread are pathologic hallmarks of advanced human pancreatic adenocarcinoma. Pancreatic tumors arising in Pdx1-Cre; LSL-KrasG12D; Ink4a/Arflox/lox animals showed extensive invasion of adjacent organs, including the duodenum, stomach, liver, and spleen (Fig. 3A,C,E; Table 1). Tumor encroachment of the retroperitoneum and diaphragm was also observed. Furthermore, invasion of the lymphatic and vascular system was frequently detected, an observation suggestive of metastatic potential of these neoplasms. Notably, invasion by both the glandular and anaplastic components of the tumors was observed, and invading tumor cells were shown to stain positively for Ck-19 (Fig. 3F).
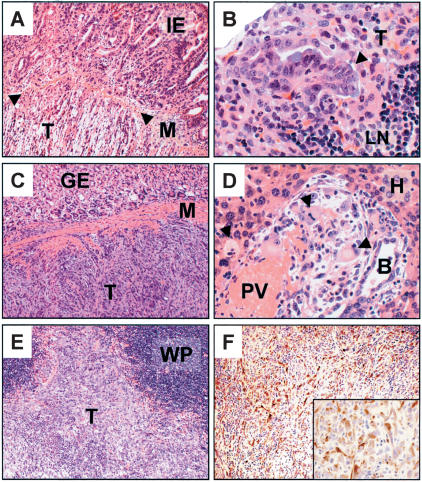
Figure 3.
Murine pancreatic tumors invade and metastasize. (A) Duodenal invasion by pancreatic ductal adenocarcinoma. Tumor (T), muscle wall (M, arrowheads), and intestinal epithelium (IE) are indicated. (B) High-magnification photomicrograph of a lymph node metastasis (T, arrowhead). LN denotes normal lymph node architecture. (C) Tumor cells (T) invading the stomach wall (M). Adjacent gastric epithelium is indicated (GE). (D) High-power micrograph of metastatic tumor cells (arrowheads) within a portal tract in the liver. Hepatocytes (H), portal vein (PV), and a reactive bile duct (B) are indicated. (E) Pancreatic tumor cells (T) invading the spleen. White pulp of the spleen is indicated (WP). (F) Immunohistochemical stain for cytokeratin-19 on tumor cells in E invading the spleen. (Inset) Higher-power image of ck-19+ invading tumor cells.
Given the invasive nature of these lesions, we conducted a systematic histologic survey of distant organs in a subset of cases. This survey revealed metastases to the lymph nodes (Fig. 3B) and occasionally to the liver (Fig. 3D), although lung metastases were not observed. Metastases were often multifocal in nature but small in size. The extensive invasion of the vasculature and lymph nodes detected in these mice make it likely that our histologic survey underestimates the true metastatic nature of these tumors. Overall, the distribution pattern of metastases is similar to that observed in the human disease that most commonly spreads to the liver and regional lymph nodes (Solcia et al. 1995).
Accelerated malignant transformation in the Pdx1-Cre; LSL-KrasG12D; Ink4a/Arflox/lox pancreas
To understand better the earlier stages of tumor development produced by KRAS activation and homozygous Ink4a/Arf deficiency, an autopsy series was performed on outwardly normal Pdx1-Cre; LSL-KrasG12D; Ink4a/Arflox/lox and Pdx1-Cre; LSL-KrasG12D; Ink4a/Arflox/+ littermates at ages 3, 4, 5, and 6 weeks. At all ages, the Pdx1-Cre; LSL-KrasG12D; Ink4a/Arflox/+ animals exhibited normal gross pancreatic histology with rare and isolated PanIN-1a lesions. At 3 weeks, Pdx1-Cre; LSL-KrasG12D; Ink4a/Arflox/lox animals had primarily normal pancreatic histology; however, low-grade ductal lesions were also observed in all mice at this time point (Fig. 4B, n = 6 mice). In addition to low-grade PanIN lesions, two of these mice also showed occasional foci of malignant ductal cells. By 4 weeks, these mice (n = 4) had an increased overall number and higher grade of pancreatic ductal lesions relative to that of littermates (Fig. 4C). Furthermore, early-stage adenocarcinomas were detected at this age, and importantly, these tumors showed both ductal and anaplastic morphologies from their earliest inception (Fig. 4D). At 5 weeks, most mice exhibited small multifocal pancreatic adenocarcinoma, although exhaustive serial sectioning revealed that some animals had only advanced PanINs and had not yet progressed to frank malignancy (Fig. 4E). Notably, in several cases advanced PanIN lesions could be found surrounded by invasive ductal and anaplastic tumor cells (Fig. 4F), an observation consistent with these tumors arising from the progression of such PanIN lesions. By 6 weeks of age, all Pdx1-Cre; LSL-KrasG12D; Ink4a/Arflox/lox mice analyzed had small pancreatic adenocarcinomas (n = 5) with histology resembling the invasive tumors arising in adult mice. Thus, it appears that once initiated, the pancreatic ductal lesions in Pdx1-Cre; LSL-KrasG12D; Ink4a/Arflox/lox mice undergo rapid histologic and clinical progression to invasive pancreatic adenocarcinoma. These features are supportive of the model in which human ductal adenocarcinoma derives from PanIN lesions with activating KRAS mutations that progress toward malignancy on loss of INK4A/ARF function.
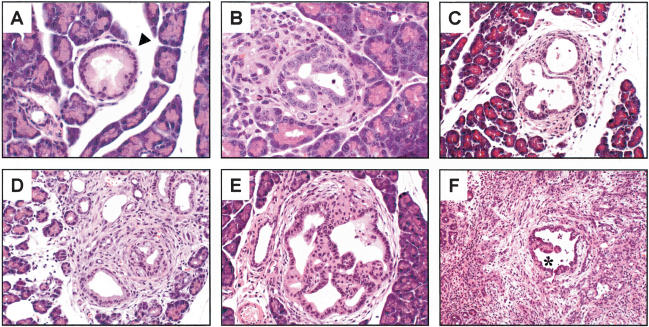
Figure 4.
Early-stage pancreatic lesions in Pdx1-Cre; LSL-KrasG12D; Ink/Arflox/lox animals. (A) High-magnification view of a low-grade PanIN lesion (arrowhead) seen in a 5-week Pdx1-Cre; LSL-KrasG12D; Ink4a/Arflox/+ animal. (B) Low-grade preinvasive ductal lesion in a 3-week-old Pdx1-Cre; LSL-KrasG12D; Ink4a/Arflox/lox mouse. (C) High-grade preinvasive ductal lesion in a Pdx1-Cre; LSL-KrasG12D; Ink4a/Arflox/lox mouse at 4 weeks. (D) Early focus of pancreatic adenocarcinoma in a 4-week-old Pdx1-Cre; LSL-KrasG12D; Ink4a/Arflox/lox mouse. Note both the ductal and anaplastic components of this early cancer. (E) High-grade PanIN lesion in a 5-week Pdx1-Cre; LSL-KrasG12D; Ink4a/Arflox/lox mouse. Serial sectioning through the entire pancreas at 10-μm intervals failed to discover any foci of adenocarcinoma in this animal. (F) High-grade PanIN lesion (asterisk) surrounded by anaplastic tumor cells in a 5-week Pdx1-Cre; LSL-KrasG12D; Ink4a/Arflox/lox mouse.
Molecular analyses of pancreatic adenocarcinomas
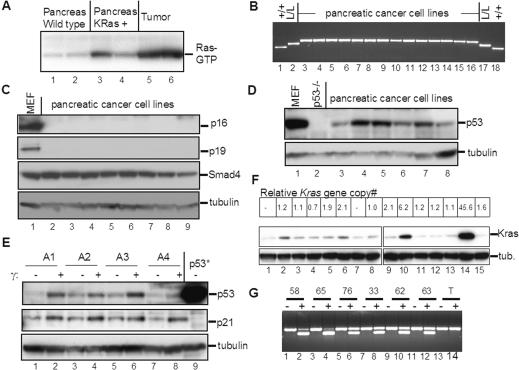
Next, we conducted a molecular analysis of the tumors to determine the status of pathways that are commonly altered in human pancreatic adenocarcinomas. Whole pancreas lysates from Pdx1-Cre; LSL-KrasG12D mice showed a modest elevation in Ras-GTP compared with that of age-matched wild-type controls (Fig. 5A), consistent with expression of the mutant constitutively active KrasG12D allele. Furthermore, levels of Ras-GTP were significantly elevated in Pdx1-Cre; LSL-KrasG12D; Ink4a/Arflox/lox tumor lysates (Fig. 5A), indicating that the activated Kras signaling pathway remains engaged and may even be further enhanced in advanced-stage tumors—although this increase could alternatively reflect the altered balance of cell types in the tumor versus normal pancreas.
Figure 5.
Molecular analysis of murine pancreatic adenocarcinomas. (A) Ras activation assay. Lysates from wild-type pancreas (lanes 1,2), Pdx1-Cre; LSL-KrasG12D pancreas (lanes 3,4), and the murine pancreatic adenocarcinomas (lanes 5,6) affinity precipitated with Raf RBD agarose (Upstate) and then subjected to immunoblot analysis with anti-Ras antibodies. (B) PCR analysis of the Ink4a/Arf locus in murine pancreatic adenocarcinoma cell lines. Multiplex PCR was performed on DNA from the pancreatic cancer cell lines (lanes 3-16) with primers that amplify the Ink4a/Arf+ (lower band), Ink4a/Arf lox (middle band), and Ink4a/Arf- (upper band) alleles. DNA from Ink4a/Arf+/+ (+/+, lanes 1,18) and Ink4a/Arflox/lox (L/L, lanes 2,17) mice served as controls. All cell lines show only the Ink4a/Arf- allele. (C) Immunoblot analysis of the tumor lysates. Membranes were immunoblotted for p16Ink4a, p19Arf, Smad4, and α-tubulin (as a loading control). Lysates from primary mouse embryonic fibroblasts (MEF, lane 1) served as a positive control. (D) Immunoblot analysis of p53 expression. Primary MEFs (lane 1) and p53-/- MEFs (lane 2), were positive and negative controls, respectively. (E) Induction of p53 and p21 in pancreatic adenocarcinoma cells by ionizing irradiation. Mouse pancreatic cancer cell lines were either untreated (-) or gamma irradiated (+; lanes 1-8). Lysates were immunoblotted for p53, p21, and α-tubulin. MEFs with a mutant p53 allele (p53*) were a control for p53 overexpression. Note that the tumors show modest expression of p53 compared with that of cells with mutant stabilized p53 and that ionizing radiation can effectively induce p53 and p21 in these tumor cells. (F) Amplification of the Kras gene and elevated Kras protein expression in a subset of pancreatic adenocarcinomas. The upper panel shows the relative Kras gene copy number as measured by quantitative real-time PCR. Wild-type specimens have a ratio of 1.0; -indicates not done. The middle and lower panels show Western blot analysis of the corresponding Kras levels and the tubulin (tub) loading control, respectively. Lane 1 is a control MEF specimen. Lanes 2-15 are tumor cell line specimens. Note that lanes 10 and 14 show both high-level Kras gene amplification and protein overexpression. (G) The mutant Kras allele is amplified in tumors showing increased Kras gene copy number. RT-PCR/RFLP analysis was performed on pancreatic adenocarcinoma cell line RNA to evaluate the whether the wild-type and KrasG12D alleles are expressed based on the KrasG12D-specific HindIII site. PCR-amplified cDNA was untreated (-) or digested with HindIII (+). Lanes 1-12 are tumor cell lines. Lanes 13 and 14 are control testes cDNA. All tumors express both alleles. Note that tumors 58 and 65 (lanes 2,4)—corresponding to lanes 10 and 14 in F—show an increased relative ratio of the lower KrasG12D allele, consistent with amplification and overexpression of this mutant allele.
Further molecular studies of tumor samples used early passage cell lines derived from the murine pancreatic adenocarcinomas (Materials and Methods) in order to avoid the presence of contaminating normal cells. As expected, the tumor cell lines showed homozygous deletion of the Ink4a/Arf locus as determined by PCR analysis (Fig. 5B) and did not express p16Ink4a or p19Arf (Fig. 5C; data not shown). Next, we assessed the status of the Smad4 and p53 tumor suppressor genes; advanced human pancreatic adenocarcinomas often sustain homozygous deletion or truncating mutations of Smad4, resulting in loss of expression, as well as p53 missense mutations, resulting in stabilization of the mutant protein. In all cases examined (n = 15), Western blots showed robust expression of full-length SMAD4 protein (Fig. 5C; data not shown), and sequence analysis of the RT-PCR-generated open reading frame revealed wild-type Smad4 sequences (see Materials and Methods). In addition, Western blot analysis revealed modest levels of p53 (Fig. 5D; data not shown) and showed induction of increased p53 and p21CIP1 levels in response to ionizing radiation (Fig. 5E; data not shown), consistent with wild-type p53 function. Accordingly, sequence analysis of the RT-PCR-generated p53 open reading frame confirmed that all specimens had a wild-type p53 status.
Gene copy number alterations at the KRAS locus are thought to contribute to the progression of some tumors harboring activating KRAS mutations. Specifically, KRAS gene amplification occurs in certain human malignancies, including human pancreatic adenocarcinomas, and in several tumor types in the mouse (Yamada et al. 1986; Mahlamaki et al. 1997; Liu et al. 1998; Schleger et al. 2000; Heidenblad et al. 2002; O'Hagan et al. 2002). In addition, loss of the wild-type RAS allele has also been shown to promote cellular transformation by activated RAS (Bremner and Balmain 1990; Finney and Bishop 1993; Zhang et al. 2001). We performed quantitative real-time PCR (QPCR) on genomic DNA derived from 15 pancreatic cancer cell lines to assess the relative Kras gene copy numbers (see Materials and Methods). These analyses revealed high-level amplifications in two specimens (Fig. 5F, upper panel, lanes 10,14; sixfold and 45-fold, respectively); these changes were also evident in the corresponding primary tumors (threefold and 5.5-fold, respectively, data not shown). In addition, approximately twofold gains were detected in three other specimens (Fig. 5F, upper panel, lanes 5,6,9) and in the corresponding primary tumors (data not shown). The reduced relative magnitude of the amplification in some primary tumor specimens may have been due to the presence contaminating stromal tissue. Immunoblot analysis revealed marked Kras expression increases in the lysates from cell lines that had high level gene amplifications and more modest increases in the lysates from lines with lower level gains (Fig. 5F, lower panels). Next, we used RT-PCR/RFLP analysis to assess whether the mutant or wild-type allele is amplified in these tumors. These analyses revealed an increased intensity of the band corresponding to the mutant KrasG12D transcript in the samples with Kras amplification (Fig. 5G, lanes 2,4), indicating that specifically the mutant allele is amplified. Finally, these data show that the expression of the wild-type Kras allele is also retained in all tumors, a result corroborated by PCR analysis of genomic DNA showing that both the wild-type and mutant Kras alleles were present (Fig. 5G; data not shown). Hence, gene amplification of activated Kras—but not loss of the wild-type allele—appears to contribute to malignant progression in the pancreas.
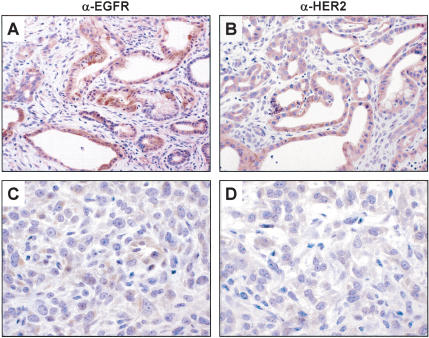
Finally, we wished to address the status of accessory signaling pathways in these tumors. The activation and extinction of EGFR and HER2/NEU are characteristic of the evolving human disease. Specifically, both EGFR and HER2/NEU are induced early in PanIN progression and remain elevated in ductal adenocarcinomas, whereas HER2/NEU expression becomes extinguished in more advanced, undifferentiated tumors (Korc et al. 1992; Day et al. 1996; Friess et al. 1996). Likewise, in the mouse model, we observed expression of both Egfr (Fig. 6A) and Her2/Neu (Fig. 6B) in the glandular component of the tumors but not in the undifferentiated sarcomatoid regions (Fig. 6C,D). In this model, the lack of Smad4 and p53 mutations, along with the activation of the growth signaling molecules, provides opportunity for the further analysis of these pathways in the biology of this disease.
Figure 6.
Expression of Egfr and Her2 in pancreatic adenocarcinomas. (A,B) Immunohistochemistry with anti-Egfr (A) or anti-Her2 (B) antibodies shows robust expression of these proteins in the glandular regions of the tumors. (C,D) Immunohistochemistry for Egfr and Her2 reveals very weak or absent expression in the poorly differentiated regions of these tumors. Note that C and D were photographed from adjacent regions of the slides depicted in A and B.
Discussion
We have shown that activation of Kras combined with Ink4a/Arf deficiency potently induces ductal adenocarcinoma, whereas either genetic lesion in isolation is not sufficient for production of advanced malignant disease. The experimental evidence in this study provides strong genetic support for the histologic progression model of ductal lesions and points to specific genetic events that regulate the genesis and critical pathological transition to lethal disease. In keeping with the detection of mutant KRAS alleles in the earliest human PanINs, Kras activation appears to be an initiating step in PanIN development in the mouse. The grossly normal pancreatic architecture despite universal KrasG12D expression in Pdx1-Cre; LSL-KrasG12D mice suggests that additional—possibly epigenetic—events are required to allow the emergence of PanINs. Equivalently, KRAS mutations have been observed in histologically normal human pancreas specimens (Luttges et al. 1999). In the presence of an intact Ink4a/Arf locus, the murine PanINs do not progress beyond the PanIN-2 stage by age 30 weeks—despite their high multiplicity—suggesting that an efficient and potent p16INK4A and/or p19ARF-mediated checkpoint mechanism restrains malignant progression in initiated lesions. Our data indicate that the Ink4a/Arf locus is critical in regulating both the progression of PanINs and the development of invasive pancreatic adenocarcinoma.
It should be noted that our work does not directly address the cellular origin of the pancreatic adenocarcinomas, and it remains possible that these tumors derive from an acinar compartment via transdifferentiation, de novo from other differentiated lineages or from pancreatic progenitors. However, given the fully penetrant onset of pancreatic adenocarcinomas by 6 weeks, the preceding rapid progression of PanINs to advanced stages and the absence of neoplastic change in other compartments provide a strong case for a PanIN-to-adenocarcinoma sequence in Pdx1-Cre; LSL-KrasG12D; Ink4a/Arflox/lox mice. Likewise, we are unable to determine whether PanINs arise from differentiated ductal cells, resident multipotent progenitor cells, or transdifferentiation of acinar or other cell types. In fact, previous data have shown that Ink4a/Arf loss permits dedifferentiation of mature astrocytes to glioblastoma in vivo (Bachoo et al. 2002), and therefore, p16INK4A and/or p19ARF could be playing a similar role in blocking progression in this model. It is of interest to note that the expression of Cre early in pancreas development through the Pdx1 promoter enables efficient activation of Kras in all pancreatic compartments, yet only ductal tumors develop. This unique association of Kras activation and pancreatic ductal tumorigenesis appears applicable to human cancer of the pancreas because KRAS mutations are detected in ductal adenocarcinomas and in another type of pancreatic ductal malignancy, intraductal papillary mucinous neoplasms (Z'Graggen et al. 1997), but not in acinar cell tumors (Terhune et al. 1994). The unbiased Pdx1-Cre system, therefore, suggests that at physiological levels, either activated Kras drives progenitor cells or differentiated cells of various lineages toward a ductal phenotype, or that this oncoprotein is singularly able to exert its transforming effects in the duct compartment. Formal resolution of these questions will likely require rigorously defined cell culture-based systems as well as crosses to Cre recombinase strains specifically targeted to more differentiated pancreatic lineages.
The absence of PanIN lesions in Pdx1-Cre; Ink4a/Arflox/lox mice suggests that p16INK4A and p19ARF do not regulate the onset of these earliest neoplastic stages. Rather, the rapid progression of PanIN lesions and development of adenocarcinomas in the Pdx1-Cre; LSL-KrasG12D; Ink4a/Arflox/lox mouse indicates that the Ink4a/Arf locus is required to restrain the malignant transformation of these initiated lesions. Such a role in progression rather than initiation fits well with the documented appearance of INK4A/ARF loss at the PanIN-2 stage in humans, and the comparable age-of-onset of pancreatic adenocarcinoma observed in a subset of families with germline INK4A mutations and observed for sporadic tumors (Moskaluk et al. 1997; Wilentz et al. 1998; Lynch et al. 2002). The mechanistic basis for the in vivo cooperative interactions of activated KRAS and INK4A/ARF deficiency remains an important issue. Until recently, a prevailing model has proposed the existence of a feedback loop whereby activated RAS mediates induction of MAPK kinase leading directly to induction of Ink4a and Arf and subsequent growth arrest (Serrano et al. 1997). However, recent studies have shown that such a direct loop is not operative in response to endogenous—as opposed to overexpressed—levels of activated RAS (Guerra et al. 2003; D. Tuveson and T. Jacks, pers. comm.). It appears that physiological expression of the Ink4a/Arf locus is tightly controlled by both positive and negative regulators, possibly modulated by such factors as integrin-extracellular matrix interactions (Plath et al. 2000; Natarajan et al. 2003) and mitogenic stimuli (Alani et al. 2001; Ohtani et al. 2001). On the basis of these findings, it is tempting to speculate that alterations in the balance of these signals occur in PanIN lesions, but not in the normal pancreas (with or without Kras activation), resulting in activation of the Ink4a/Arf locus.
Another important issue in human pancreatic adenocarcinoma is the relative pathogenic roles of INK4A versus ARF loss. In humans, INK4A mutation seems to determine disease predisposition as there are both sporadic and germline mutations that specifically target INK4A yet spare ARF (Rozenblum et al. 1997; Liu et al. 1999; Lal et al. 2000; Lynch et al. 2002). On the other hand, the loss of ARF—via homozygous deletion of the locus—occurs in ∼50% of the tumors (Rozenblum et al. 1997). Notably, a significant proportion of these ARF-deficient tumors also harbor p53 mutations (Rozenblum et al. 1997), potentially reflecting an oncogenic role of ARF loss other than in p53 regulation and/or the specific role of p53 loss on the DNA damage response (see below). The specific tumor suppressor activities contributed by Ink4a and by Arf should be resolved by crosses of the Pdx1-Cre; LSL-KrasG12D mice onto genetic backgrounds deficient for either gene of this locus. It is notable that the p53 mutations were absent in the mouse tumor model. This may reflect the need to inactivate the p19ARF-independent DNA damage-sensing function of p53 in human cancer but not in the mouse. Thus, in the Pdx1-Cre; LSL-KrasG12D; Ink4a/Arflox/lox mice, Arf deletion—which neutralizes p53 induction by other stresses such as aberrant cell cycle entry and activated oncogene expression—may effectively substitute for p53 loss in tumor progression. The basis for this divergence may relate to the cross-species differences in telomere dynamics (Maser and DePinho 2002), the prominent role of p53 (and not ARF) in the telomere checkpoint response of evolving tumors (Chin et al. 1999), and evidence of telomere dysfunction in the progression of human pancreatic adenocarcinoma (van Heek et al. 2002).
Lastly, our results suggest that, at least in this animal model, many of the classical features of malignancy in general and of pancreatic cancer in specific can be recapitulated by Ink4a/Arf loss in the setting of Kras activation. Whereas Kras activation or Ink4a/Arf loss alone does not cause local invasion and advanced local growth, the combination does. Although other unknown genetic alterations may contribute to the malignant behavior of these tumors, the rapid and stereotyped appearance of invasive cancer in these animals indicates that such changes are either unnecessary for malignant behavior or occur with such high frequency as to not be rate-limiting. The distinction between these two possibilities is of immense practical importance for the development of molecularly targeted therapeutics that would seek to inhibit the malignant growth of a tumor. We anticipate that unbiased genome-wide analyses of the tumors reported here will help to resolve these possibilities.
Materials and methods
Engineering of the conditional Ink4a/Arf mouse strain
The Ink4a/Arf locus was subcloned into the pKOII targeting vector that carried a negative selection marker for diptheria toxin (DT), a positive selection marker for neomycin acetyltransferase (Neo), Frt sites, and loxP sites (see Supplemental material). Embryonic stem (ES) cells were electroporated and selected by standard techniques. We screened clones by Southern analysis using the PstI restriction enzyme and a 3′ fragment external to the targeting construct (Supplemental Fig. 1). Blastocyst injections were carried out with targeted clones, and transmitting chimeric mice were bred to CAGG-Flpe and transgenic mice to generate the Ink4a/Arflox allele. These mice were bred onto an FVB/n background (backcrossed four generations). Mice were genotyped by Southern analysis and multiplex PCR (primers and conditions are available on request). For functional tests of the allele, these mice were crossed to the EIIA-Cre general deleter strain (Lakso et al. 1996), resulting in efficient deletion of exons 2 and 3 as assessed by Southern blot. Methods for testing the functionality of the Ink4a/Arflox allele in MEFs, including MEF isolation and culturing, Cre-mediated deletion and 3T3 assay, were performed as described (Bardeesy et al. 2002b).
Mouse colony generation
The LSL-KrasG12D knock-in strain was kindly provided by Tyler Jacks and David Tuveson (Massachusetts Institute of Technology; Jackson et al. 2001). The Pdx1-Cre transgenic strain was generously provided by Doug Melton (Harvard; Gu et al. 2002). These strains were bred to Ink4a/Arflox/lox mice to generate the genotypes, Pdx1-Cre; Ink4a/Arflox/+ and LSL-KrasG12D; Ink4a/Arflox/lox. These strains were intercrossed to produce the experimental cohorts. Mice were genotyped by slot blot and PCR (conditions available on request).
Histology and immunohistochemistry
Tissues were fixed in 10% formalin overnight and embedded in paraffin. For immunohistochemistry, slides were deparaffinized in xylene and rehydrated sequentially in ethanol. For antibodies requiring antigen retrieval, antigen unmasking solution (Vector) was used according to the manufacturer's instructions. Slides were quenched in hydrogen peroxide (0.3%-3%) to block endogenous peroxidase activity and then washed in automation buffer (Biomeda). Slides were blocked in 5% normal serum for 1 h at room temperature. Slides were incubated overnight at 4°C with primary antibody diluted in blocking buffer. The avidinbiotin peroxidase complex method (Vector) was used, and slides were counterstained with hematoxylin. Slides were dehydrated sequentially in ethanol, cleared with xylenes, and mounted with Permount (Fisher). The antibodies and dilutions were amylase, 1:500 (Calbiochem); insulin 1:100 (Dako); TROMA3 (Ck 19) 1:10 (a gift from Rolf Kemler, Max-Planck Institute of Immunobiology, Freiburg, Germany); and HER2 and EGFR, 1:50 (Cell Signaling). EGFR, HER2, and Ck-19 staining required antigen unmasking. Biotinylated DBA lectin (Vector) was used at 1:100.
Establishment and cultivation of primary pancreatic adenocarcinoma cell lines
Freshly isolated tumor specimens were minced with sterile razor blades, digested with dispase II/colagenase (4 mg/mL each) for 1 h at 37°C, and then resuspended in RPMI and 20% fetal calf serum and seeded on vitrogen/fibronectin coated plates. Cells were passaged by trypsinization. All studies were done on cells cultivated for less than seven passages.
Molecular analysis
RNA was isolated by the Trizol reagent (GIBCO), treated with RQ1 DNAse (Promega), and then purified by the RNAeasy kit (Qiagen), each according to manufacturer's instructions. RNA was reverse-transcribed by using Superscript II (GIBCO). PCR primers were designed to amplify the entire Smad4 and p53 coding regions as a series of overlapping 400-500-bp fragments. For Smad4 the primer pairs were as follows:
A, 5′-TCCAGAAATTGGAGAGTTGGA-3′;
A1, 5′-TCAATTCCAGGTGAGACAACC-3′;
B, 5′-TGACAGTGTCTGTGTGAATCCAT-3′;
B1, 5′-TTAGGTGTGTATGGTGCAGTCC-3′;
C, 5′-ACAGCACTACCACCTGGACTGG-3′;
C1, 5′-ACAAAGACCGCGTGGTCACTAA-3′;
D, 5′-TTTGGGTCAGGTGCCTTAGTGA-3′; and
D1, 5′-GTCCACCATCCTGGAAATGGT-′3.
For p53 the primer pairs were as follows:
E, 5′-GTGTCACGCTTCTCCGAAGACT-3′;
E1, 5′-CGTCATGTGCTGTGACTTCTTGT-3′;
F, 5′-GCACGTACTCTCCTCCCCTCAA-3′;
F1, 5′-AGGCACAAACACGAACCTCAAA-3′;
G, 5′-ATGAACCGCCGACCTATCCTTA-3′; and
G1, 5′-GGATTGTGCTCAGCCCTGAAGT-3′.
PCR products were subjected to direct sequencing with one of the primers used in the PCR.
RT-PCR/RFLP analysis to distinguish the wild-type Kras and KrasG12D mutant transcripts used primers KRAS1, 5′-AGGC CTGCTGAAAATGACTG-3′, and KRAS7, 5′-CCCTCCCCAG TTCTCATGTA-3′, to amplify a 243-bp product from both the wild-type and mutant transcripts. The KrasG12D allele but not the wild-type allele contains a HindIII restriction site engineered in exon 1. Thus, digestion of the 243-bp PCR product with HindIII yields 213-bp and 30-bp bands from the mutant product only.
For Western blot analyses, tissues or cell pellets were sonicated in 20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, and 1% Triton X-100 in the presence of a protease inhibitor cocktail (Roche) and a phosphatase inhibitor cocktail (kits I and II, Calbiochem). Fifty micrograms of lysate was resolved on 4%-12% Bis-Tris NuPAGE gels (Invitrogen) and transferred to PVDF membranes. Membranes were blotted for p16Ink4a (M-156, Santa Cruz), p53 (Ab-7, Oncogene Research), Smad4 (B-8, Santa Cruz), p21 (C-19, Santa Cruz), p19Arf (Ab-80, Abcam), and tubulin (DM-1A, Sigma). For p53 induction, cells were irradiated by using a cesium source (Atomic Energy Commission of Canada) at 10 Gy and harvested after 4 h. For measurement of activated Ras level, the Ras activation assay kit (Upsate) was used according to the manufacturer's instructions.
Quantitative real-time PCR
PCR primers were designed to amplify a 154-bp product of Kras genomic DNA encompassing exon 1 (KrasE1-F, 5′-tgtaag GCCTGCTGAAAATG-3′; KrasE1-R, 5′-gcacgcagactgtagagcag 3′). Quantitative PCR was performed by monitoring in real-time the increase in fluorescence of SYBR Green dye (Qiagen) with an ABI 7700 sequence detection system (Perkin Elmer Life Sciences). Data was analyzed by relative quantitation using the comparative Ct method and normalization to GAPDH.
Acknowledgments
We thank Tyler Jacks for providing the LSL-KRAS mice and for sharing unpublished data and Doug Melton for providing the Pdx1-Cre strain. We are grateful to Ralph Hruban for consultation and review of the pathology presented here. We also thank Alice Yu and Jonathan Iaconelli for technical assistance, as well as Lana Ritchie of the DFCI transgenic and gene targeting core for her technical expertise. We are grateful to Ned Sharpless for critical review of this manuscript and to Ruben Carrasco and other members of the DePinho laboratory for their insights and assistance. N.B. is supported by grants from the Barr Foundation and the Lustgarten Foundation. N.B. offers particular thanks to Jeffrey Morgan, whose impassioned patient care helped inspire this work. This work was supported by grants from the National Institutes of Health and the American Cancer Society to R.A.D and the Lustgarten Foundation to N.B. R.A.D. is an American Cancer Society Professor.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1158703.
Corresponding authors.
References
- Alani R.M., Young, A.Z., and Shifflett, C.B. 2001. Id1 regulation of cellular senescence through transcriptional repression of p16/Ink4a. Proc. Natl. Acad. Sci. 98: 7812-7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachoo R.M., Maher, E.A., Ligon, K.L., Sharpless, N.E., Chan, S.S., You, M.J., Tang, Y., DeFrances, J., Stover, E., Weissleder, R., et al. 2002. Epidermal growth factor receptor and Ink4a/Arf: Convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell 1: 269-277. [DOI] [PubMed] [Google Scholar]
- Bardeesy N. and DePinho, R.A. 2002. Pancreatic cancer biology and genetics. Nat. Rev. Cancer 2: 897-909. [DOI] [PubMed] [Google Scholar]
- Bardeesy N., Morgan, J., Sinha, M., Signoretti, S., Srivastava, S., Loda, M., Merlino, G., and DePinho, R.A. 2002a. Obligate roles for p16(Ink4a) and p19(Arf)-p53 in the suppression of murine pancreatic neoplasia. Mol. Cell. Biol. 22: 635-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardeesy N., Sinha, M., Hezel, A.F., Signoretti, S., Hathaway, N.A., Sharpless, N.E., Loda, M., Carrasco, D.R., and DePinho, R.A. 2002b. Loss of the Lkb1 tumour suppressor provokes intestinal polyposis but resistance to transformation. Nature 419: 162-167. [DOI] [PubMed] [Google Scholar]
- Brembeck F.H., Schreiber, F.S., Deramaudt, T.B., Craig, L., Rhoades, B., Swain, G., Grippo, P., Stoffers, D.A., Silberg, D.G., and Rustgi, A.K. 2003. The mutant K-ras oncogene causes pancreatic periductal lymphocytic infiltration and gastric mucous neck cell hyperplasia in transgenic mice. Cancer Res. 63: 2005-2009. [PubMed] [Google Scholar]
- Bremner R. and Balmain, A. 1990. Genetic changes in skin tumor progression: Correlation between presence of a mutant ras gene and loss of heterozygosity on mouse chromosome 7. Cell 61: 407-417. [DOI] [PubMed] [Google Scholar]
- Chin L., Pomerantz, J., Polsky, D., Jacobson, M., Cohen, C., Cordon-Cardo, C., Horner 2nd, J.W., and DePinho, R.A. 1997. Cooperative effects of INK4a and ras in melanoma susceptibility in vivo. Genes & Dev. 11: 2822-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L., Artandi, S.E., Shen, Q., Tam, A., Lee, S.L., Gottlieb, G.J., Greider, C.W., and DePinho, R.A. 1999. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell 97: 527-538. [DOI] [PubMed] [Google Scholar]
- Cubilla A.L. and Fitzgerald, P.J. 1976. Morphological lesions associated with human primary invasive nonendocrine pancreas cancer. Cancer Res. 36: 2690-2698. [PubMed] [Google Scholar]
- Day J.D., Digiuseppe, J.A., Yeo, C., Lai-Goldman, M., Anderson, S.M., Goodman, S.N., Kern, S.E., and Hruban, R.H. 1996. Immunohistochemical evaluation of HER-2/neu expression in pancreatic adenocarcinoma and pancreatic intraepithelial neoplasms. Hum. Pathol. 27: 119-124. [DOI] [PubMed] [Google Scholar]
- DiGiuseppe J.A., Hruban, R.H., Goodman, S.N., Polak, M., van den Berg, F.M., Allison, D.C., Cameron, J.L., and Offerhaus, G.J. 1994. Overexpression of p53 protein in adenocarcinoma of the pancreas. Am. J. Clin. Pathol. 101: 684-688. [DOI] [PubMed] [Google Scholar]
- Efrat S., Fleischer, N., and Hanahan, D. 1990. Diabetes induced in male transgenic mice by expression of human H-ras oncoprotein in pancreatic β cells. Mol. Cell. Biol. 10: 1779-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney R.E. and Bishop, J.M. 1993. Predisposition to neoplastic transformation caused by gene replacement of H-ras1. Science 260: 1524-1527. [DOI] [PubMed] [Google Scholar]
- Friess H., Berberat, P., Schilling, M., Kunz, J., Korc, M., and Buchler, M.W. 1996. Pancreatic cancer: The potential clinical relevance of alterations in growth factors and their receptors. J. Mol. Med. 74: 35-42. [DOI] [PubMed] [Google Scholar]
- Glasner S., Memoli, V., and Longnecker, D.S. 1992. Characterization of the ELSV transgenic mouse model of pancreatic carcinoma: Histologic type of large and small tumors. Am. J. Pathol. 140: 1237-1245. [PMC free article] [PubMed] [Google Scholar]
- Goldstein A.M., Fraser, M.C., Struewing, J.P., Hussussian, C.J., Ranade, K., Zametkin, D.P., Fontaine, L.S., Organic, S.M., Dracopoli, N.C., Clark Jr., W.H., et al. 1995. Increased risk of pancreatic cancer in melanoma-prone kindreds with p16INK4 mutations. N. Engl. J. Med. 333: 970-974. [DOI] [PubMed] [Google Scholar]
- Grippo P.J., Nowlin, P.S., Demeure, M.J., Longnecker, D.S., and Sandgren, E.P. 2003. Preinvasive pancreatic neoplasia of ductal phenotype induced by acinar cell targeting of mutant Kras in transgenic mice. Cancer Res. 63: 2016-2019. [PubMed] [Google Scholar]
- Gu G., Dubauskaite, J., and Melton, D.A. 2002. Direct evidence for the pancreatic lineage: NGN3+: Cells are islet progenitors and are distinct from duct progenitors. Development 129: 2447-2457. [DOI] [PubMed] [Google Scholar]
- Guerra C., Mijimolle, N., Dhawahir, A., Dubus, P., Barradas, M., Serrano, M., Campuzano, V., and Barbacid, M. 2003. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell 4: 111-120. [DOI] [PubMed] [Google Scholar]
- Hall P.A. and Lemoine, N.R. 1992. Rapid acinar to ductal transdifferentiation in cultured human exocrine pancreas. J. Pathol. 166: 97-103. [DOI] [PubMed] [Google Scholar]
- Heidenblad M., Jonson, T., Mahlamaki, E.H., Gorunova, L., Karhu, R., Johansson, B., and Hoglund, M. 2002. Detailed genomic mapping and expression analyses of 12p amplifications in pancreatic carcinomas reveal a 3.5-Mb target region for amplification. Genes Chromosomes Cancer 34: 211-223. [DOI] [PubMed] [Google Scholar]
- Holland E.C., Hively, W.P., DePinho, R.A., and Varmus, H.E. 1998. A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes & Dev. 12: 3675-3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruban R.H., Adsay, N.V., Albores-Saavedra, J., Compton, C., Garrett, E.S., Goodman, S.N., Kern, S.E., Klimstra, D.S., Kloppel, G., Longnecker, D.S., et al 2001. Pancreatic intraepithelial neoplasia: A new nomenclature and classification system for pancreatic duct lesions. Am. J. Surg. Pathol. 25: 579-586. [DOI] [PubMed] [Google Scholar]
- Jackson E.L., Willis, N., Mercer, K., Bronson, R.T, Crowley, D., Montoya, R., Jacks, T., and Tuveson, D.A. 2001. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes & Dev. 15: 3243-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhappan C., Stahle, C., Harkins, R.N., Fausto, N., Smith, G.H., and Merlino, G.T. 1990. TGF α overexpression in transgenic mice induces liver neoplasia and abnormal development of the mammary gland and pancreas. Cell 61: 1137-1146. [DOI] [PubMed] [Google Scholar]
- Kamijo T., Zindy, F., Roussel, M.F., Quelle, D.E., Downing, J.R., Ashmun, R.A., Grosveld, G., and Sherr, C.J. 1997. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91: 649-659. [DOI] [PubMed] [Google Scholar]
- Kamijo T., Weber, J.D., Zambetti, G., Zindy, F., Roussel, M.F., and Sherr, C.J. 1998. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc. Natl. Acad. Sci. 95: 8292-8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y., Cooper, B., Gannon, M., Ray, M., MacDonald, R.J., and Wright, C.V. 2002. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat. Genet. 32: 128-134. [DOI] [PubMed] [Google Scholar]
- Kern S., Hruban, R., Hollingsworth, M.A., Brand, R., Adrian, T.E., Jaffee, E., and Tempero, M.A. 2001. A white paper: The product of a pancreas cancer think tank. Cancer Res. 61: 4923-4932. [PubMed] [Google Scholar]
- Klein W.M., Hruban, R.H., Klein-Szanto, A.J., and Wilentz, R.E. 2002. Direct correlation between proliferative activity and dysplasia in pancreatic intraepithelial neoplasia (PanIN): Additional evidence for a recently proposed model of progression. Mod. Pathol. 15: 441-447. [DOI] [PubMed] [Google Scholar]
- Klimstra D.S. and Longnecker, D.S. 1994. K-ras mutations in pancreatic ductal proliferative lesions. Am. J. Pathol. 145: 1547-1550. [PMC free article] [PubMed] [Google Scholar]
- Korc M., Chandrasekar, B., Yamanaka, Y., Friess, H., Buchier, M., and Beger, H.G. 1992. Overexpression of the epidermal growth factor receptor in human pancreatic cancer is associated with concomitant increases in the levels of epidermal growth factor and transforming growth factor α. J. Clin. Invest. 90: 1352-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimpenfort P., Quon, K.C., Mooi, W.J., Loonstra, A., and Berns, A. 2001. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature 413: 83-86. [DOI] [PubMed] [Google Scholar]
- Lakso M., Pichel, J.G., Gorman, J.R., Sauer, B., Okamoto, Y., Lee, E., Alt, F.W., and Westphal, H. 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. 93: 5860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal G., Liu, L., Hogg, D., Lassam, N.J., Redston, M.S., and Gallinger, S. 2000. Patients with both pancreatic adenocarcinoma and melanoma may harbor germline CDKN2A mutations. Genes Chromosomes Cancer 27: 358-361. [PubMed] [Google Scholar]
- Liu M.L., Von Lintig, F.C., Liyanage, M., Shibata, M.A., Jorcyk, C.L., Ried, T., Boss, G.R., and Green, J.E. 1998. Amplification of Ki-ras and elevation of MAP kinase activity during mammary tumor progression in C3(1)/SV40 Tag transgenic mice. Oncogene 17: 2403-2411. [DOI] [PubMed] [Google Scholar]
- Liu L., Dilworth, D., Gao, L., Monzon, J., Summers, A., Lassam, N., and Hogg, D. 1999. Mutation of the CDKN2A 5′ UTR creates an aberrant initiation codon and predisposes to melanoma. Nat. Genet. 21: 128-132. [DOI] [PubMed] [Google Scholar]
- Luttges J., Schlehe, B., Menke, M.A., Vogel, I., Henne-Bruns, D., and Kloppel, G. 1999. The K-ras mutation pattern in pancreatic ductal adenocarcinoma usually is identical to that in associated normal, hyperplastic, and metaplastic ductal epithelium. Cancer 85: 1703-1710. [PubMed] [Google Scholar]
- Luttges J., Galehdari, H., Brocker, V., Schwarte-Waldhoff, I., Henne-Bruns, D., Kloppel, G., Schmiegel, W., and Hahn, S.A. 2001. Allelic loss is often the first hit in the biallelic inactivation of the p53 and DPC4 genes during pancreatic carcinogenesis. Am. J. Pathol. 158: 1677-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch H.T., Brand, R.E., Hogg, D., Deters, C.A., Fusaro, R.M., Lynch, J.F., Liu, L., Knezetic, J., Lassam, N.J., Goggins, M., et al. 2002. Phenotypic variation in eight extended CDKN2A germline mutation familial atypical multiple mole melanoma-pancreatic carcinoma-prone families: The familial atypical mole melanoma-pancreatic carcinoma syndrome. Cancer 94: 84-96. [DOI] [PubMed] [Google Scholar]
- Mahlamaki E.H., Hoglund, M., Gorunova, L., Karhu, R., Dawiskiba, S., Andren-Sandberg, A., Kallioniemi, O.P., and Johansson, B. 1997. Comparative genomic hybridization reveals frequent gains of 20q, 8q, 11q, 12p, and 17q, and losses of 18q, 9p, and 15q in pancreatic cancer. Genes Chromosomes Cancer 20: 383-391. [DOI] [PubMed] [Google Scholar]
- Martelli F., Hamilton, T., Silver, D.P., Sharpless, N.E., Bardeesy, N., Rokas, M., DePinho, R.A., Livingston, D.M., and Grossman, S.R. 2001. p19ARF targets certain E2F species for degradation. Proc. Natl. Acad. Sci. 98: 4455-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser R.S. and DePinho, R.A. 2002. Connecting chromosomes, crisis, and cancer. Science 297: 565-569. [DOI] [PubMed] [Google Scholar]
- Meszoely I.M., Means, A.L., Scoggins, C.R., and Leach, S.D. 2001. Developmental aspects of early pancreatic cancer. Cancer J. 7: 242-250. [PubMed] [Google Scholar]
- Moskaluk C.A., Hruban, R.H., and Kern, S.E. 1997. p16 and K-ras gene mutations in the intraductal precursors of human pancreatic adenocarcinoma. Cancer Res. 57: 2140-2143. [PubMed] [Google Scholar]
- Natarajan E., Saeb, M., Crum, C.P., Woo, S.B., McKee, P.H., and Rheinwald, J.G. 2003. Co-expression of p16(INK4A) and laminin 5 γ2 by microinvasive and superficial squamous cell carcinomas in vivo and by migrating wound and senescent keratinocytes in culture. Am. J. Pathol. 163: 477-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan R.C., Chang, S., Maser, R.S., Mohan, R., Artandi, S.E., Chin, L., and DePinho, R.A. 2002. Telomere dysfunction provokes regional amplification and deletion in cancer genomes. Cancer Cell 2: 149-155. [DOI] [PubMed] [Google Scholar]
- Ohtani N., Zebedee, Z., Huot, T.J., Stinson, J.A., Sugimoto, M., Ohashi, Y., Sharrocks, A.D., Peters, G., and Hara, E. 2001. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature 409: 1067-1070. [DOI] [PubMed] [Google Scholar]
- Ornitz D.M., Hammer, R.E., Messing, A., Palmiter, R.D., and Brinster, R.L. 1987. Pancreatic neoplasia induced by SV40 T-antigen expression in acinar cells of transgenic mice. Science 238: 188-193. [DOI] [PubMed] [Google Scholar]
- Plath T., Detjen, K., Welzel, M., von Marschall, Z., Murphy, D., Schirner, M., Wiedenmann, B., and Rosewicz, S. 2000. A novel function for the tumor suppressor p16(INK4a): Induction of anoikis via upregulation of the α5β1 fibronectin receptor. J. Cell. Biol. 150: 1467-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz J., Schreiber-Agus, N., Liegeois, N.J., Silverman, A., Alland, L., Chin, L., Potes, J., Chen, K., Orlow, I., Lee, H.W., et al. 1998. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell 92: 713-723. [DOI] [PubMed] [Google Scholar]
- Pour P.M., Pandey, K.K., and Batra, S.K. 2003. What is the origin of pancreatic adenocarcinoma? Mol. Cancer 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaife C.J., Pinkert, C.A., Ornitz, D.M., Palmiter, R.D., and Brinster, R.L. 1987. Pancreatic neoplasia induced by ras expression in acinar cells of transgenic mice. Cell 48: 1023-1034. [DOI] [PubMed] [Google Scholar]
- Quelle D.E., Zindy, F., Ashmun, R.A., and Sherr, C.J. 1995. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell 83: 993-1000. [DOI] [PubMed] [Google Scholar]
- Rocha S., Campbell, K.J., and Perkins, N.D. 2003. p53- and Mdm2-independent repression of NF-κB transactivation by the ARF tumor suppressor. Mol. Cell 12: 15-25. [DOI] [PubMed] [Google Scholar]
- Rooman I., Heremans, Y., Heimberg, H., and Bouwens, L. 2000. Modulation of rat pancreatic acinoductal transdifferentiation and expression of PDX-1 in vitro. Diabetologia 43: 907-914. [DOI] [PubMed] [Google Scholar]
- Rozenblum E., Schutte, M., Goggins, M., Hahn, S.A., Panzer, S., Zahurak, M., Goodman, S.N., Sohn, T.A., Hruban, R.H., Yeo, C.J., et al. 1997. Tumor-suppressive pathways in pancreatic carcinoma. Cancer Res. 57: 1731-1734. [PubMed] [Google Scholar]
- Ruas M. and Peters, G. 1998. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim. Biophys. Acta 1378: F115-F177. [DOI] [PubMed] [Google Scholar]
- Sandgren E.P., Luetteke, N.C., Palmiter, R.D., Brinster, R.L., and Lee, D.C. 1990. Overexpression of TGFα in transgenic mice: Induction of epithelial hyperplasia, pancreatic metaplasia, and carcinoma of the breast. Cell 61: 1121-1135. [DOI] [PubMed] [Google Scholar]
- Sandgren E.P., Quaife, C.J., Paulovich, A.G., Palmiter, R.D., and Brinster, R.L. 1991. Pancreatic tumor pathogenesis reflects the causative genetic lesion. Proc. Natl. Acad. Sci. 88: 93-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleger C., Arens, N., Zentgraf, H., Bleyl, U., and Verbeke, C. 2000. Identification of frequent chromosomal aberrations in ductal adenocarcinoma of the pancreas by comparative genomic hybridization (CGH). J. Pathol. 191: 27-32. [DOI] [PubMed] [Google Scholar]
- Serrano M., Lee, H., Chin, L., Cordon-Cardo, C., Beach, D., and DePinho, R.A. 1996. Role of the INK4a locus in tumor suppression and cell mortality. Cell 85: 27-37. [DOI] [PubMed] [Google Scholar]
- Serrano M., Lin, A.W., McCurrach, M.E., Beach, D., and Lowe, S.W. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88: 593-602. [DOI] [PubMed] [Google Scholar]
- Sharma A., Zangen, D.H., Reitz, P., Taneja, M., Lissauer, M.E., Miller, C.P., Weir, G.C., Habener, J.F., and Bonner-Weir, S. 1999. The homeodomain protein IDX-1 increases after an early burst of proliferation during pancreatic regeneration. Diabetes 48: 507-513. [DOI] [PubMed] [Google Scholar]
- Sharpless N.E., Bardeesy, N., Lee, K.H., Carrasco, D., Castrillon, D.H., Aguirre, A.J., Wu, E.A., Horner, J.W., and DePinho, R.A. 2001. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature 413: 86-91. [DOI] [PubMed] [Google Scholar]
- Solcia E., Capella, C., and Kloppel, G. 1995. Tumors of the pancreas. Armed Forces Institute for Pathology, Washington, DC.
- Stott F.J., Bates, S., James, M.C., McConnell, B.B., Starborg, M., Brookes, S., Palmero, I., Ryan, K., Hara, E., Vousden, K.H., et al. 1998. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 17: 5001-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto M., Kuo, M.L., Roussel, M.F., and Sherr, C.J. 2003. Nucleolar Arf tumor suppressor inhibits ribosomal RNA processing. Mol. Cell 11: 415-424. [DOI] [PubMed] [Google Scholar]
- Terhune P.G., Heffess, C.S., and Longnecker, D.S. 1994. Only wild-type c-Ki-ras codons 12, 13, and 61 in human pancreatic acinar cell carcinomas. Mol. Carcinog. 10: 110-114. [DOI] [PubMed] [Google Scholar]
- Van Dyke T. and Jacks, T. 2002. Cancer modeling in the modern era: Progress and challenges. Cell 108: 135-144. [DOI] [PubMed] [Google Scholar]
- van Heek N.T., Meeker, A.K., Kern, S.E., Yeo, C.J., Lillemoe, K.D., Cameron, J.L., Offerhaus, G.J., Hicks, J.L., Wilentz, R.E., Goggins, M.G., et al. 2002. Telomere shortening is nearly universal in pancreatic intraepithelial neoplasia. Am. J. Pathol. 161: 1541-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M., Greten, F.R., Weber, C.K., Koschnick, S., Mattfeldt, T., Deppert, W., Kern, H., Adler, G., and Schmid, R.M. 2001. A murine tumor progression model for pancreatic cancer recapitulating the genetic alterations of the human disease. Genes & Dev. 15: 286-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warshaw A.L. and Fernandez-del Castillo, C. 1992. Pancreatic carcinoma. N. Engl. J. Med. 326: 455-465. [DOI] [PubMed] [Google Scholar]
- Whelan A.J., Bartsch, D., and Goodfellow, P.J. 1995. Brief report: A familial syndrome of pancreatic cancer and melanoma with a mutation in the CDKN2 tumor-suppressor gene. N. Engl. J. Med. 333: 975-977. [DOI] [PubMed] [Google Scholar]
- Wilentz R.E., Geradts, J., Maynard, R., Offerhaus, G.J., Kang, M., Goggins, M., Yeo, C.J., Kern, S.E., and Hruban, R.H. 1998. Inactivation of the p16 (INK4A) tumor-suppressor gene in pancreatic duct lesions: Loss of intranuclear expression. Cancer Res. 58: 4740-4744. [PubMed] [Google Scholar]
- Wilentz R.E., Iacobuzio-Donahue, C.A., Argani, P., McCarthy, D.M., Parsons, J.L., Yeo, C.J., Kern, S.E., and Hruban, R.H. 2000. Loss of expression of Dpc4 in pancreatic intraepithelial neoplasia: Evidence that DPC4 inactivation occurs late in neoplastic progression. Cancer Res. 60: 2002-2006. [PubMed] [Google Scholar]
- Yamada H., Sakamoto, H., Taira, M., Nishimura, S., Shimosato, Y., Terada, M., and Sugimura, T. 1986. Amplifications of both c-Ki-ras with a point mutation and c-myc in a primary pancreatic cancer and its metastatic tumors in lymph nodes. Jpn. J. Cancer Res. 77: 370-375. [PubMed] [Google Scholar]
- Yamano M., Fujii, H., Takagaki, T., Kadowaki, N., Watanabe, H., and Shirai, T. 2000. Genetic progression and divergence in pancreatic carcinoma. Am. J. Pathol. 156: 2123-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T. and Hanahan, D. 1994. Murine pancreatic ductal adenocarcinoma produced by in vitro transduction of polyoma middle T oncogene into the islets of Langerhans. Am. J. Pathol. 145: 671-684. [PMC free article] [PubMed] [Google Scholar]
- Z'Graggen K., Rivera, J.A., Compton, C.C., Pins, M., Werner, J., Fernandez-del Castillo, C., Rattner, D.W., Lewandrowski, K.B., Rustgi, A.K., and Warshaw, A.L. 1997. Prevalence of activating K-ras mutations in the evolutionary stages of neoplasia in intraductal papillary mucinous tumors of the pancreas. Ann. Surg. 226: 491-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xiong, Y., and Yarbrough, W.G. 1998. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92: 725-734. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Wang, Y., Vikis, H.G., Johnson, L., Liu, G., Li, J., Anderson, M.W., Sills, R.C., Hong, H.L., Devereux, T.R., et al. 2001. Wildtype Kras2 can inhibit lung carcinogenesis in mice. Nat. Genet. 29: 25-33. [DOI] [PubMed] [Google Scholar]