Quinazoline 3 was discovered as a novel c-jun N-terminal kinase (JNK) inhibitor with good brain penetration and pharmacokinetic (PK) properties. A number of analogs which were potent both in the biochemical and cellular assays were discovered. Quinazoline 13a was found to be a potent JNK3 inhibitor (IC50 = 40 nM), with > 500-fold selectivity over p38, and had good PK and brain penetration properties. With these properties, 13a is considered a potential candidate for in vivo evaluation.
c-jun-N-terminal Kinase (JNK) was discovered in the early-mid 1990s1–4 and is a member of the mitogen activated protein kinase (MAP) family. In the nearly two decades of work on JNK, the enzyme has been implicated in numerous diseases ranging from cardiovascular disease5, to metabolic disorders6 and neurodegeneration7–11. There are three isoforms of JNK. JNK1 and JNK2 are ubiquitously expressed, whereas JNK3 is expressed primarily in the brain with lower levels of expression in the heart and testis12.
Because of the numerous potential clinical applications for JNK, many small molecule inhibitor programs have developed over the past five years. Compound classes that have shown nice JNK selectivity include: aminopyrazoles13, aminopyridines14, 15, pyridine carboxamides15, 16, benzothien-2-yl-amides and benzothiazol-2-yl acetonitriles 17, 18, quinoline derivatives19, and aminopyrimidines 20–22. For a recent review of all these classes see LoGrasso and Kamenecka 23. Most of these classes of compounds did not demonstrate good brain penetration, although Kamenecka et al. recently reported aminopyrimidines showing excellent brain penetration properties22.
In the current work we present a series of novel quinazolines which were potent JNK inhibitors with > 2200-fold selectivity over p38 (compound 14d). Moreover, a systematic SAR approach utilizing biochemical and cell-based assays, along with mouse and rat pharmacokinetics enabled us to develop compounds (e.g. 13a) which maintained their potency and selectivity (> 500-fold over p38), while also incorporating good brain penetration (brain: plasma ratio of 0.8:1) in mouse, and excellent pharmacokinetics in rat. With these properties, 13a is an attractive candidate for in vivo evaluation in CNS efficacy models.
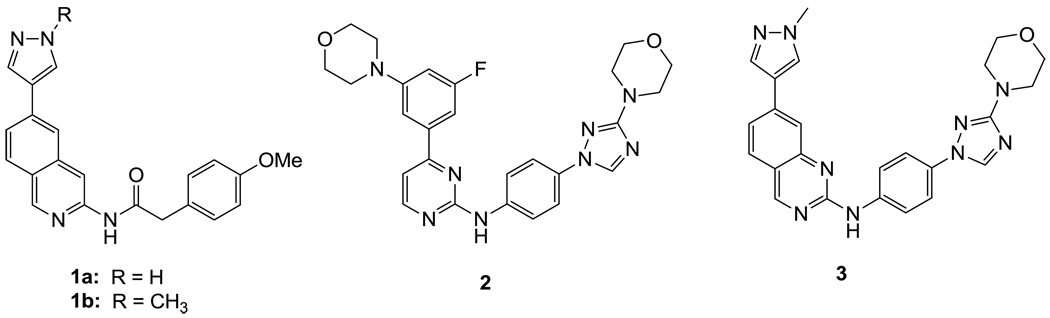
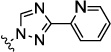
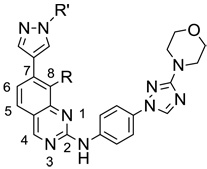
The JNK inhibitors 1a,b and 2 (figure 1) were described in the patent and primary literature,22, 24 with JNK3 IC50 = 90 nM for 1b and IC50 = 180 nM for 2 respectively 22,25. The isoquinoline 1b was only moderately potent in cells however (inhibition of c-jun phosphorylation = 1.0 µM). Compounds 1b and 2 were found to have good brain penetration and PK properties22,24. As a strategy to design a novel structural class with improved JNK3 potency and similar or improved PK and brain penetration properties, we decided to combine the amino isoquinoline of compound 1 and the amino pyrimidine scaffolds (compound 2). With this in mind we designed quinazoline 3 (Figure 1). We found that quinazoline 3 was a potent JNK inhibitor with good brain penetration (Table 1). To establish an SAR on the quinazoline ring, we first modified the 7-position (Table 1). The synthesis is outlined in scheme 1. For the syntheses of the 2-chloro quinazolines from the corresponding fluoro aldehydes we followed the procedure described by Patel et al.26 A series of pyrazole, isoxazole, morpholino, and pyridyl substitutions were assessed at the 7-position on the quinazoline ring (compounds 3-8f, Table 1). Pyrazole substitutions (compounds 3, 8a, 8g, 8h) had the lowest JNK3 IC50 values, suggesting preference for pyrazole at the 7-position (Table 1). Despite the approximate 3-fold improvement in cell-based potency of 8a over 3, the higher polar surface area of 8a caused a significant decrease in brain penetration (Table 1). The available space for modifications in this position appeared to be very limited and we concluded that the N-methyl-pyrazole moiety was optimal in terms of overall properties. Replacement of the 3-morpholino on the 1,2,4-triazole (8a) with 3-p-methyl-pyridyl on the 1,2,4-triazole (8g) decreased the cell-based IC50 by 3-fold to 40nM. However, this substitution caused brain penetration to be quite poor (8h) (Table 1), especially when compared to compound 3. Attempts to improve the potency and maintain the good pharmacological profile by replacing the morpholino-triazolo aniline moiety were unsuccessful (data not shown).
Figure 1.
JNK3 Inhibitors
Table 1.
Biochemical and Cell-based IC50 Values, and Mouse Plasma and Brain levels for a series of 2,7-substituted quinazolinesa
 | |||||||
|---|---|---|---|---|---|---|---|
| compound | R' | R | JNK3 IC50 (µM) |
JNK1 IC50 (µM) |
c-jun IC50 (µM) |
bmouse [plasma] |
(µM) [brain] |
| 3 | 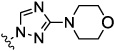 |
0.09 | 0.05 | 0.31 | 17.8 | 7.17 | |
| 8a | 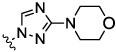 |
0.05 | 0.04 | 0.12 | 42.9 | 1.31 | |
| 8b | 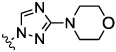 |
0.7 | nt | nt | nt | nt | |
| 8c | 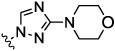 |
4.4 | nt | nt | nt | nt | |
| 8d | 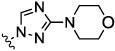 |
0.21 | nt | nt | 26.7 | 8.1 | |
| 8e | 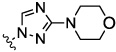 |
0.73 | nt | nt | nt | nt | |
| 8f | 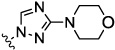 |
>20 | nt | nt | nt | nt | |
| 8g | 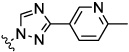 |
0.05 | nt | 0.04 | nt | nt | |
| 8h | 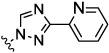 |
0.06 | 0.1 | 0.29 | 13.1 | 1.37 | |
JNK3 biochemical IC50 values are the averages of four or more experiments, and the JNK1 and cell-based IC50 values are the averages of two or more experiments. All standard deviations are ≤44% for the biochemical and ≤81% for the cell-based assays.
10 mg/kg ip 2h.
nt = not tested.
Scheme 1.
Reagents and conditions: (a) HCl in EtOH, n-BuOH, 120 °C, 2–24 h; (b) Boronic acid, Pd(PPh3)4, Na2CO3, Dioxane/water, 120 °C, 30', µW.
We next investigated the 8-position on the quinazoline ring (Table 2). We found a number of different substitutions that improved the biochemical potency. Compound 9a showed good cellular potency but unfortunately the brain penetration was poor (Table 2). Similarly, the thiazole replacement at position 8 improved the JNK3 potency by 4-fold (compare 9d to 9a), but the cell-based IC50 for 9d was 150-fold less potent than 9a suggesting decreased cell penetration for 9d. By replacing the methyl pyridine group with phenyl groups and thus reducing the polar surface area we indeed did achieve much improved brain penetration (compounds 9e/f), but unfortunately these compounds were only modestly active in cells (Table 2). Compound 9g did have both good brain penetration and potency (Table 2); however the PK properties were poor (data not shown).
Table 2.
Biochemical and Cell-based IC50 Values, and Mouse Plasma and Brain levels for a series of 2,8-substituted quinazolinesa
 | |||||||
|---|---|---|---|---|---|---|---|
| Compound | R' | R | JNK3M IC50 (µM) |
JNK1 IC50 (µM) |
c-jun IC50 (µM) |
bmouse [plasma] |
[µM] [brain] |
| 9a |  |
 |
0.03 | 0.02 | 0.02 | 2.7 | 0.15 |
| 9b | 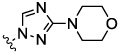 |
0.02 | 0.01 | 0.33 | 2.2 | 0.23 | |
| 9c | 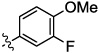 |
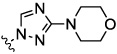 |
0.07 | 0.09 | 0.25 | 6.1 | 1.5 |
| 9d | 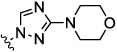 |
0.007 | 0.03 | 3.0 | nt | nt | |
| 9e | 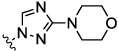 |
0.06 | 0.05 | 1.4 | 5.2 | 12.7 | |
| 9f | 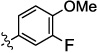 |
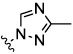 |
0.08 | nt | 0.5 | 10.3 | 7.6 |
| 9g | 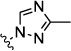 |
0.02 | 0.01 | 0.2 | 5.0 | 4.5 | |
| 9h | 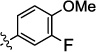 |
 |
0.09 | nt | 5.1 | nt | nt |
JNK3 biochemical IC50 values are the averages of four or more experiments, and the JNK1 and cell-based IC50 values are the averages of two or more experiments. All standard deviations are ≤44% for the biochemical and ≤81% for the cell-based assays.
10 mg/kg ip 2h.
nt = not tested.
In an effort to improve the brain penetration and also PK properties of 9a we further explored the effect of substitutions in the 2-position (Table 3). We hypothesized that by introducing groups in the 2-position that would reduce the polar surface area and molecular weight we would improve the brain penetration. Compound 10b indeed showed much improved brain to plasma ratio compared to 9a; however the plasma concentration was low presumably due to high clearance. Furthermore, the good biochemical potency did not translate into good cellular potency (Table 3). Despite extensive synthetic efforts we were unable to achieve the good PK and brain penetration properties we had with quinazoline 3 by modifying the 2-, 7- and 8- positions. Generally, the 8-substituted quinazolines led to more potent compounds but poorer PK properties. A contributing factor may be the significantly shorter half lives in microsomes as compared to the 7-substituted analogs. It became apparent that compounds with the N-methyl pyrazole group in the 7-position led to compounds with good pharmacological properties and substitutions in the 8-position improved the potency. We should also note that substitutions in the 5- and 6-positions were generally not well tolerated (data not shown). We therefore focused on the synthesis of analogs bearing the N-methyl pyrazole group in the 7-position and different substitutions in the 8-position (Table 4). All compounds shown in Table 4 were synthesized as outlined in scheme 2, except for compounds 14b and 14f, which were made in five steps starting from formylation of 3-fluoro-2-(trifluoromethyl)bromobenzene and 2-bromo-6-fluoroanisole respectively. Compound 14d showed the best JNK3 potency with IC50 = 9 nM and cell-based IC50 = 40 nM, and had great selectivity over p38 (IC50 > 20 µM), but unfortunately had very poor brain penetration (Table 4). The great selectivity of compound 14d for JNK3 over p38 can potentially be attributed to a better fit in the smaller active site for JNK3 and/or a planar structure to the molecule. Compound 14f was found to be very potent (IC50 = 4 nM vs JNK3) with good brain penetration; however its oral bio-availability was low (%F = 8) and therefore not a good candidate for further advancement. The same was true for many of the other compounds with good brain penetration in the 14 series. We decided to introduce a chloro substitution at position 8 on the quinazoline ring and identified compound 13a, which had excellent brain penetration and good PK properties as well (Table 4 and Table 5). Moreover, the compound had > 500-fold selectivity over p38. Compound 13b had a superior brain:plasma ratio of 1.7:1 compared to 13a, but suffered from a 6-fold less potent cell-based activity compared to 13a (Table 4). The PK properties of compounds 3 and 13a are summarized in Table 5. Compound 13a is currently under further investigation in animal models.
Table 3.
Biochemical and Cell-based IC50 Values, and Mouse Plasma and Brain Concentrations for 2-position Substitutions Designed to Reduce Polar Surface Area and Molecular Weighta
 | ||||||
|---|---|---|---|---|---|---|
| compound | R | JNK3 IC50 (µM) |
c-jun IC50 (µM) |
bpolar surface area |
cmouse [plasma] |
[µM] [brain] |
| 9a | 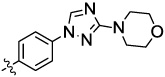 |
0.03 | 0.02 | 90 | 2.72 | 0.15 |
| 10a | 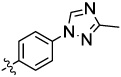 |
0.03 | nt | 77 | 2.31 | 0.23 |
| 10b | 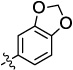 |
0.06 | 0.63 | 68 | 0.78 | 1.06 |
| 10c | 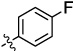 |
0.17 | nt | 49 | nt | nt |
| 10d | 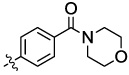 |
0.09 | nt | 79 | 0 | 0 |
| 10e | 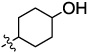 |
0.05 | 0.93 | 69 | nt | nt |
| 10f | 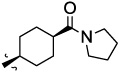 |
0.03 | 3.0 | 69 | nt | nt |
JNK3 biochemical IC50 values are the averages of four or more experiments, and the JNK1 and cell-based IC50 values are the averages of two or more experiments. All standard deviations are ≤44 % for the biochemical and ≤81 % for the cell-based assays.
Polar surface area calculated by ChemBioDraw Ultra 11.0.
10 mg/kg ip 2h.
Table 4.
Biochemical and Cell-based IC50 Values, and Mouse Plasma and Brain levels for a series of 2,7,8-substituted quinazolinesa
 | ||||||
|---|---|---|---|---|---|---|
| compound | R | R' | JNK3 IC50 (µM) |
c-jun IC50 (µM) |
bmouse [plasma] |
[µM] [brain] |
| 13a | Cl | CH3 | 0.04 | 0.10 | 10.4 | 8.3 |
| 13b | Cl | 0.063 | 0.62 | 4.3 | 7.2 | |
| 14a | CH3 | CH3 | 0.01 | 0.16 | 7.4 | 7.6 |
| 14b | CF3 | CH3 | 0.19 | nt | 8.3 | 5.6 |
| 14c | CH3 | 0.02 | 0.13 | 10.6 | 5.0 | |
| 14d | 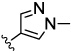 |
CH3 | 0.009 | 0.04 | 6.3 | 0.22 |
| 14e | 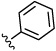 |
CH3 | 0.01 | 0.04 | 6.4 | 1.1 |
| 14f | OMe | CH3 | 0.004 | 0.05 | 9.4 | 4.9 |
The JNK3 biochemical IC50 values are the averages of four or more experiments, the JNK1 and cell-based IC50 values are the averages of two or more experiments. All standard deviations are ≤44 % for the biochemical and ≤81 % for the cell-based assays.
10 mg/kg ip 2h.
nt = not tested.
Scheme 2.
Reagents and conditions: (a) HCl in EtOH, n-BuOH, 120 °C, 2 –24 h; (b) Boronic acid, Pd(PPh3)4, Na2CO3, Dioxane/water, 120 °C, 30', µW; (c) Boronic acid, Pd(PPh3)4, Na2CO3, Dioxane/water, 100–140 °C 12–16h, µW.
Table 5.
PK properties of the two lead compounds
| arat pk | bmouse | [µM] | ||||||
|---|---|---|---|---|---|---|---|---|
| compound | Clp (mL/ min/kg) |
t1/2 (h) |
Vd (L/kg) |
oral AUC (µM*h) |
oral Cmax (µM) |
%F | [plasma] | [brain] |
| 3 | 3 | 3 | 0.8 | 6.3 | 0.7 | 28 | 17.8 | 7.2 |
| 13a | 6 | 3 | 1.4 | 2.3 | 0.3 | 19 | 10.4 | 8.3 |
l mg/kg IV, 2 mg/kg po;
10 mg/kg ip 2h.
In summary, we discovered a series of novel quinazoline compounds as JNK inhibitors with good potency in biochemical and cellular assays. Compound 13 improved upon compound 3 by 3-fold in cell potency and had 80% brain penetration compared to 40% for compound 3. In contrast, compound 3 had a greater systemic exposure and oral bioavailability suggesting that compound 3 may be preferred for non-central nervous system (CNS) indications and compound 13 may be preferred for CNS indications.
Acknowledgements
This work was supported by NIH grant U01NS057153 awarded to P.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- 1.Derijard B, Hibi M, Wu IH, Barrett T, Su B, Deng T, Karin M, Davis R. J. Cell. 1994;76:1025. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 2.Hibi M, Lin A, Smeal T, Minden A, Karin M. Genes Dev. 1993;7:2135. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 3.Kyriakis JM, Banerjee P, Nikolakaki E, Dai T, Rubie EA, Ahmad MF, Avruch J, Woodgett JR. Nature. 1994;369:156. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 4.Mohit AA, Martin JH, Miller CA. Neuron. 1995;14:67. doi: 10.1016/0896-6273(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 5.Ferrandi C, Ballerio R, Gaillard P, Giachetti C, Carboni S, Vitte PA, Gotteland JP, Cirillo R. Br J Pharmacol. 2004;142:953. doi: 10.1038/sj.bjp.0705873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirosumi J, Tuncman G, Chang L, Gorgun CZ, Uysal KT, Maeda K, Karin M, Hotamisligil GS. Nature. 2002;420:333. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- 7.Borsello T, Clarke PG, Hirt L, Vercelli A, Repici M, Schorderet DF, Bogousslavsky J, Bonny C. Nat Med. 2003;9:1180. doi: 10.1038/nm911. [DOI] [PubMed] [Google Scholar]
- 8.Hunot S, Vila M, Teismann P, Davis RJ, Hirsch EC, Przedborski S, Rakic P, Flavell RA. Proc Natl Acad Sci U S A. 2004;101:665. doi: 10.1073/pnas.0307453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carboni S, Antonsson B, Gaillard P, Gotteland JP, Gillon JY, Vitte PA. J Neurochem. 2005;92:1054. doi: 10.1111/j.1471-4159.2004.02925.x. [DOI] [PubMed] [Google Scholar]
- 10.Carboni S, Boschert U, Gaillard P, Gotteland JP, Gillon JY, Vitte PA. Br J Pharmacol. 2008;153:157. doi: 10.1038/sj.bjp.0707574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carboni S, Hiver A, Szyndralewiez C, Gaillard P, Gotteland JP, Vitte PA. J Pharmacol Exp Ther. 2004;310:25. doi: 10.1124/jpet.103.064246. [DOI] [PubMed] [Google Scholar]
- 12.Martin JH, Mohit AA, Miller CA. Brain Res Mol Brain Res. 1996;35:47. doi: 10.1016/0169-328x(95)00181-q. [DOI] [PubMed] [Google Scholar]
- 13.Kamenecka T, Habel J, Duckett D, Chen W, Ling YY, Frackowiak B, Jiang R, Shin Y, Song X, LoGrasso P. J Biol Chem. 2009;284:12853. doi: 10.1074/jbc.M809430200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swahn BM, Xue Y, Arzel E, Kallin E, Magnus A, Plobeck N, Viklund J. Bioorg Med Chem Lett. 2006;16:1397. doi: 10.1016/j.bmcl.2005.11.039. [DOI] [PubMed] [Google Scholar]
- 15.Szczepankiewicz BG, Kosogof C, Nelson LT, Liu G, Liu B, Zhao H, Serby MD, Xin Z, Liu M, Gum RJ, Haasch DL, Wang S, Clampit JE, Johnson EF, Lubben TH, Stashko MA, Olejniczak ET, Sun C, Dorwin SA, Haskins K, Abad-Zapatero C, Fry EH, Hutchins CW, Sham HL, Rondinone CM, Trevillyan JM. J Med Chem. 2006;49:3563. doi: 10.1021/jm060199b. [DOI] [PubMed] [Google Scholar]
- 16.Zhao H, Serby MD, Xin Z, Szczepankiewicz BG, Liu M, Kosogof C, Liu B, Nelson LT, Johnson EF, Wang S, Pederson T, Gum RJ, Clampit JE, Haasch DL, Abad-Zapatero C, Fry EH, Rondinone C, Trevillyan JM, Sham HL, Liu G. J Med Chem. 2006;49:4455. doi: 10.1021/jm060465l. [DOI] [PubMed] [Google Scholar]
- 17.Angell RM, Atkinson FL, Brown MJ, Chuang TT, Christopher JA, Cichy-Knight M, Dunn AK, Hightower KE, Malkakorpi S, Musgrave JR, Neu M, Rowland P, Shea RL, Smith JL, Somers DO, Thomas SA, Thompson G, Wang R. Bioorg Med Chem Lett. 2007;17:1296. doi: 10.1016/j.bmcl.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Gaillard P, Jeanclaude-Etter I, Ardissone V, Arkinstall S, Cambet Y, Camps M, Chabert C, Church D, Cirillo R, Gretener D, Halazy S, Nichols A, Szyndralewiez C, Vitte PA, Gotteland JP. J Med Chem. 2005;48:4596. doi: 10.1021/jm0310986. [DOI] [PubMed] [Google Scholar]
- 19.Jiang R, Duckett D, Chen W, Habel J, Ling YY, LoGrasso P, Kamenecka TM. Bioorg Med Chem Lett. 2007;17:6378. doi: 10.1016/j.bmcl.2007.08.054. [DOI] [PubMed] [Google Scholar]
- 20.Alam M, Beevers RE, Ceska T, Davenport RJ, Dickson KM, Fortunato M, Gowers L, Haughan AF, James LA, Jones MW, Kinsella N, Lowe C, Meissner JW, Nicolas AL, Perry BG, Phillips DJ, Pitt WR, Platt A, Ratcliffe AJ, Sharpe A, Tait LJ. Bioorg Med Chem Lett. 2007;17:3463. doi: 10.1016/j.bmcl.2007.03.078. [DOI] [PubMed] [Google Scholar]
- 21.Humphries PS, Lafontaine JA, Agree CS, Alexander D, Chen P, Do QQ, Li LY, Lunney EA, Rajapakse RJ, Siegel K, Timofeevski SL, Wang T, Wilhite DM. Bioorg Med Chem Lett. 2009;19:2099. doi: 10.1016/j.bmcl.2009.03.023. [DOI] [PubMed] [Google Scholar]
- 22.Kamenecka T, Jiang R, Song X, Duckett D, Chen W, Ling YY, Habel J, Laughlin JD, Chambers J, Figuera-Losada M, Cameron MD, Lin L, Ruiz CH, LoGrasso PV. J. Med. Chem. 2010;53:419. doi: 10.1021/jm901351f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LoGrasso P, Kamenecka T. Mini Rev Med Chem. 2008;8:755. doi: 10.2174/138955708784912120. [DOI] [PubMed] [Google Scholar]
- 24.Huang LLS, Lunney EA, Planken SP. WO 2007/125405. 2007
- 25.The biochemical and cellular potency as well as the PK properties and brain penetration of all compounds (including the reference compounds 1a, 1b and 2) were tested at Scripps Florida. The biochemical IC50s for JNK1 and JNK3 were determined using HTRF. Briefly, final assay concentrations of JNK1, JNK3, biotinylated-ATF2 and ATP were 0.1 nM, 0.3 nM, 0.4 nM and 1 mM respectively. In each assay the phosphor-Thr71-ATF-2 product was detected using Europium-cryptate labeled anti-phospho-THR71-ATF-2 antibody. Streptavidin-allophycocyanin-XL was used as the acceptor. A 10-point dose-response curve for each compound was generated in duplicate and data was fit to a four parameter logistic. See Reference 19 for details of cell-based assays.
- 26.Patel VFKJL, Geuns-Meyer SD, Chaffee SC, Cee VJ, Hodous BL, Bellon S, Harmange J-C, Olivieri PR, Thaman MC, Dimauro EF, Buchanan JL, McGowan DC, Albrecht BK, Deak HL, Bemis JE, White R, Martin MW, Habgood GJ, Tempest PA, Masse CE, Buckner WH, Herberich BJ, Graceffa R, Zhang D, Xu S, Sham K, Rzasa RM, Falsey JR, Chakrabarti PP, Cao G-Q, Tomlinson SA, Pettus LH, Smith LA, Paras NA, Liu G, Demorin FF, Tasker A, Reed A. WO 2006/039718. 2006