Abstract
What specific genes and regulatory sequences contribute to the organization and functioning of brain circuits that support social behavior? How does social experience interact with information in the genome to modulate these brain circuits? Here we address these questions by highlighting progress that has been made in identifying and understanding two key “vectors of influence” that link genes, brain, and social behavior: 1) social information alters gene readout in the brain to influence behavior; and 2) genetic variation influences brain function and social behavior. We also briefly discuss how evolutionary changes in genomic elements influence social behavior and outline prospects for a systems biology of social behavior.
Genes and social behavior have long had a tempestuous relationship in both science and society, and the “nature-nurture” debate is alive and well (1). This controversy persists because the relationships between genes, brain and social behavior have complex entanglements across many different timeframes, ranging from organismal development and physiology all the way to evolutionary time (Fig. 1). Genes do not specify behavior directly, but rather encode molecular products that build and govern the functioning of the brain through which behavior is expressed. Brain development, brain activity and behavior depend on both inherited and environmental influences, and there is increasing appreciation that social information can in turn impact brain gene expression and behavior. Furthermore, variation in behavior shapes the evolution of genomic elements that influence social behavior through the feedback of natural selection.
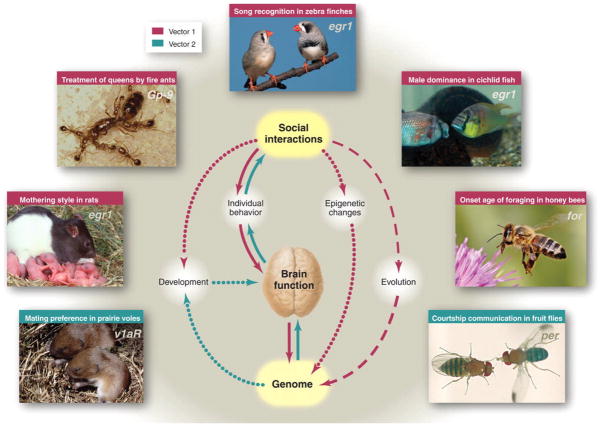
Figure 1. Complex relationships connect genes, the brain, and social behavior.
These relationships operate over three time scales: (i) physiological time via effects on brain activity (solid lines), (ii) developmental time via slower effects on brain development and genome modification (dotted lines), and (iii) evolutionary time via the processes of natural selection (dashed line). Arrow colors refer to Figs. 2 and 3 (pink, Fig. 2; blue, Fig. 3), which provide details about the nature of these interactions. Images depict some of the animals and genes featured in this review, clockwise from top: zebra finch (T. guttata), cichlid fish (A. burtoni), honey bee (A. mellifera), fruit fly (D. melanogaster), prairie vole (M. ochrogaster), rat (R. norvegicus), and fire ant (S. invicta). The genes listed (in italics on the photographs) are responsive to social interactions as described in the text.
What is social behavior? Animals perform many activities during the course of their lives to survive and reproduce: they find food, find mates, defend themselves, and in many cases care for their offspring or other relatives. These activities become “social” when they involve interactions among members of the same species in a way that influences immediate or future behavior. One of the fundamentals of social behavior is communication (2). A common denominator across diverse social behaviors is the production, reception and interpretation of signals that influence the behavior of the individual depending on the social context.
Given the diversity and complexity of social behavior, is it realistic to anticipate that conserved mechanisms and general principles operate to control social behavior at the level of genes and genomes? We believe so. Although specific behavioral outcomes vary widely from species to species, the biological needs that drive these behaviors are deeply shared. Social behavior clearly evolved multiple times, but probably within a framework of conserved neural mechanisms. All systems of social behavior share the following features: 1) They are acutely sensitive and responsive to social and environmental information; 2) This information is transduced within individual organisms by one or more primary sensory pathways; 3) The transduced neural signals are processed and integrated in specific circuits of the brain via conserved signal transduction and neuromodulatory systems; and 4) The resulting internal state of the animal ultimately controls behavioral activity.
Understanding the relationships between genes and social behavior is especially challenging because methods of experimental genetics have not been developed for animal species with the most compelling social lives, such as songbirds, cichlid fish, social insects (featured in this review) and voles, discussed in Donaldson and Young’s review in this issue (42). Fortunately, through progress in whole-genome sequencing and comparative genomics, “model social” species are taking their place alongside the classic model genetic species in molecular analyses of behavior (4). It is now possible to compare model social species that vary in behavior and in brain activity using one or more large scale ‘omics technologies (e.g., genomics, epigenomics, proteomics, metabolomics). Results can be readily translated to organisms with sequenced genomes allowing sophisticated genetic manipulations in model genetic species such as the mouse, Mus musculus, or the fruit fly, Drosophila melanogaster (5,6). Biologists no longer have to choose to study either a model genetic or a model social species, but can choose both. This underscores the importance of actively pursuing research on diverse organisms that can capture the full richness and range of social behaviors.
In this article we present selected findings that illustrate the relationships between genes and social behavior. As an organizational heuristic, we highlight two “vectors of influence” that describe the relationships between genes, brain and social behavior (Figs. 2 and 3) as follows: Vector 1 (Fig. 2): From social information to changes in brain gene expression, brain function and social behavior; and Vector 2 (Fig. 3): From variation in gene expression to variation in social behavior.
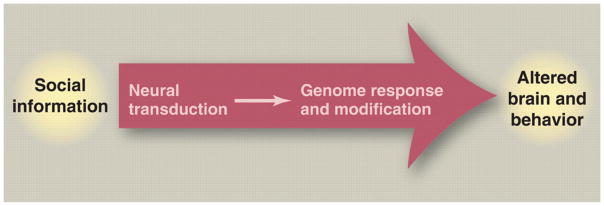
Figure 2. Vector 1: From social information to changes in brain function and behavior.
Social information is perceived by sensory systems and transduced into responses in the brain. Social information leads to developmental influences often mediated by parental care, as well as acute changes in gene expression that cause diverse effects (e.g., changes in metabolic states, synaptic connections, and transcriptional networks). Social information also can cause epigenetic modifications in the genome. Variation in both environment (VE) and genotype (VG) influences how social information is received and transduced and how these factors themselves interact (VE × VG).
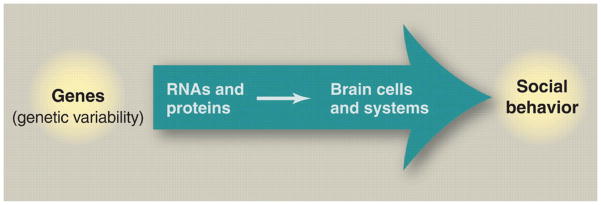
Figure 3. Vector 2: From genes to social behavior.
Genes influence the social behavior of an individual through their effects on brain development and physiology. This linkage is sensitive to both genetic (VG) and environmental (VE) variation and to their interactions (VG × VE).
Social influences on brain gene expression (Vector 1)
The genome was once thought to be a relatively passive blueprint guiding organismal development. Recent results show that genomes in fact remain highly responsive throughout life to a variety of stimuli associated with social behavior. Social information can lead to changes in brain and behavior via effects on the genome (Fig. 2).
The first demonstrations of gene responses to social stimuli focused on a handful of immediate early genes (7), and one of these has proven especially useful. Referred to now as egr1 (8), this transcription factor-encoding gene was discovered and named (ngfi-a, zif-268, krox-24, tis8, zenk) independently in different species. A specific link to social behavior was first suggested by studies in songbirds (9). Songbirds engage in rich social interactions that are mediated by learned vocal signals (birdsongs). The structure of songbird society varies by species, ranging from colonial to territorial, but in all cases songbirds appear to recognize and discriminate individual conspecifics according to their vocalizations. In the zebra finch (Taeniopygia guttata), the sound of another bird singing induces egr1 expression in a specific subregion of the auditory forebrain (9). Not simply an auditory response, egr1 expression in the this region is specifically linked to the social significance of the signal. Pure tones or white noise are ineffective stimuli and the response varies with familiarity to the particular song (10). The response is also enhanced when the bird is listening in the presence of conspecifics compared to when he is alone. This provides a neuro-molecular analog of the “audience effect,” a phenomenon in which an individual’s performance depends on whether he or she is alone or with others (11). Other social interactions trigger egr1 responses in other regions of the songbird brain, and the magnitude of the response can vary according to the intrinsic sociality of the species (12) and the immediate context of the experience (13).
egr1 was also the focus of another striking demonstration of brain gene responses resulting from recognition of social opportunity in a highly social cichlid fish (Astatotilapia burtoni) (14). In many animal societies, dominance hierarchies structure all social interactions; position in the hierarchy governs access to necessary resources and determines who reproduces and how often. A. burtoni has an elaborate dominance hierarchy, reinforced both by aggressive fighting and the ability of dominant males to ascertain relative rank by observation alone, using transitive inference to infer which male in a group is most dominant (15). Subordinate males have reduced fertility, and when the alpha male is removed from a group, a subordinate male assumes dominance within minutes. In this social ascent he displays dramatic changes in body coloration and behavior. Within minutes of starting his ascent, egr1 is induced specifically in the hypothalamic anterior preoptic area in neurons containing gonadotropin-releasing hormone (GNRH), a peptide critical for reproduction. These neurons increase in size and degree of dendritic arborization, while exhibiting an increase in GNRH mRNA and protein expression. This molecular response depends on the recognition of a social opportunity and ascension to dominance: it is not elicited in individuals who are already dominant. Since egr1is a transcription factor, it is likely that these effects on the GNRH neurons are direct, but this has not yet been demonstrated.
Although egr1 is only one of many socially responsive genes (see below), its molecular and cellular character provides insights of general significance. First, egr1 can be induced by brief experiences and its expression reaches a peak 20–60 min later, in a metaphorical “genomic action potential” (7). Second, it can immediately suppress or enhance the transcription of other genes depending on which proteins it interacts with in different cell types (16). The example of egr1 suggests how social experience might trigger changes in larger gene networks in the brain. Indeed, through the application of high-throughput technologies for measuring the expression of many genes simultaneously, it is now becoming apparent that responses to social stimuli can be massive, involving hundreds or thousands of genes and perhaps many different brain regions at once.
One of the first such studies used microarrays to measure brain gene expression patterns in the honey bee (Apis mellifera) at distinct life stages, and found expression differences in thousands of genes (17). Worker honey bees change jobs as they age; they spend the first 2–3 weeks of their adult life working in the hive caring for the brood, maintaining the nest and other activities, and then shift to collecting nectar and pollen outside the hive on behalf of their hive for the remainder of their 4–6 week life. Despite this fixed pattern of behavioral maturation, precisely when a bee redirects its energies from work in the hive to foraging depends on perceptions of its colony’s needs, which are communicated to it in part by pheromones. For example, if a bee colony loses a large fraction of its foraging force, some of the younger bees can speed up their rate of maturation and become “precocious” foragers. This can occur because of a decrease in the availability of inhibitory pheromones that are produced by older, foraging-age bees. In other words, although it looks like a bee foraging on a flower is a solitary affair, the onset age of foraging is subject to strong social influence. Perception of bee pheromones alters the expression of hundreds of genes in the bee brain over a period of days to weeks (18). Particularly affected are genes encoding transcription factors (18) and metabolic proteins (19).
Shifts in the expression of large populations of genes during social experience are also now being observed in microarray-based experiments using fish and songbirds, and in these cases the shifts are both large and rapid. In the swordtail fish, Xiphophorus nigrensis, different social experiences quickly induce different patterns of genomic response measured at the level of the whole brain (22). For example, some genes are turned on in females as they interact with attractive males, but are off when they interact with other females, and vice versa. In zebra finch song recognition experiments, thousands of other RNAs in addition to egr1 have now been found to increase or decrease within 30 min of onset of a novel song stimulus (21). A day after the song has been entrained by repetition, the same now-familiar song no longer induces any immediate response and a new and different baseline state has now emerged.
These observations suggest that social information can have large global effects on gene expression in the brain, perhaps best described as shifts in “neurogenomic states” rather than simple activation of particular genes in local neural circuits. A future challenge will be to confront the anatomical complexity of the brain, to describe and understand these genomic states at both finer and coarser scales of anatomy and time. For example, a single neuron may exist in different functional states as a result of modulation of synaptic proteins, which can alter the efficiency by which transient synaptic signals are consolidated into stable lasting cellular changes (7). In neuronal circuits and ensembles, changes in the expression of ion channel proteins could affect how quickly the cell can respond and lead to changes in network function (23). At the whole-brain level, the concept of a global neurogenomic state in the brain is exemplified by the familiar example of the daily sleep-waking cycle, which involves massive changes in gene expression throughout the brain as a function of behavioral state (24).
Social influences on brain gene expression: epigenetic effects
Social signals, in addition to initiating genomic state changes, can also trigger lasting epigenetic modifications of the genome, which have been defined as heritable changes in the expression of specific genes that are not due to changes in DNA sequence (Fig. 2). The first link between epigenetic regulation and social behavior was discovered in the transgenerational transmission of mothering style in rats (Rattus norvegicus) (25). Female rats that lick, groom, and nurse their pups extensively have offspring that are less responsive to stress and more responsive to their own pups. By contrast, pups that received less attention from their mothers are more easily stressed and show reduced responsiveness to their offspring.
These differences in responsiveness to stress can be passed from generation to generation, so they were assumed to be inherited via traditional genetics. Instead it was discovered that they stem from the fact that frequent mother-pup contact triggers at least two known epigenetic changes in DNA methylation and very likely many more such events. Methylation of the promoter region of the stress hormone glucocortocoid receptor gene allows NGFI-A, the protein product of the egr1 gene discussed above, to upregulate glucocorticoid expression, especially in the hippocampus (26). In addition, methylation of the α1b promoter region of the estrogen receptor gene results in the upregulation of estrogen receptors in the hypothalamus (27). These results indicate that multiple genes are differentially methylated in response to differential mothering. Together with the results presented above for songbirds and fish, they also demonstrate that social experience, acting via transcription factors such as NGFI-A, can induce both transient and lasting changes in brain gene expression.
To date epigenetic effects associated with social behavior have been studied at only a few genetic loci, but it is likely that many genes are similarly affected, especially in gene regulatory networks in the hypothalamic-pituitary-gonadal axis, known to be important for the regulation of a variety of forms of vertebrate social behavior (12, 28). Genome-wide assays of epigenetic changes, assessing different regions and cell types in the brain, are necessary to fully understand how specific epigenetic modifications influence social behavior.
Genotype-environment interactions and social behavior
The effects of social information on brain function and social behavior differ among individuals as a result of genetic variation between individuals (Fig. 2) (29). Interactions between genotype and environment must always be accounted for in molecular analyses of social behavior. Study of these interactions has become an important focus in medicine, as psychiatric geneticists have been searching for genotype-environment interactions that might have explanatory power for understanding a wide spectrum of psychiatric disorders; some, like autism and depression, involve social behavior gone awry. Evidence for such interactions has been reported but is controversial (30, 31, Abbot).
Clear evidence of genotype-environment interactions in social behavior and gene expression has been obtained in studies of the fire ant Solenopsis invicta (32). Fire ants, like honey bees, live in colonies with thousands of workers, but while honey bee colonies have just a single queen, fire ant colonies can have one or more. The tendency to have either one or more queens has a genetic basis in fire ants. A genetic locus has been identified, General Protein 9 (Gp-9), which is involved in regulating a key aspect of fire ant social organization, namely, the treatment of queens by the workers. Homozygous “BB” queens are larger and more fecund than Bb queens, and BB workers will only accept a single BB queen, resulting in one-queen colonies. Bb workers will accept multiple Bb queens, resulting in larger, and more ecologically invasive, multi-queen colonies. Cross-fostering showed that BB workers in a Bb colony become tolerant of multiple Bb queens, and also take on a Bb gene expression profile. In fact, worker gene expression profiles were more strongly affected by colony genotype than their own genotype.
From genes to social behavior (Vector 2)
Gene or behavioral variants, either within populations of the same species or between species, offer a chance to understand how genetic information influences the development and function of brain circuits that mediate social behavior (Fig. 3). With the explosion in genome sequencing, detection and analysis of genomic variation is becoming more and more routine even in species without extensive histories of genetic analysis. However, such a comparative approach is particularly effective when direct experimental manipulation of genes or molecular pathways can be incorporated in the analysis to study behavioral consequences. A now-classic precedent is seen in the study of courtship communication in fruit flies, which is described extensively in this issue by Dickson (REF). Species with relatively solitary lifestyles, such as fruit flies, can show social behavior in the context of courtship, even if it occurs for only a few fleeting moments. During courtship, animals communicate through multiple sensory modalities to collect and process information about species, gender, receptivity, and quality of a potential mate.
Drosophila has a courtship “song” produced by the wing that is characterized by species-specific temporal coding. Comparison of two Drosophila species led to identification of a specific difference in the period gene correlated with these temporal differences in song structure. Transferring a small piece of the period gene from D. melanogaster to D. simulans caused the melanogaster males to produce the simulans call, rather than melanogaster call (41). Thus manipulation of even a single gene can have profound effects on behaviors associated with reproductive success.
Species differences also were exploited to study the molecular basis of courtship in the monogamous prairie vole, (Microtus ochrogaster) in comparison with the polygamous montane vole (M. montanus). As discussed in this issue (42), sequence variation in the 5′ region of the vasopressin receptor gene v1aR causes differences both in where this gene is expressed in the brain and in mating preferences. Recent findings showing no such relationship between genetic variation and monogamy in other vole species (43) provide an excellent opportunity to explore how changes in different components of signaling pathways might result in similar changes in social behavior.
Behavioral variants within populations of Drosophila melanogaster led to discovery of another gene involved in social behavior, but in honey bees. Regulatory polymorphisms in for are implicated in inter-individual differences in foraging behavior; flies with higher levels of for expression are more active while foraging than are flies with lower levels of for expression (36). Foraging in Drosophila is not a social behavior, but these findings led to studies of for orthologs in social insects. In honey bees, brain for expression is higher in foragers than in hive bees, and socially induced precocious foragers show a precocious increase in for brain expression. Furthermore, pharmacological treatment that activates the for pathway causes precocious foraging (35). The role of for may be widespread in social insects (35).
Much like for but studied in vertebrates, variation in the forkhead box P2 gene (foxp2) influences behaviors that have important social roles in multiple species, including human speech (37) and other forms of animal communication (38–40). for encodes guanosine 3′,5′-monophosphate-dependent protein kinase (PKG) and foxp2 encodes a developmentally important transcription factor. Genes like for and foxp2 may function as elements in a developmental or neural toolkit for building the circuits and systems underlying diverse socially embedded behaviors (35) – even if they do not encode social behavior in any mechanistic sense.
For genes like for and foxp2, the link between gene and behavior may be appreciated best by considering the timeframe of evolution (Fig. 1, dotted line). Through selection, genes may evolve according to their effects on a social behavior – even if their mechanistic roles in the neural expression of that behavior are subtle and indirect. Effects of selection may be detected at several different levels in comparative genome sequence data, including statistical variations in amino acid codon frequencies, regulatory sequences, and gene copy number (33). As sequence information becomes available for more and more species, it should be possible to use molecular evolution algorithms to determine whether genes such as for and foxp2 have been subjected to positive selection in particular lineages (JensenEtAl07), thus strengthening the connection between a social behavior and its evolutionary roots. Such genes will provide important tools for understanding the evolution of genes and other genomic elements that influence social behavior.
Prospects
Some progress has been made in understanding the specific relationships between genes and social behavior in a few species, but this enterprise is still in the formative stages (see also papers by Dickson and Donaldson and Young in this issue). Understanding the molecular basis of social behavior presents a formidable intellectual challenge for several reasons. First, because behavior is diverse, diverse species must be used to extract the general principles that govern the molecular bases of social behavior. Dissecting a complex behavior into its components can help identify root similarities across distantly related species (4). But even if deep molecular conservation is found among diverse species, one important issue remains. How can molecular pathways involved in behavior be conserved even when species show major differences in brain structure and the overall organization of the nervous system?
The second challenge is that the molecular path linking genes and behavior is invariably complicated (44): there are many levels of neural and neuroendocrine regulation that lie between the genome and a social behavior, including transcription, translation, post-translational modifications, epigenetic changes, brain metabolism, neural (electrochemical) activity, and neuromodulation. Moreover, this regulation occurs in complex and dispersed temporal and spatial patterns within the brain, over real time, developmental time, and lifespan time. The study of social behavior adds an additional tier of complexity because it depends on interactions and communication among individuals. In most cases, social behavior must be studied in a natural context in which the full repertoire of environmental influences and behaviors are expressed.
Despite the challenges, genetic and genomic approaches hold great promise for elucidating the molecular basis of social behavior. We have reasonably detailed knowledge of the two physical substrates responsible for behavior: the brain and the genome. We have a strong and growing arsenal of ‘omic technologies and increasingly sophisticated methods of systems biology to profile changes in the brain during social behaviors. The time is ripe to combine this knowledge and these tools to aim for a comprehensive understanding of social behavior in molecular terms.
Acknowledgments
We thank J. Desjardins, A. Fernald, K.A. Hughes, D.B. Kelley, K. Maruska, C. Olin, M.B. Sokolowski, LJ. Stubbs, members of the Clayton and Robinson laboratories and two anonymous reviewers for reviews of this manuscript and C. Harrell for graphical assistance. Research by the authors cited here was supported by the following grants: NIH R01 NS051820 and NS045264 (D.F.C); NIH NS34950 Javits Award and United States-Israel Binational Science Foundation 200596 (R.D.F); and NIH R01 GM073644, NSF Frontiers in Biological Research EF04-25852, U.S. Department of Agriculture AG2003-35302-13490, and a Burroughs Wellcome Fund Innovation Award (G.E.R).
References and Notes
- 1.Robinson GE. Science. 2004;304:397–399. doi: 10.1126/science.1095766. [DOI] [PubMed] [Google Scholar]
- 2.Floreano D, Mitri S, Magnenat S, Keller L. Curr Biol. 2007;17:514–591. doi: 10.1016/j.cub.2007.01.058. [DOI] [PubMed] [Google Scholar]
- 3.Ruan H, Wu CF. Proc Natl Acad Sci. 2008;105:7506–7510. doi: 10.1073/pnas.0711127105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson GE, Grozinger CM, Whitfield CW. Nature Rev Gen. 2005;6:1–15. doi: 10.1038/nrg1575. [DOI] [PubMed] [Google Scholar]
- 5.Young LJ, Nilsen R, Waymire KG, MacGregor GR, Insel TR. Nature. 1999;400:766–768. doi: 10.1038/23475. [DOI] [PubMed] [Google Scholar]
- 6.Ben-Shahar Y, Robichon A, Sokolowski MB, Robinson GE. Science. 2002;296:742–744. doi: 10.1126/science.1069911. [DOI] [PubMed] [Google Scholar]
- 7.Clayton DF. Neurobiol Learn Mem. 2000;74:185–216. doi: 10.1006/nlme.2000.3967. [DOI] [PubMed] [Google Scholar]
- 8.Eyre TA, et al. Nucleic Acids Res. 2006 Jan 1;34:D319. doi: 10.1093/nar/gkj147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mello CV, Vicario DS, Clayton DF. PNAS. 1992;89:6818–6822. doi: 10.1073/pnas.89.15.6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong S, Clayton DF. Genes, Brain and Behavior. doi: 10.1111/j.1601-183X.2008.00423.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vignal C, Andru J, Mathevon N. Eur J Neurosci. 2005;22:949–955. doi: 10.1111/j.1460-9568.2005.04254.x. [DOI] [PubMed] [Google Scholar]
- 12.Goodson JL. Horm Behav. 2005;48:11–22. doi: 10.1016/j.yhbeh.2005.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarvis ED, Scharff C, Grossman MR, Ramos JA, Nottebohm F. Neuron. 1998;21:775–788. doi: 10.1016/s0896-6273(00)80594-2. [DOI] [PubMed] [Google Scholar]
- 14.Burmeister SS, Jarvis ED, Fernald RD. PloS Biology. 2005;3:0001–0009. doi: 10.1371/journal.pbio.0030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grosenick L, Clement TS, Fernald RD. Nature. 2007;445:429–432. doi: 10.1038/nature05511. [DOI] [PubMed] [Google Scholar]
- 16.O’Donovan KJ, Tourtellotte WG, Milbrandt J, Baraban JM. Trends Neurosci. 1999;22:167–173. doi: 10.1016/s0166-2236(98)01343-5. [DOI] [PubMed] [Google Scholar]
- 17.Whitfield CW, Cziko AM, Robinson GE. Science. 2003;302:296–299. doi: 10.1126/science.1086807. [DOI] [PubMed] [Google Scholar]
- 18.Grozinger CM, Sharabash N, Whitfield CW, Robinson GE. PNAS. 2003;2:14519–25. doi: 10.1073/pnas.2335884100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ament SA, Corona M, Pollack HS, Robinson GE. PNAS. 2008;105:4226–4231. doi: 10.1073/pnas.0800630105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruse AA, Stripling R, Clayton DF. Neurobiol Learn Mem. 2000;74:179–184. doi: 10.1006/nlme.2000.3968. [DOI] [PubMed] [Google Scholar]
- 21.Replogle K, et al. BMC Genomics. 2008;9:131. doi: 10.1186/1471-2164-9-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cummings ME, Larkins-Ford J, Reilly CRL, Wong RY, Ramsey M, Hofmann HA. Proc R Soc B. 2008;275:393–402. doi: 10.1098/rspb.2007.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H, Wu MM, Zakon HH. Dev Neurobiol. 2007;67:1289–1304. doi: 10.1002/dneu.20404. [DOI] [PubMed] [Google Scholar]
- 24.Cirelli C, Gutierrez CM, Tononi G. Neuron. 2004;41:35–43. doi: 10.1016/s0896-6273(03)00814-6. [DOI] [PubMed] [Google Scholar]
- 25.Champagne FA, Francis DD, Mar A, Meaney MJ. Phys Behav. 2003;79:359–371. doi: 10.1016/s0031-9384(03)00149-5. [DOI] [PubMed] [Google Scholar]
- 26.Weaver IC, et al. Nat Neurosci. 2004;7:847–54. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 27.Champagne DL, Bagot RC, van Hasselt F, Ramakers G, Meaney MJ, de Kloet ER, Joëls M, Krugers H. J Neurosci. 2008;28:6037–6045. doi: 10.1523/JNEUROSCI.0526-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfaff DW. Drive. MIT Press; 1999. [Google Scholar]
- 29.Falconer DS, Mackay TFC. Intro Quant Gen. 4. Longmans Green; Harlow Essex, UK: 1996. [Google Scholar]
- 30.Caspi A, Moffitt TE. Nature Revs Neurosci. 2006;7:583–590. doi: 10.1038/nrn1925. [DOI] [PubMed] [Google Scholar]
- 31.Lahiri DK, Maloney B. Nature Rev Neurosci. Vol. 7. 2006. [Google Scholar]
- 32.Wang J, Ross KG, Keller L. PLoS Genet. 2008;4:e1000127. doi: 10.1371/journal.pgen.1000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sebat J, et al. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krebs JR, Davies NB. Blackwell Science. 1997. Behavioural Ecology: An Evolutionary Approach. [Google Scholar]
- 35.Toth AL, Robinson GE. Trends in Genetics. 2007;23:334–341. doi: 10.1016/j.tig.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 36.Sokolowski MB. Nature Rev Gen. 2001;2:879–890. doi: 10.1038/35098592. [DOI] [PubMed] [Google Scholar]
- 37.Enard W, Przeworski M, Fisher S, Lai C, Wiebe V, Kitano T, Monaco A, Pääbo S. Nature. 2002;418:869–872. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- 38.Shu W, Cho JY, Jiang Y, Zhang M, Weisz D, Elder GA, Schmeidler J, De Gasperi R, Sosa MA, Rabidou D, Santucci AC, Perl D, Morrisey E, Buxbaum JD. PNAS. 2005;102:9643–9648. doi: 10.1073/pnas.0503739102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haesler S, Rochefort C, Georgi B, Licznerski P, Osten P, Scharff C. PLoS Biol. 2007;5:e321. doi: 10.1371/journal.pbio.0050321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li G, Wang J, Rossiter SJ, Jones G, Zhang S. PloS ONE. 2(9):e900. doi: 10.1371/journal.pone.0000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wheeler DA, Kyriacou CP, Greenacre ML, Yu Q, Rutila JE, Rosbash M, Hall JC. Science. 1991;251:1082–1085. doi: 10.1126/science.1900131. [DOI] [PubMed] [Google Scholar]
- 42.Young, this issue
- 43.Fink S, Excoffier L, Heckel G. PNAS. 2006;103:10956–10960. doi: 10.1073/pnas.0602380103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hall JC. J Neurogenet. 2003;17:1–90. [PubMed] [Google Scholar]