Abstract
This study aimed to describe a short term ex vivo assay to predict response to epidermal growth factor receptor (EGFR) targeted therapy (gefitinib) in adenocarcinoma patients. Four patients with locally advanced esophageal adenocarcinoma were treated with gefitinib (250 mg/day) for 14 days and pharmacokinetic (PK) studies were conducted to monitor plasma drug concentrations. Tumor cells were sampled by endoscopic biopsy prior to (baseline, day 0) and at the completion of (day 14) treatment. Cells obtained at baseline were exposed to gefitinib in short-term cell culture conditions (ex vivo assay). Western blot analyses with phospho-specific antibodies were performed to evaluate activation and biochemical response to therapy of EGFR and its downstream signaling components ERK and AKT ex vivo and in vivo. The in vivo profiles were correlated with the gefitinib-mediated alteration in Proliferating Cell Nuclear Antigen (PCNA) expression, a marker of cell proliferation. The correlation between EGFR expression and ERK activity was also investigated by immunohistochemical analysis in pretreatment biopsies. Mutational status of the genes encoding EGFR, K-RAS, and PI3KCA (the phosphoinositide 3-kinase catalytic subunit p110) as well as expression levels of PTEN protein were tested in order to investigate potential confounders of the gefitinib effect. All patients completed the gefitinib therapy. PK studies demonstrated constant gefitinib concentrations during the treatment, confirming persistent exposure of target tissue to the drug at sufficient levels to achieve EGFR blockade. Ex vivo culture with gefitinib resulted in distinct response patterns representing various states of activity of the ERK and AKT pathways. The results of the ex vivo studies correctly predicted the pharmacodynamic (PD) effects of the agents in tumor tissue in vivo. PCNA expression correlated with ERK pathway inhibition, but not with gefitinib-mediated inhibition of EGFR activity alone. Immunohistochemical analysis performed on pretreatment biopsies correlated with Western blot analysis of EGFR and phospho-ERK expression. No mutations were identified in exons 18–21 of EGFR, exons 2 and 3 of K-RAS or exons 9 and 22 of PI3KCA. Levels of PTEN were comparable across tumors. The novel pharmacodynamic approach described In this proof of principle study may be useful to refine the patient selection to maximize the potential benefits of drugs and design individualized rational therapies for cancer patients.
Keywords: Gefitinib, targeted agents, esophageal carcinoma, chemosensitivity assay
INTRODUCTION
The modern revolution in molecular biology has led to the characterization of important signaling pathways in cancer, the elements of which have emerged as candidate targets for novel therapies (1). The specificity of targeted agents raises the possibility of therapy tailored to the specific biologically relevant molecular aberrations for individual cases. Early development schemes for these agents, however, continue to be largely empirical rather than direct knowledge of target activity within a given patient’s tumor. Recent experience with targeted agents in solid tumors, while still promising, has yielded modest results (2–6). Notably, retrospective analyses of clinical trials are consistently revealing that differences in treatment effect between subgroups of patients can be linked to specific molecular profiles (7–13). These findings suggest the potential for a more rational approach to trial design, and eventually, to individualized cancer therapy.
Among many targeted therapeutic agents, compounds targeting EGFR have shown promise in cancer treatment. The EGFR signaling pathway plays a key role in the regulation of cell proliferation, survival and differentiation. EGFR is a plasma membrane glycoprotein composed of extracellular ligand-binding, transmembrane and intracellular tyrosine kinase domains (14). The binding of extracellular ligands to EGFR causes autophosphorylation of multiple tyrosine residues within the cytoplasmic domain of EGFR, which consequently activates the downstream signaling molecules involved in the RAS-RAF-MEK-ERK and PI3K/AKT signaling cascades that contribute to cell proliferation (15). In some tumors, however, other membrane tyrosine kinase receptors may redundantly activate these pathways, in which case EGFR blockade may not have its desired effect (16–18).
EGFR is expressed in the large majority of carcinomas and its dysregulation has been implicated in resistance to conventional chemotherapy and poor clinical outcome (19) EGFR expression is detected in about 50% of non–small cell lung cancers and about 30% to 70% of esophageal carcinomas (20,21).Therefore, EGFR became an attractive target for anticancer therapy in aerodigestive carcinomas. In recent years, a number of new compounds including small molecule tyrosine kinase inhibitors gefitinib (ZD1839, Iressa; AstraZeneca, Wilmington, DE) and erlotinib (Tarceva, OSI-774) and monoclonal antibodies such as cetuximab (IMC-C225) targeting the EGFR signaling have been developed and their therapeutic effects have been extensively evaluated in clinical studies (22,23). Although EGFR inhibitors have significant efficacy and less toxicity than conventional chemotherapeutics, cancer patients show highly variable responses to these agents. Hence, there is a pressing need to develop clinically useful tools that may potentially optimize patient selection and therapeutic efficacy.
The incorporation of tumor-based pharmacodynamic (PD) markers may permit improved understanding of mechanisms of sensitivity and resistance to different agents in different tumor types and among individual patients. However, thus far, this concept has not been incorporated into clinical trials in a systematic or uniform fashion.
We have previously described a simple and reliable short term ex vivo chemosensitivity assay to explore pharmocodynamic predictors and indicators of response to biologically targeted agents in pre-clinical animal models (24,25). In that work, we demonstrated that cancer cells obtained by tumor fine-needle aspiration biopsy can be used to predict the efficacy of targeted drugs prior to systemic treatment and to correlate target pathway inhibition in vivo with antitumor response. In the present study, we showed that tumor cells obtained by endoscopic biopsy prior to initiation of therapy can be successfully assayed ex vivo to predict the in vivo pharmacodynamic effects of gefitinib in patients with locally advanced esophageal cancer.
MATERIALS AND METHODS
Eligibililty Criteria
Patients with histologically confirmed invasive adenocarcinoma of the distal esophagus (below 20 cm from the incisors) or gastroesophageal junction (<2 cm extension into the gastric cardia) were enrolled and treated at the Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins Hospital. All patients were newly diagnosed and with no prior treatment, greater than 18 years of age and with an ECOG performance status of 0 or 1. Disease was limited to the primary and regional nodes, though celiac nodal involvement (M1a) was permitted for primary tumors in the distal esophagus or gastroesophageal junction, as long as the disease could be encompassed in a single radiation port. The treatment protocol and human subject studies were approved by the Institutional Review Board at the Johns Hopkins University, and all patients provided informed consent.
Patients received gefitinib (AstraZeneca, Wilmington, DE) 250 mg/day for 14 days. Endoscopic biopsies were obtained at the beginning (day 0) and at the end of the 14 day period.
Endoscopic Biopsy and Tissue Handling
Endoscopic forceps biopsies of esophageal tumors were carried out by a single board certified gastroenterologist (SJ) following standard procedures. Separate informed consent was obtained for these procedures. Touch preps of fresh tissue were immediately evaluated by cytologic stain for the presence of tumor cells, and all evaluations were done by a single cytopathologist (SA). Portions of each sample were used for the ex vivo chemosensitivity assay while the remainder of the tissue was used to prepare paraffin blocks.
Pharmacokinetics
Trough concentrations of gefitinib were determined pre-treatment and on days 8 and 14 of the run-in period. Blood samples were collected in heparinized tubes at these three time points. The blood samples were immediately placed in an ice bath and then centrifuged at 1000 g at 4°C for 10 minutes. The plasma was stored at −20°C until analyzed. Quantitation of gefitinib in total and unbound plasma concentrations was performed using a validated LC-MS-MS analytical assay (for total concentrations) and equilibrium dialysis (for unbound concentrations) (26).
Immunohistochemistry
The expression of total-EGFR, and phospho-ERK, in tumor tissues was determined as described (25). Briefly, 5 µm sections of the paraffin blocks were deposited onto positively charged glass slides. Slides were deparaffinized and rehydrated in graded alcohols before antigen retrieval in citrate buffer (pH 6.0) for 20 minutes at 100C. The sides were then cooled for 20 minutes before they were washed in 1× TBST (DAKO Corp. Carpinteria, CA). Slides were incubated in 3% H2O2 for 10 minutes, followed by the primary monoclonal antibody phospho-ERK1/2 (Cell Signaling Technology, Beverly, MA) in 1:50 dilution or polyclonal total-EGFR antibody (Cell Signaling Technology, Beverly, MA) for 60 minutes. Tris-HCl [0.2 mol/L (pH 7.5); Quality Biological, Inc., Gaithersburg,MD] was used as the antibody diluent solution. Negative controls were incubated for 60 minutes with the antibody diluent solution [0.2 mol/L Tris-HCl (pH 7.5); Quality Biological]. Staining was developed using the DAKO LSAB+ System (DAKO). Slides were washed using 1× TBST after incubation with each reagent and with distilled H2O following incubation with 3,3'-diaminobenzidine.
Ex vivo studies
Tumor samples were obtained by endoscopic biopsy before the start of treatment, as described in Figure 1. All samples were confirmed to be enriched in cancer cells by microscopic assessment of a staff cytopathologist (SA). Tumor cells were prepared by mincing the tumor tissue in sterile prewarmed complete RPMI-1640 culture medium containing 10% FBS, penicillin (200 µg/ml), and streptomycin (200 µg/ml). Tissue was disaggregated by filtering through mesh and cells were incubated with 0.04% trypan blue (Sigma) dissolved in PBS (9.1 mM Na2HPO4, 1.7 mM NaH2PO4, and 150 mM NaCl, pH 7.4) to assess viability. The viable (membrane-intact) and dead cells were then counted and the total viable cell count was used to calculate final working volumes. Ex vivo drug treatment was performed as described before (25). Briefly, viable tumor cells were treated with vehicle (control) or gefitinib (10 µM) (AstraZeneca, Wilmington,DE) in a humidified 5% CO2 incubator at 37°C for 18 hours. No fibroblast or endothelial cell growth was observed. Following treatment, cells were collected, washed, lysed in lysis buffer (50 mM Tris-HCl, 0.25 M NaCl, 0.1% (v/v) Triton X-100, 1 mM EDTA, 50 mM NaF, and 0.1 mM Na3VO4, pH 7.4) containing protease and phosphatase inhibitors (Sigma Chemical Co., St. Louis, MO) and analyzed by Western blot.
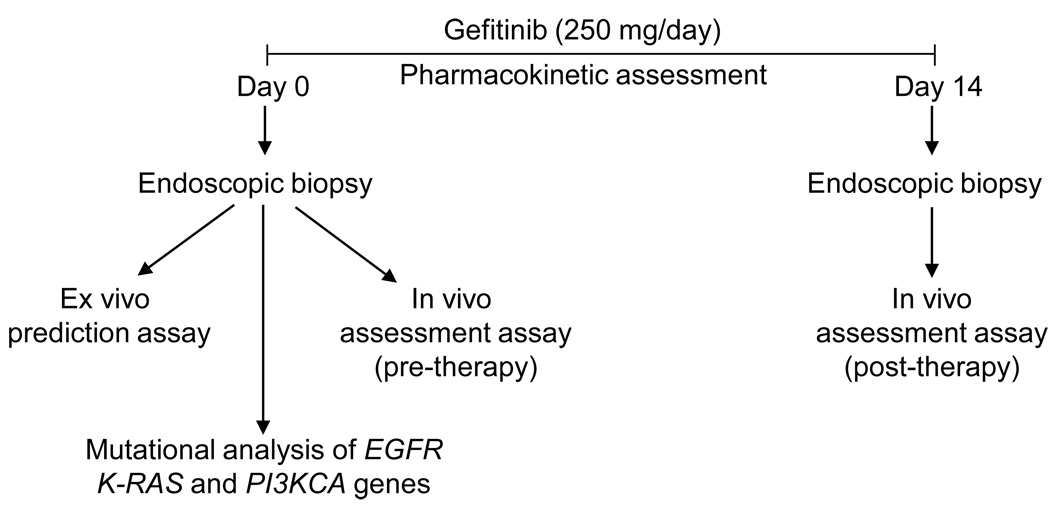
Figure 1.
Schematic representation of the experimental design. Tumor samples were collected by endoscopy prior to (day 0) and after (day 14) the initiation of gefitinib therapy. Ex vivo and in vivo assays were performed to predict and assess the efficacy of therapy in esophageal cancer patients. Mutational analysis of EGFR, K-RAS and PI3KCA genes were performed in pre-therapy tumor samples. Plasma concentrations of gefitinib were monitored in all patients during the therapy.
In vivo studies
Imprint cytology (touch prep) technique was used to prepare air dried/Diff Quik (AD/DQ)-stained glass slides from endoscopic biopsy samples obtained before (day 0) and at the end (day 14) of gefitinib therapy (Figure 1) and whole cell lysates were prepared from tumor cells as described previously (24,25). The cellular composition of the touch prep imprints was assessed for adequate sampling under the microscope prior to protein extraction.
Western blot analysis
Protein concentrations obtained from endoscopic tumor samples were quantified before each experiment. Western blot analyses were performed as previously described (24,25). Briefly, protein extracts (25 µg) electrophoresed on a 10% (w/v) SDS-polyacrylamide gel were electrotransfered to Immobilon-P membranes (Millipore Corporation, Bedford, MA), membranes were blocked and incubated with primary antibodies. The antibodies tested were total-and phospho-EGFR, total-and phospho-ERK1/2, total-and phospho-AKT, PTEN and PCNA. All primary antibodies were obtained from the Cell Signaling Technology, Beverly, MA. Antibody binding was visualized using enhanced chemiluminescence (SuperSignal West Pico, Pierce, Rockford, IL) and autoradiography after treatment of membranes with horseradish peroxidase (HRP)-conjugated secondary antibodies, rabbit IgG-HRP (Cell Signaling Technology, Beverly, MA), or mouse IgG-HRP (Cell Signaling Technology, Beverly, MA).
DNA isolation, PCR and sequencing
Total genomic DNA was isolated from AD/DQ-stained tumor smear samples after microscopic assessment using QIAamp DNA Micro Kit (Qiagen Inc., Valencia, CA), according to the manufacturer’s protocol. Nested PCR reactions were carried out using the primers described previously (27,28). Primers were synthesized by Integrated DNA Technologies (Coralville, IA). Amplification was carried out in 25µl reaction containing 40ng DNA, 1µM each primer, 0.15µM each dNTPs (Life Technologies, Rockville, MD), and either 10mM Tris-HCl, pH 8.3, 50mM KCl, 2.5mM MgCl2, and 0.5U Taq Gold DNA Polymerase (Perkin Elmer, Foster City, CA) or HotStar1X PCR buffer and 0.5U HotStarTaq DNA Polymerase (Qiagen Inc, Valencia, CA). Thermocycling was carried out in a Thermo Hybaid MBS 0.2S (Needham Heights, MA). Samples were denatured for 12 minutes at 94°C followed by 35 cycles of 94°C for 20 seconds, annealing (see chart) for 30 seconds, and 72°C for 30 seconds followed by a final extension at 72°C for 10 minutes. The 25µl PCR products were treated with 0.2 µl Exonuclease I (P/N 70073Z, USB) and 2 µl Shrimp Alkaline Phosphatase (P/N 70092x, USB). Products were incubated for 1 hour at 37°C followed by 15 min at 80°C. 5 µl of the treated products were electrophoresed on 1% agarose/1% NuSieve gels to check for amplification. For sequencing, 8–10 µl of treated product, depending on amplification intensity, was combined with 2 µl 10µM of the forward and reverse PCR primers in separate tubes. Sequencing reactions and sequence analysis was carried out by the DNA Analysis Facility. PCR products were sequenced using fluorescent dideoxy terminator method of cycle sequencing. Reactions were run on a 3730xl DNA Analyzer (Applied Biosystems Division, Foster City, CA) following Applied Biosystems protocols. Sequence data was analyzed using Sequencher Software (Gene Codes, Ann Arbor, MI), followed by manual review.
RESULTS
Patient Population
In this exploratory study of the pharmacokinetics and pharmacodynamics of gefitinib, four patients (all male, ages 39, 54, 63, 69) with locally advanced esophageal adenocarcinoma were treated for 14 days with gefitinib alone. Each patient underwent both pre- and post- therapy biopsies, and the pharmacokinetic data were obtained during the treatment (Figure 1). No toxicities other than diarrhea and rash were noted.
Pharmacokinetics
Given the tumor location in the upper gastrointestinal tract, there is a theoretical concern regarding altered absorption of oral agents by these patients. In order to measure pharmacodynamic effects at the drug-target interface, one must ensure adequate uptake, distribution and tumor tissue penetration of drug. In the unlikely setting of no PD effect, it is possible that the result is due to low concentration of drug at the target site. In this analysis, no patient took a drug that impacted the CYP3A4 enzyme. PK analysis revealed a constant trough gefitinib plasma concentration of 100 ng/ml (data not shown), which is adequate to block EGFR activity and achieve antitumor effect in tumor cells (29,30).
Prediction of tumor response to gefitinib ex vivo
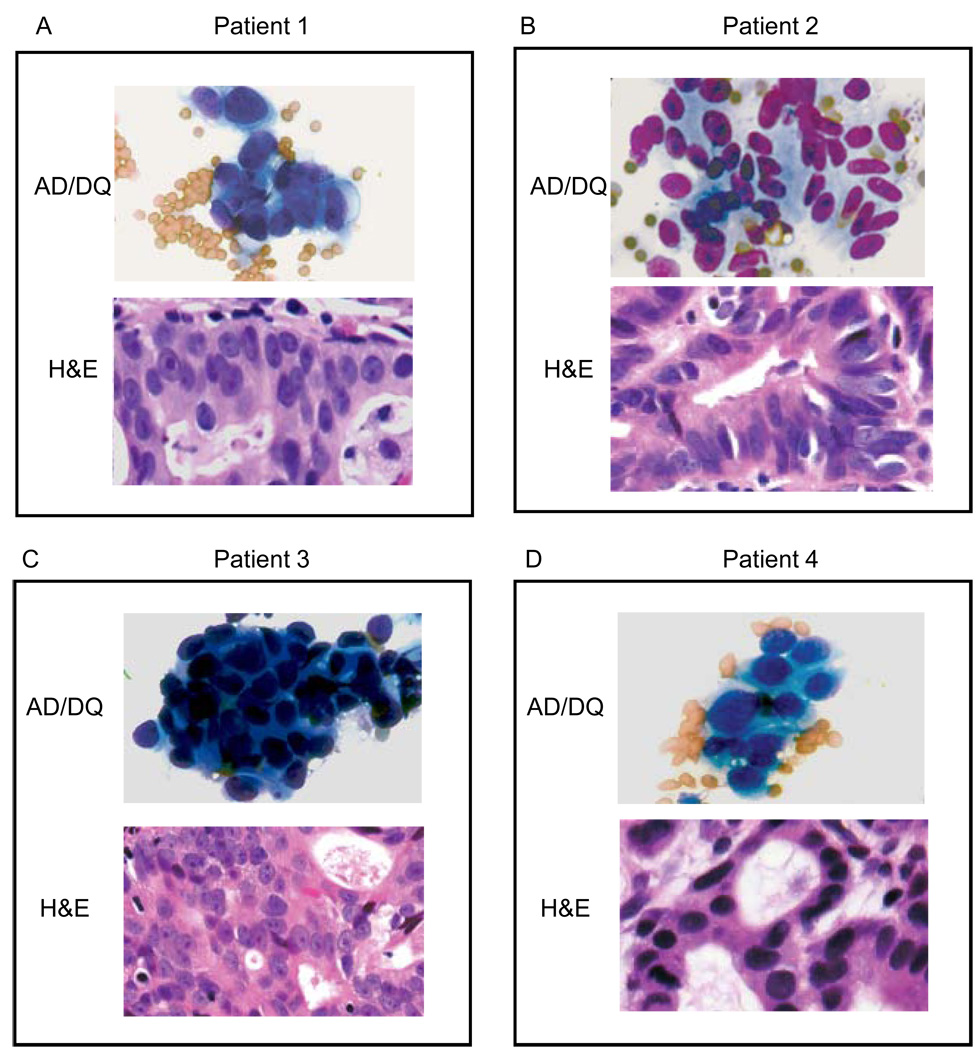
To determine whether the short term ex vivo chemosensitivity assay could predict gefitinib treatment effects in vivo, cancer cells were harvested by endoscopic biopsy prior to initiation of systemic treatment (Figure 1) and treated with gefitinib ex vivo, after which EGFR signaling pathway inhibition was analyzed by Western blot. Prior to analysis by Western blot, AD/DQ-stained smears were prepared from endoscopic biopsy samples as previously described. Morphologic assessment of the cytologic smears demonstrated that, on average, 90% of the cells were neoplastic, with some red blood cells and a negligible amount of connective tissue fragments in the background (Figures 2A–D). Under these cell culture conditions, no fibroblast or endothelial cell growth was detected. No significant apoptosis or necrosis was detected in tumor cells obtained before or after gefitinib treatment.
Figure 2.
A–D) Upper picture, tumor touch prep (AD/DQ), lower picture, paraffin sections of tumor tissue (H&E) obtained by esophageal endoscopy prior to gefitinib therapy. AD/DQ: Air-dried and Diff-Quik stain; H&E: Hematoxylin and Eosin stain.,
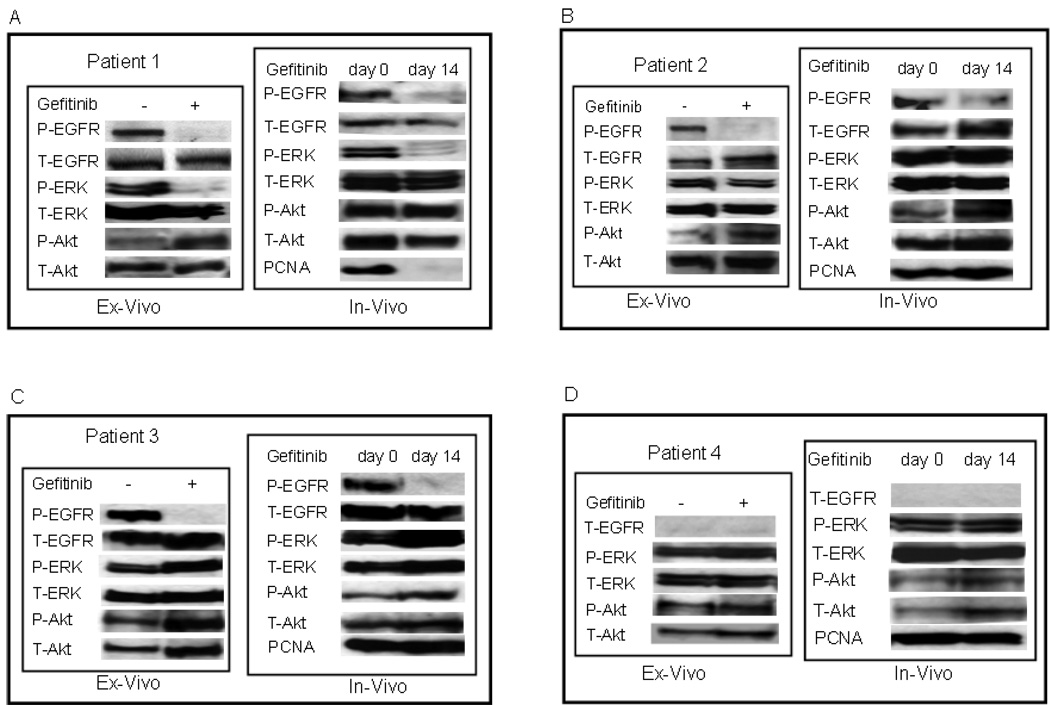
As shown in Figure 3A, ex vivo treatment of tumor cells with gefitinib completely blocked EGFR phosphorylation in patient 1. Analysis of downstream signaling pathways revealed that drug treatment inhibited phosphorylation of ERK, but not of AKT. This finding suggests that in this patient’s tumor cells the ERK pathway was under the control of EGFR, however, the PI3K/AKT pathway was constitutively activated independent of EGFR activity. In patients 2 and 3 (Figures 3B and C, respectively), ex vivo gefitinib therapy dramatically inhibited EGFR phosphorylation; however, there was no concurrent inhibition in phosphorylation of the downstream ERK and AKT. This demonstrates that activation of the downstream ERK and AKT pathways is not dependent on EGFR. As illustrated in Figure 3D, in patient 4 no EGFR protein and accordingly no P-ERK or P-AKT inhibition was identified by Western blot analysis.
Figure 3.
A–D) Ex vivo and in vivo studies with gefitinib in four esophageal cancer patients. Ex vivo studies were performed with tumor cells collected from endoscopic tumor biopsies obtained before the systemic gefitinib therapy (day 0). For the in vivo assessment analysis, tumor samples were collected from the same patient before (day 0) and 14 days after therapy with gefitinib. Cell lysates were prepared from AD/DQ-stained touch prep slides to determine protein phosphorylation (P-) and/or total expression (T-) levels of EGFR ERK, AKT and PCNA proteins on Western blot analysis.
Assessment of response to gefitinib in tumor cells in vivo
To analyze the pharmacodynamic efficacy of gefitinib in vivo, AD/DQ-stained smears were prepared from tumor samples obtained by esophageal endoscopy prior to initiation (day 0) and at the end (day 14) of treatment (Figure 1). Following morphologic evaluation, whole cell extracts were prepared from AD/DQ-stained tumor touch prep samples and the phosphorylation status and/or expression levels of EGFR, ERK and AKT were determined on Western blot analysis. Additionally, to correlate target inhibition in vivo with tumor growth inhibition at the molecular level, we also measured gefitinib mediated changes in the expression levels of PCNA, a marker of cell proliferation, in patient samples obtained before and after 14 days of gefitinib therapy. As summarized in Table 1, overall, the pharmacodynamic effect of gefitinib ex vivo was concordant with in vivo target effect for all four patients. The phosphorylation of EGFR was inhibited in patients 1, 2, and 3, (Figure 3A–C), whereas no EGFR expression was detected in patient 4 (Figure 3D). Only in patient 1 did gefitinib therapy cause inhibition of the P-ERK, but the level of P-AKT was unaffected. In this patient, gefitinib led to strong inhibition of PCNA levels. In all other patients, gefitinib therapy did not inhibit downstream signaling pathway activities and PCNA expression in tumor cells in vivo, suggesting that PCNA expression correlates with ERK pathway inhibition. These data show that the ex vivo assays can predict the pharmacodynamic effects of gefitinib in esophageal tumors prior to in vivo treatment.
Table 1.
Correlation between the ex vivo and in vivo response to gefitinib therapy in esophageal cancer patients.
| Gefitinib response | |||||||
|---|---|---|---|---|---|---|---|
| P-EGFR | P-ERK | P-AKT | PCNA in vivo |
||||
| Ex vivo | In vivo | Ex vivo | In vivo | Ex vivo | In vivo | ||
| PT 1 | off1 | off | off | off | on2 | on | off3 |
| PT 2 | off | off | on | on | on | on | on |
| PT 3 | nd4 | nd | on | on | on | on | on |
| PT 4 | off | off | on | on | on | on | on |
off, effective pathway inhibition by gefitinib;
on, persistent pathway activity after gefitinib treatment.
off, PCNA expression downregulated by gefitinib;
on, PCNA expression remained high in tumor cells in vivo upon gefitinib therapy;
nd, not detected. PT 1–4: patient numbers.
Mutation Analysis of EGFR, K-RAS and PI3KCA
Acquired activating mutations of the intracellular tyrosine kinase domain of the EGFR are known to drive the neoplastic behavior of non-small cell lung cancer (31). Furthermore, cancers with these mutations are much more sensitive to the inhibitory effects of the EGFR inhibitors erlotinib and gefitinib (32). In addition, also in lung cancer, cells that become resistant to these therapies often harbor a second EGFR mutation or a point mutation in the K-RAS oncogene (28,32). There are reports of similar EGFR mutations in esophageal adenocarcinoma; however, their impact on the biology and response to therapy of these tumors is unknown (33–35).
Mutations in EGFR and/or the downstream signaling proteins K-RAS and PI3KCA have been implicated in differential inhibition of the ERK and AKT pathways in tumor cells (27). Furthermore, deletion mutation of the PTEN tumor suppressor protein can lead to constitutive activation of AKT, independent of mitogenic stimuli (36). Given the observed differential response of these four esophageal cancer patients to gefitinib we measured PTEN protein expression in tumor cells and also performed genomic sequencing analysis of mutational hotspots in exons 18–21 of EGFR, exons 2 and 3 of K-RAS and exons 9 and 22 of PI3KCA. As summarized in Table 2, no mutations were identified in mutation hot spots in the EGFR, K-RAS and PI3KCA genes and all tumors expressed comparable amounts of PTEN protein. These results suggest that the differential sensitivity of ERK and AKT pathways to gefitinib in patient 1 is not due to gain of function mutations in PI3KCA or deletion mutations in PTEN genes. Furthermore, failure of gefitinib to block activation of signaling pathways downstream to EGFR in patients 2 and 3 are likely not due to EGFR or K-RAS mutations.
Table 2.
Mutational analysis of EGFR, K-RAS and PI3KCA genes and expression levels of PTEN protein in tumor cells.
| Gene mutation (exons) | PTEN protein expression relative to β-actin1 |
|||
|---|---|---|---|---|
|
EGFR (18–21) |
PIK3CA (9+21) |
K-RAS (1+2) |
||
| PT 1 | WT | WT | WT | 0.54 |
| PT 2 | WT | WT | WT | 0.45 |
| PT 3 | WT | WT | WT | 0.64 |
| PT 4 | WT | WT | WT | 0.62 |
WT, wild-type;
PTEN protein expression levels relative to β-actin, analyzed by Western blot.
Immunohistochemical analysis of total EGFR and phospho-ERK expressions in tumor tissue samples
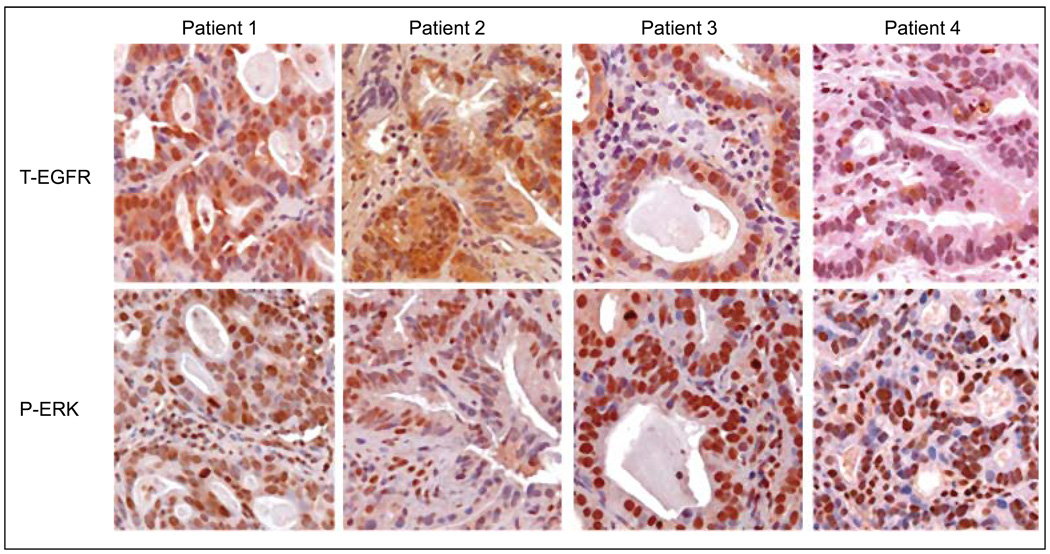
To corroborate the results observed in the in vivo assays, tumor samples obtained before gefitinib treatment were used to determine protein expression levels of EGFR and phospho-ERK by immunohistochemical staining. As illustrated in Figure 4, interestingly, EGFR displayed weak to moderate cytoplasmic and nuclear expression patterns in patients 1, 2 and 3, as described previously (37); whereas strong membranous EGFR staining was observed in positive control sections (data not shown). No EGFR expression was observed in patient 4 (Figure 4). However, in all patients phospho-ERK showed strong nuclear staining regardless expression of total-EGFR. The expression patterns of both EGFR and phospho-ERK closely correlated with Western blot analysis shown above (Figure 3), confirming that the results are representative of tumor cells.
Figure 4.
Representative immunohistochemical staining of total EGFR (T-EGFR) and phospho-ERK (P-ERK) in patients’ tumors obtained by endoscopic biopsy prior to initiation of gefitinib therapy.
DISCUSSION
The results of this study show that small samples of tumor cells obtained by endoscopic biopsy prior to initiation of therapy can be used for pharmacodynamic assays to predict how tumors will respond to an EGFR inhibitor. In patients with locally advanced esophageal adenocarcinoma, we found a strong correlation between the pharmacodynamic effects of gefitinib on activation of EGFR and its downstream targets in ex vivo and in vivo conditions. Pharmacokinetic studies performed in esophageal cancer patients demonstrated that plasma concentrations of gefitinib were adequate to achieve anti-tumor effect (29,30).
No consistent correlation was found between blockade of EGFR phosphorylation and the effect on downstream signaling pathways. For example, phospho-EGFR was blocked in three patients expressing high EGFR activity, but in only one of these were ERK pathway activity and expression of PCNA downregulated. These results suggest that differential regulation of these signaling pathways is often independent of EGFR. Examples of alternative regulation are widely known and include: PTEN deletion/deactivation to activate AKT, independent activation of RAS and MAP kinase pathways and upregulation of other membrane tyrosine kinase receptors such as MET, HER-2, PDGFR (15, 18–20,27,36,38). While we identified no mutations in the known mutational hotspots of exons of EGFR, K-RAS and PI3KCA genes and no deletion of PTEN, this does not exclude other bypass mechanisms or the presence of other infrequent mutations within these genes.
Thus far, the implementation of pharmacodynamic studies in clinical trials with targeted therapeutics has been limited. To date, most predictive studies have relied on analysis of static determinants of tumor response, such as receptor expression, gene amplification, and target mutations in tumor materials (39–41). Despite examples of success in some cases including trastuzumab for HER2 overexpressing breast cancer, imatinib for CML and GIST and rituximab for CD20 expressing lymphomas, there is accumulating evidence that the detection of target protein expression in pre-treatment samples may be inadequate to predict the activity of targeted drugs. This is especially true since the effect of a given agent may depend upon alterations in the activity of signaling pathway components both upstream and downstreamof the target protein (11,15). This fact is best illustrated in studies correlating EGFR expression with tumor response to anti-EGFR therapy conducted by different groups. In esophageal, head and neck and non-small cell lung cancers, EGFR overexpression has been reported to correlate with worse prognosis (42–44). However, although overexpression of EGFR detected by fluorescence in situ hybridization was found to be predictive of tumor reponse (45), EGFR expression demonstrated by immunohistochemistry has not been accepted as a reliable predictor of responsiveness in most studies of EGFR inhibitors (23, 45–47). We propose that the pharmacodynamic ability of a drug to inhibit the target pathway is more important as a predictor of efficacy than the expression or activation of the target per se. Thus, the assays described herein provide a more direct, mechanism-based approach for determining both prognostic and predictive markers in tumor cells. This approach allows testing drug-mediated changes in the activity of specific pharmacodynamic markers of EGFR signaling and correlates these effects with tumor response at the molecular level.
In the previous drug sensitivity assays growth inhibition or cell death has been used to predict tumor sensitivity to conventional chemotherapeutic agents (48–51). However, due to poor tumor growth under assay conditions, and lack of reliable criteria for defining tumor sensitivity, these chemosensitivity assays have not gained wide acceptance in clinical oncology. We recently presented a novel short term biological (ex vivo) assay to predict the efficacy of targeted drugs by analyzing pharmacodynamic markers specific to the molecular pathways targeted by individual agents (25). Our results herein demonstrate that cytologically confirmed endoscopic tumor samples can yield viable tumor cells and sufficient protein quantities for analysis of the efficacy of targeted drugs prior to (ex vivo) and during (in vivo) systemic treatment. Microscopic evaluation of air-dried/Diff-Quik–stained cytologic slides prepared from endoscopic tumor samples rendered an immediate assessment of specimen adequacy and diagnosis. This on-site sample evaluation also enabled avoidance of necrotic samples and demonstrated that there were no apoptotic changes after treatment with gefitinib.
While these results should be considered preliminary due to our small sample size, this is the first study to our knowledge that describes utilization of a short term ex vivo assay than can predict the PD effect of a targeted agent in cancer patients. Despite the lack of patient outcome data, our correlative results support our hypothesis that the ex vivo assays described herein may have potential to evaluate the sensitivity or resistance of a patient’s tumor to targeted therapeutics prior to initiation of therapy. The implementation of this novel approach may allow enrichment of clinical trials and help to facilitate the development of targeted therapeutics. Furthermore, simultaneous profiling of multiple targets and pathways may permit determination of resistance mechanisms in tumor cells. This may ultimately lead to designing the most efficient drug combination regimens for individual patients while sparing patients unlikely to respond to a drug.
REFERENCES
- 1.Krause DS, Van Etten RA. Tyrosine kinases as targets for cancer therapy. N Engl J Med. 2005;353:172–187. doi: 10.1056/NEJMra044389. [DOI] [PubMed] [Google Scholar]
- 2.Rothenberg ML, LaFleur B, Levy DE, et al. Randomized phase II trial of the clinical and biological effects of two dose levels of gefitinib in patients with recurrent colorectal adenocarcinoma. J Clin Oncol. 2005;23:9265–9274. doi: 10.1200/JCO.2005.03.0536. [DOI] [PubMed] [Google Scholar]
- 3.Fukuoka M, Yano S, Giaccone G, et al. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (The IDEAL 1 Trial) [corrected] J Clin Oncol. 2003;21:2237–2246. doi: 10.1200/JCO.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 4.Giaccone G, Herbst RS, Manegold C, et al. Gefitinib in combination with gemcitabine and cisplatin in advanced non-small-cell lung cancer: a phase III trial--INTACT 1. J Clin Oncol. 2004;22:777–784. doi: 10.1200/JCO.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Herbst RS, Prager D, Hermann R, et al. TRIBUTE: a phase III trial of erlotinib hydrochloride (OSI-774) combined with carboplatin and paclitaxel chemotherapy in advanced non-small-cell lung cancer. J Clin Oncol. 2005;23:5892–5899. doi: 10.1200/JCO.2005.02.840. [DOI] [PubMed] [Google Scholar]
- 6.Kris MG, Natale RB, Herbst RS, et al. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: a randomized trial. Jama. 2003;290:2149–2158. doi: 10.1001/jama.290.16.2149. [DOI] [PubMed] [Google Scholar]
- 7.Bell DW, Lynch TJ, Haserlat SM, et al. Epidermal growth factor receptor mutations and gene amplification in non-small-cell lung cancer: molecular analysis of the IDEAL/INTACT gefitinib trials. J Clin Oncol. 2005;23:8081–8092. doi: 10.1200/JCO.2005.02.7078. [DOI] [PubMed] [Google Scholar]
- 8.Cappuzzo F, Hirsch FR, Rossi E, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97:643–655. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- 9.Haas-Kogan DA, Prados MD, Tihan T, et al. Epidermal growth factor receptor, protein kinase B/Akt, and glioma response to erlotinib. J Natl Cancer Inst. 2005;97:880–887. doi: 10.1093/jnci/dji161. [DOI] [PubMed] [Google Scholar]
- 10.Hirsch FR, Varella-Garcia M, McCoy J, et al. Increased epidermal growth factor receptor gene copy number detected by fluorescence in situ hybridization associates with increased sensitivity to gefitinib in patients with bronchioloalveolar carcinoma subtypes: a Southwest Oncology Group Study. J Clin Oncol. 2005;23:6838–6845. doi: 10.1200/JCO.2005.01.2823. [DOI] [PubMed] [Google Scholar]
- 11.Mellinghoff IK, Wang MY, Vivanco I, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 12.Ogino S, Meyerhardt JA, Cantor M, et al. Molecular alterations in tumors and response to combination chemotherapy with gefitinib for advanced colorectal cancer. Clin Cancer Res. 2005;11:6650–6656. doi: 10.1158/1078-0432.CCR-05-0738. [DOI] [PubMed] [Google Scholar]
- 13.Pao W, Wang TY, Riely GJ, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 15.Brivanlou AH, Darnell JE., Jr Signal transduction and the control of gene expression. Science. 2002;295:813–818. doi: 10.1126/science.1066355. [DOI] [PubMed] [Google Scholar]
- 16.Anderson MR, Harrison R, Atherfold PA, et al. Met receptor signaling: a key effector in esophageal adenocarcinoma. Clin Cancer Res. 2006;12:5936–5943. doi: 10.1158/1078-0432.CCR-06-1208. [DOI] [PubMed] [Google Scholar]
- 17.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 18.Wang SE, Narasanna A, Perez-Torres M, et al. HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer Cell. 2006;10:25–38. doi: 10.1016/j.ccr.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 19.Mendelsohn J, Baselga J. Status of epidermal growth factor receptor antagonists in the biology and treatment of cancer. J Clin Oncol. 2003;21:2787–2799. doi: 10.1200/JCO.2003.01.504. [DOI] [PubMed] [Google Scholar]
- 20.Karamouzis MV, Grandis JR, Argiris A. Therapies directed against epidermal growth factor receptor in aerodigestive carcinomas. JAMA. 2007;298:70–82. doi: 10.1001/jama.298.1.70. [DOI] [PubMed] [Google Scholar]
- 21.Pande AU, Iyer RV, Rani A, et al. Epidermal growth factor receptor-directed therapy in esophageal cancer. Oncology. 2007;73:281–289. doi: 10.1159/000132393. [DOI] [PubMed] [Google Scholar]
- 22.Baselga J, Arteaga CL. Critical update and emerging trends in epidermal growth factor receptor targeting in cancer. J Clin Oncol. 2005;23:2445–2459. doi: 10.1200/JCO.2005.11.890. [DOI] [PubMed] [Google Scholar]
- 23.Minna JD, Gazdar AF, Sprang SR, et al. Cancer. A bull's eye for targeted lung cancer therapy. Science. 2004;304:1458–1461. doi: 10.1126/science.1099578. [DOI] [PubMed] [Google Scholar]
- 24.Hidalgo M, Amador ML, Jimeno A, et al. Assessment of gefitinib- and CI-1040-mediated changes in epidermal growth factor receptor signaling in HuCCT-1 human cholangiocarcinoma by serial fine needle aspiration. Mol Cancer Ther. 2006;5:1895–1903. doi: 10.1158/1535-7163.MCT-05-0525. [DOI] [PubMed] [Google Scholar]
- 25.Rubio-Viqueira B, Mezzadra H, Nielsen ME, et al. Optimizing the development of targeted agents in pancreatic cancer: tumor fine-needle aspiration biopsy as a platform for novel prospective ex vivo drug sensitivity assays. Mol Cancer Ther. 2007;6:515–523. doi: 10.1158/1535-7163.MCT-06-0388. [DOI] [PubMed] [Google Scholar]
- 26.Jones HK, Stafford LE, Swaisland HC, et al. A sensitive assay for ZD1839 (Iressa) in human plasma by liquid-liquid extraction and high performance liquid chromatography with mass spectrometric detection: validation and use in Phase I clinical trials. J Pharm Biomed Anal. 2002;29:221–228. doi: 10.1016/s0731-7085(02)00014-6. [DOI] [PubMed] [Google Scholar]
- 27.Janmaat ML, Rodriguez JA, Gallegos-Ruiz M, et al. Enhanced cytotoxicity induced by gefitinib and specific inhibitors of the Ras or phosphatidyl inositol-3 kinase pathways in non-small cell lung cancer cells. Int J Cancer. 2006;118:209–214. doi: 10.1002/ijc.21290. [DOI] [PubMed] [Google Scholar]
- 28.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 29.Ranson M, Hammond LA, Ferry D, et al. ZD1839, a selective oral epidermal growth factor receptor-tyrosine kinase inhibitor, is well tolerated and active in patients with solid, malignant tumors: results of a phase I trial. J Clin Oncol. 2002;20:2240–2250. doi: 10.1200/JCO.2002.10.112. [DOI] [PubMed] [Google Scholar]
- 30.Wakeling AE, Guy SP, Woodburn JR, et al. ZD1839 (Iressa): an orally active inhibitor of epidermal growth factor signaling with potential for cancer therapy. Cancer Res. 2002;62:5749–5754. [PubMed] [Google Scholar]
- 31.Kosaka T, Yatabe Y, Endoh H, et al. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 32.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo M, Liu S, Lu F. Gefitinib-sensitizing mutations in esophageal carcinoma. N Engl J Med. 2006;354:2193–2194. doi: 10.1056/NEJMc052698. [DOI] [PubMed] [Google Scholar]
- 34.Mimori K, Ogawa K, Okamoto M, et al. Clinical significance of enhancer of zeste homolog 2 expression in colorectal cancer cases. Eur J Surg Oncol. 2005;31:376–380. doi: 10.1016/j.ejso.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Sudo T, Mimori K, Nagahara H, et al. Identification of EGFR mutations in esophageal cancer. Eur J Surg Oncol. 2006 doi: 10.1016/j.ejso.2006.10.034. [DOI] [PubMed] [Google Scholar]
- 36.Kokubo Y, Gemma A, Noro R, et al. Reduction of PTEN protein and loss of epidermal growth factor receptor gene mutation in lung cancer with natural resistance to gefitinib (IRESSA) Br J Cancer. 2005;92:1711–1719. doi: 10.1038/sj.bjc.6602559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Psyrri A, Yu Z, Weinberger PM, et al. Quantitative determination of nuclear and cytoplasmic epidermal growth factor receptor expression in oropharyngeal squamous cell cancer by using automated quantitative analysis. Clin Cancer Res. 2005;11:5856–5862. doi: 10.1158/1078-0432.CCR-05-0420. [DOI] [PubMed] [Google Scholar]
- 38.Magne N, Fischel JL, Dubreuil A, et al. Influence of epidermal growth factor receptor (EGFR), p53 and intrinsic MAP kinase pathway status of tumour cells on the antiproliferative effect of ZD1839 ("Iressa") Br J Cancer. 2002;86:1518–1523. doi: 10.1038/sj.bjc.6600299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han SW, Kim TY, Hwang PG, et al. Predictive and prognostic impact of epidermal growth factor receptor mutation in non-small-cell lung cancer patients treated with gefitinib. J Clin Oncol. 2005;23:2493–2501. doi: 10.1200/JCO.2005.01.388. [DOI] [PubMed] [Google Scholar]
- 40.Moroni M, Veronese S, Benvenuti S, et al. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 2005;6:279–286. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 41.Scartozzi M, Bearzi I, Berardi R, et al. Epidermal growth factor receptor (EGFR) status in primary colorectal tumors does not correlate with EGFR expression in related metastatic sites: implications for treatment with EGFR-targeted monoclonal antibodies. J Clin Oncol. 2004;22:4772–4778. doi: 10.1200/JCO.2004.00.117. [DOI] [PubMed] [Google Scholar]
- 42.Chung CH, Ely K, McGavran L, et al. Increased epidermal growth factor receptor gene copy number is associated with poor prognosis in head and neck squamous cell carcinomas. J Clin Oncol. 2006;24:4170–4176. doi: 10.1200/JCO.2006.07.2587. [DOI] [PubMed] [Google Scholar]
- 43.Gibson MK, Abraham SC, Wu TT, et al. Epidermal growth factor receptor, p53 mutation, and pathological response predict survival in patients with locally advanced esophageal cancer treated with preoperative chemoradiotherapy. Clin Cancer Res. 2003;9:6461–6468. [PubMed] [Google Scholar]
- 44.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, et al. Molecular predictors of outcome with gefitinib in a phase III placebo-controlled study in advanced non-small-cell lung cancer. J Clin Oncol. 2006;24:5034–5042. doi: 10.1200/JCO.2006.06.3958. [DOI] [PubMed] [Google Scholar]
- 45.Tsao MS, Sakurada A, Cutz JC, et al. Erlotinib in lung cancer - molecular and clinical predictors of outcome. N Engl J Med. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 46.Parra HS, Cavina R, Latteri F, et al. Analysis of epidermal growth factor receptor expression as a predictive factor for response to gefitinib ('Iressa', ZD1839) in non-small-cell lung cancer. Br J Cancer. 2004;91:208–212. doi: 10.1038/sj.bjc.6601923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perez-Soler R, Chachoua A, Hammond LA, et al. Determinants of tumor response and survival with erlotinib in patients with non--small-cell lung cancer. J Clin Oncol. 2004;22:3238–3247. doi: 10.1200/JCO.2004.11.057. [DOI] [PubMed] [Google Scholar]
- 48.Andreotti PE, Cree IA, Kurbacher CM, et al. Chemosensitivity testing of human tumors using a microplate adenosine triphosphate luminescence assay: clinical correlation for cisplatin resistance of ovarian carcinoma. Cancer Res. 1995;55:5276–5282. [PubMed] [Google Scholar]
- 49.Gerhardt RT, Perras JP, Sevin BU, et al. Characterization of in vitro chemosensitivity of perioperative human ovarian malignancies by adenosine triphosphate chemosensitivity assay. Am J Obstet Gynecol. 1991;165:245–255. doi: 10.1016/0002-9378(91)90075-3. [DOI] [PubMed] [Google Scholar]
- 50.Kern DH, Weisenthal LM. Highly specific prediction of antineoplastic drug resistance with an in vitro assay using suprapharmacologic drug exposures. J Natl Cancer Inst. 1990;82:582–588. doi: 10.1093/jnci/82.7.582. [DOI] [PubMed] [Google Scholar]
- 51.Sharma S, Neale MH, Di Nicolantonio F, et al. Outcome of ATP-based tumor chemosensitivity assay directed chemotherapy in heavily pre-treated recurrent ovarian carcinoma. BMC Cancer. 2003;3:19. doi: 10.1186/1471-2407-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]