Abstract
Chromatin plays a fundamental role in eukaryotic genomic regulation, and the increasing awareness of the importance of epigenetic processes in human health and disease emphasizes the need for understanding the structure and function of the nucleosome. Recent advances in chromatin structural studies, including the first structures of nucleosomes containing the Widom 601 sequence and the structure of a chromatin protein-nucleosome assembly, have provided new insight into stretching of nucleosomal DNA, nucleosome positioning, binding of metal ions, drugs and therapeutic candidates to nucleosomes, and nucleosome recognition by nuclear proteins. These discoveries ensure promising future prospects for unravelling structural attributes of chromatin.
Keywords: Nucleosome, Chromatin, DNA structure, Histone structure, Nuclear protein factors, Anticancer agents, Drug targets, Metal cations
Introduction
As the basic repeating subunit of chromatin, the nucleosome assembly of histone proteins and DNA is a key participant in genetic processes [1–3]. For many years, the main function of the histone packaging of DNA was considered to be to compact the double helix into a repressive structure, with the more interesting aspects of gene activation occurring when the DNA was freed of its protein wardens. We now understand that the nucleosome is an active participant in both the repression and activation of eukaryotic genes, and that the proper coordination of gene expression involves the dynamic action of hundreds of chromatin protein factors.
Nucleosomes consist of a nucleosome core, entailing 145 to 147 bp of DNA in complex with a histone octamer of H2A, H2B, H3 and H4 proteins, in addition to a variable length of linker DNA that can be associated with linker histones. The positioning of histone octamers on the double helix in vivo is influenced by DNA sequence and other nuclear factors [2]. Genome-wide mapping of nucleosomes in several organisms has shown that a nucleosome-free region precedes a well-defined nucleosome positioned at the transcription start site of genes, which is followed by a series of positioned nucleosomes in the body of the gene [4]. Beyond nucleosome positioning, gene activity is further regulated by alteration of nucleosome composition through histone variant substitutions, DNA methylases and histone modifying enzymes, and by dynamic rearrangement of nucleosomes through the activity of chromatin remodelling factors [5–7]. Lastly, the nature of higher order interactions between nucleosomes in the chromatin fibre can determine gene activation status, such as highly compact states associated with silenced regions [8,9]. Therefore, three outstanding questions in chromatin biology and eukaryotic gene regulation are: what is the basis for the non-random (non-statistical) positioning of nucleosomes, how do chromatin enzymes, factors and small molecules recognize and act upon their nucleosome substrates and what higher order structures are relevant to which chromatin states?
Since the first crystallographic studies of the nucleosome core particle (NCP) in the mid to late 1970s [10–12], structural investigations of the nucleosome have provided a wealth of information to understand chromatin biology. Here we focus on exciting progress and insights provided by recent structural characterization of nucleosomes and nucleosome assemblies. These findings explain how a chromatin factor binds to the nucleosome, provide critical insight into the contribution of DNA sequence to nucleosome positioning and structure and illuminate how small molecules, including metal ions and therapeutic agents, interact with the nucleosome.
Assemblies with Protein Factors
In contrast to the richness of data available for the NCP s internal organization, we currently possess relatively sparse information about how the nucleosome is recognized by an increasing number of chromatin enzymes and factors being discovered. In the 13 years since the 2.8 Å resolution structure of the NCP [13], only synthetic DNA minor groove ligands had been cocrystallized with the nucleosome (Table 1) [14,15]. Fortunately, the technical problems of cocrystallizing a chromatin protein in complex with the NCP have recently been overcome
Table 1.
Nucleosomal X-ray crystal structures
| Variant/Ligand | Histone species | DNA length | DNA sequence | References |
|---|---|---|---|---|
| NCPs differing in histone content | ||||
| Major histones | Frog | 146 | α-satellite | [13,25] |
| Major histones | Chicken | 146 | α-satellite | [55] |
| Major histones | Yeast | 146 | α-satellite | [56] |
| Major histones | Human | 146 | α-satellite | [57] |
| Major histones | Fly | 147 | α-satellite | [58] |
| H2A.Z | Frog/Mouse | 146 | α-satellite | [59] |
| macroH2A | Mouse/Human | 146 | α-satellite | [60] |
| H3T | Human | 146 | α-satellite | [61] |
| Sin mutants | Frog | 146 | α-satellite | [62] |
| Methylated H3/H4 | Frog | 146 | α-satellite | [63] |
| H3K56Q/E | Frog | 146 | α-satellite | [64] |
| NCPs differing in DNA fragment | ||||
| Distinct sequence | Frog | 146 | α-satellite (type 2) | [25] |
| Length | Frog | 147 | α-satellite | [25,26] |
| Length | Frog | 145 | α-satellite | [27] |
| A16, MRE | Frog | 147 | α-satellite-based* | [65] |
| TTTAA elements | Frog | 147 | α-satellite-based | [39] |
| Strong positioning | Frog | 145 | 601* | [24] |
| NCP in complex with small molecules | ||||
| Polyamides | Frog | 146 | α-satellite | [14] |
| Polyamide dimer | Frog | 146 | α-satellite | [15] |
| LANA peptide | Frog | 146 | α-satellite | [20] |
| Platinum drugs | Frog | 147 | α-satellite | [41] |
| Intercalating agent | Frog | 145 | α-satellite | [29] |
| MnCl2, MnSO4 | Frog | 147 | α-satellite | [49,50] |
| Co2+, Ni2+ | Frog | 147 | α-satellite | [44] |
| Rb+, Cs+ | Frog | 147 | α-satellite | [44] |
| NCP in complex with protein factors | ||||
| RCC1 | Frog | 147 | 601* | [19] |
| Higher order structure | ||||
| Tetranucleosome | Frog | 694 | 601* | [53] |
Non-palindromic DNA fragments (*).
RCC1 is the guanine-exchange factor for the Ran small GTPase [16]. RCC1 recruits Ran to chromosomes by directly binding to nucleosomes, and activates Ran s nucleotide exchange activity to create a concentration gradient around the chromosomes of Ran in its GTP bound state. This RanGTP gradient is a critical nuclear signal for mitotic spindle formation and for nucleocytoplasmic transport of macromolecules. The crystal structures of the RCC1 β-propeller on its own and in complex with Ran determined previously had explained how RCC1 activated Ran s enzymatic activity [17,18], but the structural basis for how RCC1 interacted with the nucleosome was not known.
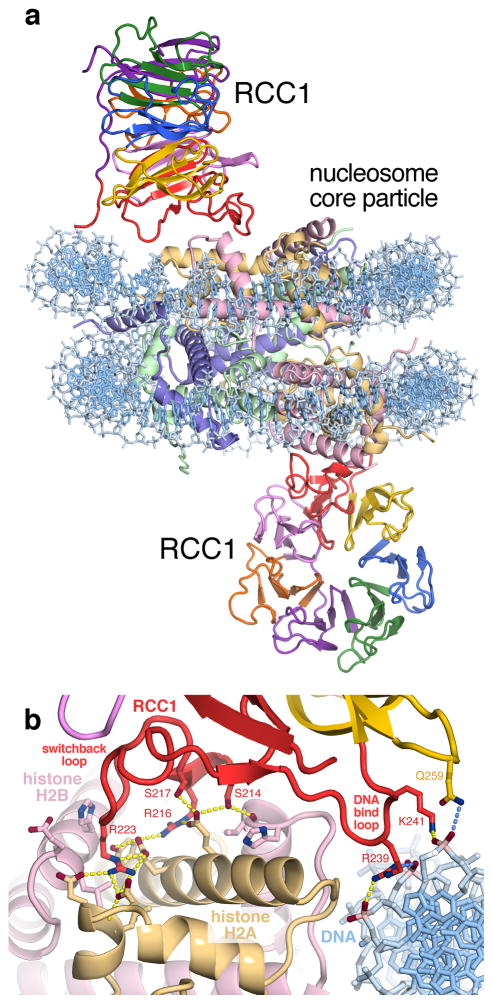
Makde et al have determined the crystal structure of the 300 kDa RCC1-NCP complex at 2.9 Å, providing the first atomic view of how a chromatin protein interacts with the nucleosome (Figure 1a) [19]. The structure shows that RCC1 recognizes the architecture of the nucleosome by interacting with both histone and DNA components. As expected, one RCC1 binds to either side of the nucleosome disk. However, contrary to expectation, RCC1 uses loops along its β-propeller edge and not its broader βpropeller face to bind to the nucleosome.
Figure 1.
Crystal structure of the RCC1-nucleosome complex (PDB: 3MVD). (a) Overview of RCC1-nucleosome complex shows that RCC1 uses loops and its N-terminal arm to engage the nucleosome. (b) RCC1 uses its switchback loop to bind to the histone dimer acidic patch and an adjacent DNA-binding loop to interact with nucleosomal DNA.
RCC1 s interaction with the H2A/H2B dimer component of the nucleosome accounts for the majority of the intermolecular interactions (Figure 1b) [19]. RCC1 employs Arg and Ser residues in its switchback loop to make an intricate network of hydrogen bonds with a prominently acidic patch on the histone dimer surface; the same acidic patch that the herpesvirus LANA peptide binds to [20] and the same acidic patch proposed to mediate chromatin higher order structure together with the histone H4 N-terminal tail [21,22]. This intriguing coincidence raises the possibility that additional chromatin proteins will target this histone dimer acidic patch for recognition. This RCC1 switchback loop-histone dimer interaction had been anticipated by biochemical studies by England et al [23], but RCC1 s additional interactions with the DNA phosphate backbone by an adjacent loop region were not expected.
Besides showing how a chromatin protein recognizes the nucleosome assembly, the RCC1-nucleosome crystal structure also provides the first crystal structure of the NCP in the absence of divalent metal ions [19]. The overall structures of the NCP on its own (Mn2+ used as a crystallizing agent) and in the RCC1-nucleosome complex (in the absence of divalent metal ions) are highly similar, suggesting that the structure of the NCP is not significantly affected by either the presence or absence of divalent metals. The RCC1-nucleosome crystal structure also provides a first structure of a Widom 601 DNA sequence NCP [19], together with the structure of a Widom 601 NCP on its own by Vasudevan et al [24] and by Koerber et al (manuscript in preparation; see below).
Nucleosomal DNA Structure
The precise conformation of DNA as it wraps around the histone octamer is fundamental to understanding the structure and function of the nucleosome. The 1.94 Å resolution NCP structure containing a 147 bp human α-satellite fragment showed how the double helix conformation depends on the interplay of the DNA sequence with its precise positioning on the histone octamer [25,26]. DNA sequence dependent nucleosome structure and thermodynamics arise from the oscillation in DNA groove orientation with respect to the histone binding surface, whereby the major groove faces inward at integral SHL (SuperHelix Location = number of double helical turns from the nucleosome center) values and the minor groove faces inward at one-half SHL values (Figure 2a and 2b; major groove-inward regions, black/grey bases, minor groove-inward regions, orange/white bases). Moreover, the identity and configuration of specific histone binding motifs can influence DNA structure and promote stretching of the double helix (Figure 2b) [26].
Figure 2.
DNA binding and stretching in the nucleosome core. (a) Comparison of DNA sequence and histone-DNA register in NCP crystal structures (PDB: 1KX5, 2NZD, 1KX3, 2CV5, 1KX4, 3LEL, 3LZ0/3LZ1, 3MVD, top to bottom). All particles are with Xenopus laevis histones, with the exception of hNCP146, which contains histones from Homo sapiens. Asterisks denote severe kinks associated with regions of stretching (underlined in magenta). (b) View of the NCP147 crystal structure down the DNA superhelix axis for approximately one half of the particle (PDB: 1KX5). Numbers correspond to SHL, and alphanumeric entities refer to the DNA-binding histone motifs (α1, alpha helix 1; L1, loop 1; L2, loop 2). Long magenta arrows indicate potential regions of DNA stretching around SHL ±2 and ±5. Histone proteins are colored blue for H3, green for H4, gold for H2A, and red for H2B. (c) The special histone binding site at SHL ±1.5, which selects for or imposes an extremely narrow minor groove via the sugar clamp motif (space filling; NCP-TA; PDB: 3LEL).
Until recently, all NCP structures contained human α-satellite DNA fragments, mostly the same 146 bp symmetric sequence utilized in the original structure reported at 2.8 Å resolution (Table 1) [13]. That structure (NCP146) showed that the DNA was stretched by one base pair in one half of the pseudosymmetric complex around SHL 2 (Figure 2a). DNA stretching in the nucleosome is achieved by elevated dinucleotide twist and rise parameters, which cause a shift in the histone-DNA register by a single base pair increase in the effective length (the number of base pairs plus the number of stretching incidences) of the double helix [24,27].
Subsequent to the determination of the original structure, other NCPs containing α-satellite DNA of varying length and sequence have displayed one base pair stretching around SHL ±2 and SHL ±5 but not at other positions (Figure 2a). SHL ±2 and ±5 are flanked, respectively, by a unique configuration of H3–H4 and H2A-H2B DNA-binding motifs (Figure 2b). It appears that the double helix is strained or distorted at these positions and must choose between being compressed and underwound in the unstretched state or expanded and overwound in the stretched state [24,27]. The evidence suggests that both long-range positioning preferences and local sequence dependent DNA structure within the potential stretched region determine the outcome.
It was originally assumed that stretching was an artifact of the crystalline state since the DNA termini from adjacent NCPs stack end-to-end in the crystal, and this could potentially modulate the effective DNA length [13]. However, the phenomenon of DNA stretching occurs in the solution state [28,29], in most and in diverse NCP crystal structures [24] and even in the RCC1-NCP co-crystal structure, where direct NCP-NCP crystal contacts are completely absent [19]. DNA stretching is not unique to nucleosomes containing α-satellite DNA sequences since it is also observed in the RCC1-NCP structure [19] and a separate NCP only structure [24], both of which contain a distinct nucleosome positioning DNA fragment—the Lowary and Widom 601 sequence discovered through selection from synthetic random sequences [30]. Despite different DNA lengths (145 and 147 bp) and very different crystal packing, these two 601 NCPs show DNA stretching around SHL ±5 (Figure 2a). Further evidence for stretching is provided by analyses of the average DNA surface periodicity within the nucleosome core of genomic samples [31–34], which generally yield local reference frame double helix twist values in agreement with those determined from NCP crystal structures that display DNA stretching [24,26,27]. Therefore, we conclude that DNA stretching is an intrinsic, site-specific property of the nucleosome.
DNA stretching in the nucleosome bears both biological and methodological significance. In vivo, stretching likely influences protein factor and small molecule recognition and modulates nucleosome positioning and chromatin compaction. In vitro, it appears to be the single most important factor determining NCP crystal diffraction quality. If one considers NCP only crystals that contain Xenopus histones, all display an effective DNA length of 147 bp once stretching is accounted for, and thus all allow for similar DNA end-to-end crystal packing [24]. This observation has emerged as a key principle behind the design of diverse DNA sequences for NCP structural characterization.
The importance of highly flexible base pair steps in nucleosome positioning was apparent early on from analyses of the highest affinity histone octamer-binding sequences discovered in vitro and in vivo. In this regard, the occurrence of the most flexible dinucleotide type, TA [35–37], is paramount, and these bp steps tend to be enriched in the highest affinity DNA sequences, such as the 601, at locations where the double helix is forced to undergo maximal minor groove bending/compression (Figure 2a, center of orange sequence elements). Notably, this mode of DNA deformation generally requires greater energetic input compared to bending into the major groove [38].
Although all of the core histone-DNA binding sites apparently contribute to nucleosome positioning, the special positioning power of TTTAA elements in vitro and TATAA sequences in vivo may arise from the imposition of an extremely narrow minor groove at SHL ±1.5 via a unique, conserved histone motif that clamps the DNA (Figure 2c) [39]. Such DNA sequences are seemingly ideal at this location since they intrinsically adopt a narrow minor groove, while they simultaneously contain a TA dinucleotide where the DNA tends to be maximally distorted. It is interesting to note that the only sequence element common to both halves of the Widom 601 strong nucleosome positioning fragment is TTTAA at SHL ±1.5 [19,24] and further that the DNA conformations in all nucleosome crystal structures align particularly well within the region between SHL 1.5 and SHL 1.5.
Complexes with Small Molecules
The potential for developing molecular probes or therapeutic agents that act on the nucleosome has been explored by Luger, Dervan, Gottesfeld and colleagues with their work characterizing the site-specific DNA-binding of pyrrole-imidazole polyamides [14,15,40]. Compared to naked DNA, the nucleosome offers additional features such as the histone proteins and juxtaposed double helix components as binding targets. For instance, a hairpin polyamide dimer has been cleverly designed to associate with two adjacent minor groove sections (the so-called “supergroove”) on the nucleosomal superhelix [15].
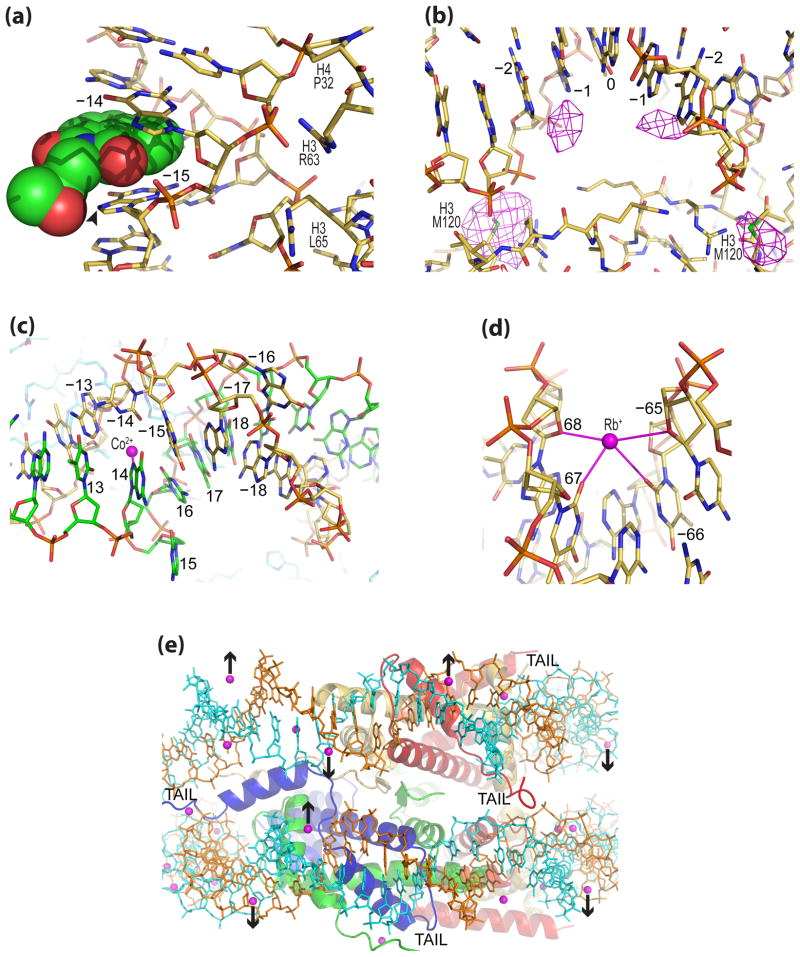
Recent work has identified other unique features of the nucleosome to target. Notably, DNA stretching can be associated with extreme kinking, in which two consecutive base pairs are unstacked at either the major (negative roll) or minor groove edge (positive roll) [24,27]. The severe unstacking provides a localized increase in dinucleotide rise, underlying the DNA stretch . In a 145 bp NCP construct displaying stretching around both SHL 2 and SHL 2, a massive kink into the minor groove (roll, 55°) can occur at SHL ±1.5 [27], which serves as a recognition site for an intercalating alkylative antitumor agent (Figure 3a) [29]. The kink promotes intercalation at the opened major groove edge, and since it coincides with a GG dinucleotide, it predisposes the guanine N7 atom to alkylation by the epoxide group. This may have general implications for the localization of intercalating compounds in the cell and opens up new possibilities for the design of more site selective intercalating species.
Figure 3.
Crystallographic studies of therapeutic candidate, drug and metal cation association with the NCP. (a) Facilitated intercalation of an alkylating antitumor compound (space-filling) between guanine bases at the site of stretch-associated extreme minor groove kinking at SHL ±1.5 in NCP145 creates a hotspot for DNA alkylation by the epoxide group (arrowhead; PDB: 3KUY). (b) Anticancer drug cisplatin adduct formation observed around the nucleosome center in NCP147 (PDB: 3B6F). An anomalous difference map (3.4σ, magenta) shows platinum atom locations between purine bases and at methionine sulfur groups. (c) Co2+ binding (magenta) promotes expulsion of two bases ( 16 and 15) from the double helical stack at SHL 1.5 in NCP147 (PDB: 3MGP). (d) A narrow minor groove favors monovalent cation (Rb+) coordination (magenta) at AT dinucleotide elements in NCP147 (PDB: 3MGR). (e) View along the nucleosomal DNA supergroove of NCP147 showing that minor groove-divalent metal cation (magenta spheres) hydrate binding can influence DNA wrapping (inward-pointing arrows) and nucleosome-nucleosome interactions (outward-pointing arrows; PDB: 3LJA).
Another class of nucleosome binding small molecules with therapeutic potential is heavy metal cations. The high electron density and anomalous scattering properties of heavy metals facilitate characterization of their binding sites by X-ray crystallography. Recent work on platinum anticancer drugs has yielded new insight into principles that may govern the chromatin site selectivity of these agents [41]. In particular, it was found that platinum adducts can be accommodated without large conformational changes in the nucleosome core, even at central locations where histone-DNA contacts are strongest (Figure 3b). Supporting footprinting studies demonstrated that histone octamer association can alter the drug adduct formation profile, relative to naked DNA; an exciting prospect because it suggests that one may be able to improve site selectivity by exploiting such nucleosome-based modulation. Elucidating the basic mechanistic features of platinum drug-DNA reaction and cross-link formation will be important next steps.
Beyond studies of therapeutic metal compounds, recent nucleosome crystallographic work on heavy metal cation interactions has shed light both on their pathological potential as well as the essential function that ubiquitous cations in the nucleus serve (Figure 3c,d,e). Ni2+ and Co2+ are significant industrial pollutants, especially in developing nations, and their carcinogenic properties likely relate to epigenetic alterations as well as DNA damage [42,43]. These divalent cations were found to bind extensively to guanine bases and histidine and carboxylate sites in the NCP [44]. In particular, the presence of Ni2+ or Co2+ in NCP crystals was able to induce an unprecedented distortion in the DNA, whereby stretching in one particle half was coupled with flipping out of two bases in the opposing particle half (Figure 3c). The displacement of two bases from the double helical stack occurred at SHL 1.5, which is apparently not coincidently the site with the greatest DNA distorting and positioning potential in the nucleosome [26,27,39,44–48]. This special location could turn out to be a therapeutic hot spot and may preside over yet undiscovered mechanical functions in the cell.
Other recent NCP-metal studies include investigations of analogues for ubiquitous cation species in the nucleus. Although earlier work demonstrated how histone-imposed DNA conformation can dictate Mn2+ binding [49], a more recent investigation using soft X-rays to tune to the manganese absorption edge allowed identification of many more divalent metal binding sites [50]. This new work confirms the great similarity in DNA binding behavior of Mn2+ compared to Mg2+ and Ca2+ and suggests that nucleosome and chromatin solubility and compaction behavior is influenced by an interplay of competition and cooperatively between the histone N-terminal tails and divalent metal hydrates in associating with the same sites in the minor groove (Figure 3e). Lastly, a study on Rb+ and Cs+ binding in NCP crystals appears to confirm long-standing assertions, based on oligonucleotide DNA studies, that Na+ and K+ coordination stabilizes minor groove narrowing over specific sequence elements in the genome (Figure 3d) [44,51].
Future Directions & Challenges
The recent studies examining different DNA sequences, metals and medicinal agents have yielded new insight into double helix structure and molecular recognition in the nucleosome. At the same time, these works are providing the basis for future investigations and novel applications; for instance, in the further development of nucleosome targeting and drug development platforms. Nonetheless, much work remains to be done to fully understand the determinants of DNA positioning and structure in the nucleosome. Considering the level of histone- and DNA sequence-dependent structure that is observed from a generous handful of NCP structures available (Table 1), and given that there are an estimated ~15 million unique nucleosomes in the genome, we would ultimately need high resolution structures for many different nucleosomal constructs in order to have a comprehensive perspective. On a related note, our understanding of nucleosome dynamics and the mechanism of chromatin remodeling enzymes is modest, and therefore it is likely that we will need to harness a variety of techniques, including single molecule methodologies, to tackle such issues.
The first atomic structure of a chromatin protein interacting with the nucleosome reported recently is timely, and we expect that the RCC1-nucleosome structure will be just the first of many chromatin factor-nucleosome complexes to come. We are beginning to understand how histone modification binding modules such as bromodomains, chromodomains, tudor domains and PHD fingers interact with acetylated and methylated histone peptides [52]. These pioneering studies will need to be complemented by structures showing how such epigenetic mark readers recognize their histone substrates in the context of chromatin and how other epigenetic mark writers install these marks onto histones incorporated into nucleosomes. If this is not challenging enough, there is still the perennial question of the precise nature of chromatin higher order structure(s) [53,54].
These are exciting times for chromatin biologists interested in structure. We have a firm foundation to stand on, and so much still remains to be discovered.
Acknowledgments
We are grateful to the Davey and the Tan laboratories for stimulating discussions. This work was supported by Academic Research Council grant 19/08 from the Ministry of Education (Singapore) to C.A.D. and U.S. National Institutes of Health grant GM088236 to S.T.
Footnotes
Conflict of interest statement: The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116:259–272. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 2.Segal E, Widom J. What controls nucleosome positions? Trends Genet. 2009;25 :335–343. doi: 10.1016/j.tig.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 4.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 6.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 8.Tremethick DJ. Higher-order structures of chromatin: the elusive 30 nm fiber. Cell. 2007;128:651–654. doi: 10.1016/j.cell.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 9.Woodcock CL, Ghosh RP. Chromatin higher-order structure and dynamics. Cold Spring Harb Perspect Biol. 2010;2:a000596. doi: 10.1101/cshperspect.a000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finch JT, Lutter LC, Rhodes D, Brown RS, Rushton B, Levitt M, Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977;269:29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- 11.Finch JT, Klug A. X-ray and electron microscope analyses of crystals of nucleosome cores. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):1–9. doi: 10.1101/sqb.1978.042.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Rhodes D. Nucleosome cores reconstituted from poly (dA-dT) and the octamer of histones. Nucleic Acids Res. 1979;6:1805–1816. doi: 10.1093/nar/6.5.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13**.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. First atomic structure of the nucleosome—a classic paper. [DOI] [PubMed] [Google Scholar]
- 14.Suto RK, Edayathumangalam RS, White CL, Melander C, Gottesfeld JM, Dervan PB, Luger K. Crystal structures of nucleosome core particles in complex with minor groove DNA-binding ligands. J Mol Biol. 2003;326:371–380. doi: 10.1016/s0022-2836(02)01407-9. [DOI] [PubMed] [Google Scholar]
- 15*.Edayathumangalam RS, Weyermann P, Gottesfeld JM, Dervan PB, Luger K. Molecular recognition of the nucleosomal “supergroove”. Proc Natl Acad Sci U S A. 2004;101:6864–6869. doi: 10.1073/pnas.0401743101. Targeting unique features of the nucleosome with ligands designed by structure-based principles. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clarke PR, Zhang C. Spatial and temporal coordination of mitosis by Ran GTPase. Nat Rev Mol Cell Biol. 2008;9:464–477. doi: 10.1038/nrm2410. [DOI] [PubMed] [Google Scholar]
- 17.Renault L, Nassar N, Vetter I, Becker J, Klebe C, Roth M, Wittinghofer A. The 1.7 A crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature. 1998;392:97–101. doi: 10.1038/32204. [DOI] [PubMed] [Google Scholar]
- 18.Renault L, Kuhlmann J, Henkel A, Wittinghofer A. Structural basis for guanine nucleotide exchange on Ran by the regulator of chromosome condensation (RCC1) Cell. 2001;105:245–255. doi: 10.1016/s0092-8674(01)00315-4. [DOI] [PubMed] [Google Scholar]
- 19**.Makde RD, England JR, Yennawar HP, Tan S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature. 2010;467:562–566. doi: 10.1038/nature09321. First crystal structure of a protein factor-nucleosome complex and, together with Vasudevan et al, the first atomic view of a 601 NCP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20**.Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, Kaye KM. The nucleosomal surface as a docking station for Kaposi’s sarcoma herpesvirus LANA. Science. 2006;311:856–861. doi: 10.1126/science.1120541. The first structure of a nucleosome-peptide complex. Shows how the viral protein specifically recognizes the H2A–H2B dimer acidic cleft. [DOI] [PubMed] [Google Scholar]
- 21.Dorigo B, Schalch T, Bystricky K, Richmond TJ. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J Mol Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 22.Chodaparambil JV, Barbera AJ, Lu X, Kaye KM, Hansen JC, Luger K. A charged and contoured surface on the nucleosome regulates chromatin compaction. Nat Struct Mol Biol. 2007;14:1105–1107. doi: 10.1038/nsmb1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.England JR, Huang J, Jennings MJ, Makde RD, Tan S. RCC1 uses a conformationally diverse loop region to interact with the nucleosome: a model for the RCC1-nucleosome complex. J Mol Biol. 2010;398:518–529. doi: 10.1016/j.jmb.2010.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24*.Vasudevan D, Chua EY, Davey CA. Crystal structures of nucleosome core particles containing the ‘601’ strong positioning sequence. J Mol Biol. 2010;403:1–10. doi: 10.1016/j.jmb.2010.08.039. First atomic view of 601 NCPs together with Makde et al. Detailed crystal structure analysis provides guidelines for obtaining NCP structures with diverse DNA sequences. [DOI] [PubMed] [Google Scholar]
- 25*.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 A resolution. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. Highest resolution structures of the NCP to date. Superb details of solvent structure. [DOI] [PubMed] [Google Scholar]
- 26*.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. Detailed analysis of DNA conformation in the nucleosome core. [DOI] [PubMed] [Google Scholar]
- 27.Ong MS, Richmond TJ, Davey CA. DNA stretching and extreme kinking in the nucleosome core. J Mol Biol. 2007;368:1067–1074. doi: 10.1016/j.jmb.2007.02.062. [DOI] [PubMed] [Google Scholar]
- 28.Edayathumangalam RS, Weyermann P, Dervan PB, Gottesfeld JM, Luger K. Nucleosomes in solution exist as a mixture of twist-defect states. J Mol Biol. 2005;345:103–114. doi: 10.1016/j.jmb.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 29.Davey GE, Wu B, Dong Y, Surana U, Davey CA. DNA stretching in the nucleosome facilitates alkylation by an intercalating antitumour agent. Nucleic Acids Res. 2010;38:2081–2088. doi: 10.1093/nar/gkp1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 31.Satchwell SC, Drew HR, Travers AA. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 32.Travers AA, Klug A. The bending of DNA in nucleosomes and its wider implications. Philos Trans R Soc Lond B Biol Sci. 1987;317:537–561. doi: 10.1098/rstb.1987.0080. [DOI] [PubMed] [Google Scholar]
- 33.Gale JM, Nissen KA, Smerdon MJ. UV-induced formation of pyrimidine dimers in nucleosome core DNA is strongly modulated with a period of 10.3 bases. Proc Natl Acad Sci U S A. 1987;84:6644–6648. doi: 10.1073/pnas.84.19.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayes JJ, Tullius TD, Wolffe AP. The structure of DNA in a nucleosome. Proc Natl Acad Sci U S A. 1990;87:7405–7409. doi: 10.1073/pnas.87.19.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krueger A, Protozanova E, Frank-Kamenetskii MD. Sequence-dependent basepair opening in DNA double helix. Biophys J. 2006;90:3091–3099. doi: 10.1529/biophysj.105.078774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balasubramanian S, Xu F, Olson WK. DNA sequence-directed organization of chromatin: structure-based computational analysis of nucleosome-binding sequences. Biophys J. 2009;96:2245–2260. doi: 10.1016/j.bpj.2008.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morozov AV, Fortney K, Gaykalova DA, Studitsky VM, Widom J, Siggia ED. Using DNA mechanics to predict in vitro nucleosome positions and formation energies. Nucleic Acids Res. 2009;37:4707–4722. doi: 10.1093/nar/gkp475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dickerson RE. DNA bending: the prevalence of kinkiness and the virtues of normality. Nucleic Acids Res. 1998;26:1906–1926. doi: 10.1093/nar/26.8.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39*.Wu B, Mohideen K, Vasudevan D, Davey CA. Structural insight into the sequence dependence of nucleosome positioning. Structure. 2010;18:528–536. doi: 10.1016/j.str.2010.01.015. Structural analysis of DNA distorting and positioning potential of SHL ±1.5. Characterization of special histone motif and DNA sequence preference at this site. Provides informative overview of sequence-dependent DNA structure. [DOI] [PubMed] [Google Scholar]
- 40.Gottesfeld JM, Melander C, Suto RK, Raviol H, Luger K, Dervan PB. Sequence-specific recognition of DNA in the nucleosome by pyrrole-imidazole polyamides. J Mol Biol. 2001;309:615–629. doi: 10.1006/jmbi.2001.4694. [DOI] [PubMed] [Google Scholar]
- 41.Wu B, Droge P, Davey CA. Site selectivity of platinum anticancer therapeutics. Nat Chem Biol. 2008;4:110–112. doi: 10.1038/nchembio.2007.58. [DOI] [PubMed] [Google Scholar]
- 42.Kasprzak KS. Oxidative DNA and protein damage in metal-induced toxicity and carcinogenesis. Free Radic Biol Med. 2002;32:958–967. doi: 10.1016/s0891-5849(02)00809-2. [DOI] [PubMed] [Google Scholar]
- 43.Arita A, Costa M. Epigenetics in metal carcinogenesis: nickel, arsenic, chromium and cadmium. Metallomics. 2009;1:222–228. doi: 10.1039/b903049b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mohideen K, Muhammad R, Davey CA. Perturbations in nucleosome structure from heavy metal association. Nucleic Acids Res. 2010;38:6301–6311. doi: 10.1093/nar/gkq420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richmond TJ, Finch JT, Rushton B, Rhodes D, Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984;311:532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- 46.Hogan ME, Rooney TF, Austin RH. Evidence for kinks in DNA folding in the nucleosome. Nature. 1987;328:554–557. doi: 10.1038/328554a0. [DOI] [PubMed] [Google Scholar]
- 47.Fitzgerald DJ, Anderson JN. DNA distortion as a factor in nucleosome positioning. J Mol Biol. 1999;293:477–491. doi: 10.1006/jmbi.1999.3171. [DOI] [PubMed] [Google Scholar]
- 48.Fernandez AG, Anderson JN. Nucleosome positioning determinants. J Mol Biol. 2007;371:649–668. doi: 10.1016/j.jmb.2007.05.090. [DOI] [PubMed] [Google Scholar]
- 49.Davey CA, Richmond TJ. DNA-dependent divalent cation binding in the nucleosome core particle. Proc Natl Acad Sci U S A. 2002;99:11169–11174. doi: 10.1073/pnas.172271399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu B, Davey CA. Using soft X-rays for a detailed picture of divalent metal binding in the nucleosome. J Mol Biol. 2010;398:633–640. doi: 10.1016/j.jmb.2010.03.038. [DOI] [PubMed] [Google Scholar]
- 51.McFail-Isom L, Sines CC, Williams LD. DNA structure: cations in charge? Curr Opin Struct Biol. 1999;9:298–304. doi: 10.1016/S0959-440X(99)80040-2. [DOI] [PubMed] [Google Scholar]
- 52.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53**.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. The first crystallographic analysis of higher order chromatin structure. [DOI] [PubMed] [Google Scholar]
- 54.Routh A, Sandin S, Rhodes D. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc Natl Acad Sci U S A. 2008;105:8872–8877. doi: 10.1073/pnas.0802336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harp JM, Hanson BL, Timm DE, Bunick GJ. Asymmetries in the nucleosome core particle at 2.5 A resolution. Acta Crystallogr D Biol Crystallogr. 2000;56:1513–1534. doi: 10.1107/s0907444900011847. [DOI] [PubMed] [Google Scholar]
- 56.White CL, Suto RK, Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. Embo J. 2001;20:5207–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsunaka Y, Kajimura N, Tate S, Morikawa K. Alteration of the nucleosomal DNA path in the crystal structure of a human nucleosome core particle. Nucleic Acids Res. 2005;33:3424–3434. doi: 10.1093/nar/gki663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clapier CR, Chakravarthy S, Petosa C, Fernandez-Tornero C, Luger K, Muller CW. Structure of the Drosophila nucleosome core particle highlights evolutionary constraints on the H2A–H2B histone dimer. Proteins. 2008;71:1–7. doi: 10.1002/prot.21720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59*.Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Biol. 2000;7:1121–1124. doi: 10.1038/81971. First structure of the nucleosome composed of a histone variant. Shows alterations in the histone-histone interfaces of the octamer that could influence nucleosome stability and histone composition. [DOI] [PubMed] [Google Scholar]
- 60.Chakravarthy S, Gundimella SK, Caron C, Perche PY, Pehrson JR, Khochbin S, Luger K. Structural characterization of the histone variant macroH2A. Mol Cell Biol. 2005;25:7616–7624. doi: 10.1128/MCB.25.17.7616-7624.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tachiwana H, Kagawa W, Osakabe A, Kawaguchi K, Shiga T, Hayashi-Takanaka Y, Kimura H, Kurumizaka H. Structural basis of instability of the nucleosome containing a testis-specific histone variant, human H3T. Proc Natl Acad Sci U S A. 2010;107:10454–10459. doi: 10.1073/pnas.1003064107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muthurajan UM, Bao Y, Forsberg LJ, Edayathumangalam RS, Dyer PN, White CL, Luger K. Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions. Embo J. 2004;23:260–271. doi: 10.1038/sj.emboj.7600046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lu X, Simon MD, Chodaparambil JV, Hansen JC, Shokat KM, Luger K. The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure. Nat Struct Mol Biol. 2008;15:1122–1124. doi: 10.1038/nsmb.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Watanabe S, Resch M, Lilyestrom W, Clark N, Hansen JC, Peterson C, Luger K. Structural characterization of H3K56Q nucleosomes and nucleosomal arrays. Biochim Biophys Acta. 2010;1799:480–486. doi: 10.1016/j.bbagrm.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bao Y, White CL, Luger K. Nucleosome core particles containing a poly(dA. dT) sequence element exhibit a locally distorted DNA structure. J Mol Biol. 2006;361:617–624. doi: 10.1016/j.jmb.2006.06.051. [DOI] [PubMed] [Google Scholar]