Abstract
Human and animal model evidence suggests that CD4+ T cells play a critical role in the control of chronic hepatitis C virus (HCV) infection. However, despite their importance, the mechanism behind the failure of such populations in chronic disease is not understood and the contribution of viral mutation is not known. To address this, this study defined the specificity and virological footprint of CD4+ T cells in chronic infection. CD8+ T-cell-depleted peripheral blood mononuclear cells from 61 HCV genotype 1-infected patients were analysed against a panel of peptides covering the HCV genotype 1 core – a region where CD4+ T-cell responses may be reproducibly obtained. In parallel, the core region and E2 protein were sequenced. Gamma interferon-secreting CD4+ T-cell responses directed against multiple epitopes were detected in 53 % of individuals, targeting between one and four peptides in the HCV core. Viral sequence evaluation revealed that these CD4+ T-cell responses were associated with mutants in 2/21 individuals. In these two cases, the circulating sequence variant was poorly recognized by host CD4+ T cells. Bioinformatics analyses revealed no overall evidence of selection in the target epitopes and no differences between the groups with and without detectable CD4+ T-cell responses. It was concluded that sustained core peptide-specific CD4+ T-cell responses may be reproducibly measured during chronic HCV infection and that immune escape may occur in specific instances. However, overall the virological impact of such responses is limited and other causes for CD4+ T-cell failure in HCV must be sought.
INTRODUCTION
Hepatitis C virus (HCV) is a major cause of liver disease globally (Alter, 2007). The virus is able to evade host innate and adaptive immune responses in immunocompetent adults and to set up persistent infection in the majority of people. Those persistently infected are at risk of progressive liver fibrosis, cirrhosis and cancer. A minority of those infected, however, clear the virus spontaneously, a feature that is reproduced in animal models (Bowen & Walker, 2005a).
The immunological response to HCV has an important bearing not only on the acute outcome (i.e. persistent infection versus spontaneous resolution), but also potentially on the long-term outcome in chronic carriers, although our understanding of the immune responses in acute outcome is more complete. Successful outcome is associated with the maintenance of broadly directed CD4+ and CD8+ T-cell responses, with maintained functionality (Bowen & Walker, 2005a). Specific HLA class I and II molecules are associated with a successful response (McKiernan et al., 2004; Neumann-Haefelin et al., 2006), and depletion studies in animals strongly support the critical role of both CD4+ and CD8+ T-cell subsets in acute control (Grakoui et al., 2003; Shoukry et al., 2003).
The responses in those with established persistent infection are typically described as highly attenuated, especially in the blood (Bowen & Walker, 2005a; Klenerman & Hill, 2005; Lauer et al., 2004; Lechner et al., 2000; Rehermann et al., 1996). Chronically infected human and chimpanzee studies based on proliferation responses in blood, or analysis of T-cell lines after stimulation, have typically found either no or only very limited responses (Diepolder et al., 1995; Missale et al., 1996).
In contrast to these studies, recent ex vivo analyses based around gamma interferon (IFN-γ) secretion have indicated some CD4+ T-cell responsiveness in those with persistent infection: we and others have found responses to pools of core peptides in a range of patients with chronic disease, associated with low proliferative capacity and loss of interleukin (IL)-2 secretion (Barnes et al., 2002; Harcourt et al., 2006; Ruys et al., 2008; Semmo et al., 2005, 2007); the responses obtained were reproducible and robust and were not detected in healthy normal controls. Responses using an identical assay against antigens from genotype 1 NS3–NS5b typically yield minimal responses (undetectable or weak) and are difficult to study further. Loss of CD4+ T-cell responses to these peptides is seen in the context of human immunodeficiency virus (HIV) infection, associated with an increased viral load (Harcourt et al., 2006).
The mechanism behind the failure of CD4+ T-cell responses to contain virus in chronic infection has not been established, nor has the mechanism underlying their low levels in blood (Bowen & Walker, 2005a, b; Klenerman & Hill, 2005). Individual reports have indicated that immune escape may occur in epitopes targeted by HCV-specific CD4+ T cells (Eckels et al., 1999, 2000; Puig et al., 2006; Wang & Eckels, 1999; Wang et al., 2003). Variation within epitopes has also been described as affecting T-cell cytokine secretion (Wang & Eckels, 1999; Wang et al., 2003). It is well established that escape from HCV-specific CD8+ T cells can occur and may be critical for persistence (Cox et al., 2005b; Dazert et al., 2009; Erickson et al., 2001; Ray et al., 2005; Weiner et al., 1995). However, as CD4+ T-cell responses have been considered difficult to detect during chronicity, this issue has not been systematically addressed for T helper populations.
To address this, we analysed CD4+ T-cell responses in a large, well-defined cohort of individuals with chronic HCV genotype 1 infection. We systematically analysed the ex vivo CD4+ T-cell response and defined the fine specificity of this response in relation to the sequence of autologous virus. Surprisingly, we found a persistent IFN-γ response in the majority of donors, often targeting multiple epitopes, including a newly defined immunodominant peptide in the core. We evaluated the relationship between T-cell responses targeting individual peptides and the sequence variation within those peptides, and defined the functional consequences of viral mutation and the impact of CD4+ T-cell escape in this cohort.
RESULTS
Ex vivo CD4+ T-cell responses are detectable in the majority of donors
In this study, we excluded CD8+ T-cell responses at the outset by the use of CD8-depleted peripheral blood mononuclear cells (PBMCs). We measured ex vivo responses in an IFN-γ enzyme-linked immunosorbent spot (ELISPOT) assay using established robust methods and identified a response against the pool of genotype 1 core peptides in 32/61 genotype 1-positive donors. These donors were collected prospectively and independently of previous immunological or clinical data (Table 1). Alanine transaminase (ALT) levels, viral loads, age and prior treatment status did not differ significantly between the two groups. This frequency of response is very similar to that defined in two previous studies on HCV+ chronically infected cohorts using whole undepleted PBMCs and peptide pools (Semmo et al., 2005, 2007).
Table 1.
Clinical features of the study cohort
na, Not available.
| Patient no. | M/F | Age (years) | ALT level (U l−1) | Viral load (IU ml−1) | Core response | Core region sequenced? |
|---|---|---|---|---|---|---|
| 106 | F | 71 | 135 | 110 925 | Positive | Yes |
| 112 | M | 53 | 62 | 5 017 | Positive | Yes |
| 117 | M | 45 | 39 | 61 929 806 | Positive | Yes |
| 141 | M | 28 | 31 | 26 183 | Positive | Yes |
| 160 | M | 50 | 34 | na | Positive | No |
| 168 | M | 59 | 40 | 17 913 | Negative | Yes |
| 171 | M | 45 | 39 | 51 739 | Negative | No |
| 172 | M | 50 | 38 | 478 192 | Positive | Yes |
| 182 | F | 52 | 38 | 71 729 | Negative | Yes |
| 183 | M | 48 | 199 | 2 920 108 | Negative | No |
| 188 | F | 48 | 29 | 7 172 883 | Negative | Yes |
| 191 | M | 56 | 168 | na | Negative | No |
| 193 | M | 46 | 103 | 685 931 | Positive | Yes |
| 201 | M | 52 | 78 | 235 960 | Positive | Yes |
| 205 | M | 56 | 111 | 56 642 | Negative | Yes |
| 208 | M | 28 | 59 | 52 131 | Negative | Yes |
| 210 | M | 43 | 443 | 5 056 294 | Negative | No |
| 215 | M | 49 | 77 | 1 046 535 | Negative | Yes |
| 252 | M | 44 | 17 | 92 895 | Negative | Yes |
| 263 | F | 50 | 36 | 999 500 | Positive | No |
| 269 | M | 73 | 26 | 489 951 | Negative | Yes |
| 287 | M | 54 | 32 | 1 152 365 | Negative | Yes |
| 289 | F | 49 | 25 | 960 304 | Negative | No |
| 303 | M | 45 | 34 | 27 681 | Positive | Yes |
| 304 | F | 47 | 24 | 111 709 | Positive | Yes |
| 305 | F | 41 | 67 | 132 091 | Negative | Yes |
| 306 | M | 54 | 56 | 89 367 | Positive | No |
| 308 | F | 62 | 33 | 10 504 549 | Positive | No |
| 310 | F | 50 | 186 | 169 719 | Positive | Yes |
| 311 | F | 47 | 47 | 1 226 837 | Positive | Yes |
| 317 | F | 46 | 34 | 15 992 | Positive | Yes |
| 318 | F | 42 | 21 | 603 620 | Negative | Yes |
| 319 | M | 61 | 327 | 486 031 | Negative | Yes |
| 320 | F | 50 | 15 | 9 995 | Positive | Yes |
| 323 | F | 44 | 50 | 127 425 | Positive | Yes |
| 324 | F | 40 | 76 | 333 559 | Positive | Yes |
| 328 | M | 38 | 96 | 36 139 | Negative | Yes |
| 338 | M | 47 | 94 | 97 990 | Positive | Yes |
| 342 | F | 40 | 33 | 93 679 | Negative | Yes |
| 343 | M | 47 | 57 | 86 231 | Positive | Yes |
| 348 | M | 27 | 51 | 11 092 491 | Negative | Yes |
| 349 | M | 52 | 40 | 23 674 | Negative | Yes |
| 350 | F | 71 | 81 | 82 704 | Positive | Yes |
| 365 | F | 55 | 115 | 175 206 | Positive | Yes |
| 369 | M | 55 | 91 | 148 553 | Negative | Yes |
| 370 | M | 45 | 101 | 33 669 | Positive | No |
| 371 | F | 40 | 58 | 1 046 535 | Negative | Yes |
| 375 | F | 59 | 130 | 228 121 | Positive | Yes |
| 376 | M | 58 | 134 | 41 940 | Positive | No |
| 379 | M | 50 | 47 | 226 553 | Positive | Yes |
| 382 | F | 54 | 115 | 153 649 | Negative | Yes |
| 384 | M | 52 | 40 | 212 051 | Negative | Yes |
| 386 | M | 45 | 85 | 635 | Positive | No |
| 388 | M | 36 | 12 | 5 997 000 | Negative | Yes |
| 393 | M | 54 | 230 | 587 941 | Negative | Yes |
| 396 | M | 47 | 24 | 438 996 | Positive | Yes |
| 401 | M | 49 | 50 | 3 484 532 | Positive | Yes |
| 402 | M | 40 | 167 | na | Positive | No |
| 403 | F | 52 | 115 | na | Negative | No |
| 404 | M | 53 | 34 | na | Positive | No |
| 405 | M | 57 | 27 | na | Negative | No |
Fine specificity of CD4+ T-cell responses in persistent infection
In this study, we wished initially to define the peptide targets of the observed CD4+ T-cell responses. The results of this are illustrated in Table 2 and Fig. 1(a). Table 2 shows the raw data for individual responses to single peptides derived from the core peptide sequence, as well as responses to a pool of the 18 core peptides (core pool). Only data from those responding to one or more peptides are shown (n=32). Individuals responded to diverse peptides across the sequence [spot forming cells (s.f.c.) per 106 CD8-depleted PBMCs, mean 170; range 50–732]. If responses to two adjacent peptides were considered as a single target (a conservative estimate), we observed, on average, responses to two epitopes per subject (median 2; 15 subjects with one response, 12 subjects with two responses, four subjects with three responses and one subject with four responses).
Table 2.
HCV core CD4+ IFN-γ ELISPOT responses of 32 positive subjects
Results are given as s.f.c. per 106 CD8-depleted PBMCs.
| Patient no. | Core pool | Peptide | No. of epitopes | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1–20 | 11–30 | 21–40 | 31–50 | 41–60 | 51–70 | 61–80 | 71–90 | 81–100 | 91–110 | 101–120 | 111–131 | 121–140 | 131–150 | 141–160 | 151–170 | 161–180 | 171–190 | |||
| 106 | 195 | 190 | 1 | |||||||||||||||||
| 112 | 160 | 135 | 1 | |||||||||||||||||
| 117 | 283 | 150 | 1 | |||||||||||||||||
| 141 | 60 | 65 | 1 | |||||||||||||||||
| 160 | 250 | 120 | 120 | 2 | ||||||||||||||||
| 172 | 250 | 280 | 175 | 150 | 3 | |||||||||||||||
| 193 | 56 | 350 | 560 | 2 | ||||||||||||||||
| 201 | 100 | 320 | 1 | |||||||||||||||||
| 263 | 350 | 170 | 1 | |||||||||||||||||
| 303 | 100 | 65 | 60 | 2 | ||||||||||||||||
| 304 | 297 | 77 | 110 | 77 | 220 | 173 | 3 | |||||||||||||
| 306 | 870 | 313 | 140 | 2 | ||||||||||||||||
| 308 | 140 | 124 | 159 | 1 | ||||||||||||||||
| 310 | 169 | 97 | 150 | 2 | ||||||||||||||||
| 311 | 114 | 198 | 1 | |||||||||||||||||
| 317 | 291 | 80 | 80 | 90 | 127 | 4 | ||||||||||||||
| 320 | 166 | 155 | 145 | 2 | ||||||||||||||||
| 323 | 60 | 182 | 1 | |||||||||||||||||
| 324 | 110 | 80 | 1 | |||||||||||||||||
| 338 | 335 | 180 | 268 | 2 | ||||||||||||||||
| 343 | 137 | 168 | 1 | |||||||||||||||||
| 350 | 120 | 125 | 1 | |||||||||||||||||
| 365 | 250 | 405 | 175 | 732 | 254 | 3 | ||||||||||||||
| 370 | 105 | 70 | 1 | |||||||||||||||||
| 375 | 185 | 128 | 85 | 2 | ||||||||||||||||
| 376 | 772 | 675 | 1 | |||||||||||||||||
| 379 | 294 | 109 | 80 | 2 | ||||||||||||||||
| 386 | 50 | 50 | 1 | |||||||||||||||||
| 396 | 100 | 66 | 50 | 2 | ||||||||||||||||
| 401 | 100 | 150 | 80 | 2 | ||||||||||||||||
| 402 | 250 | 110 | 90 | 125 | 3 | |||||||||||||||
| 404 | 85 | 90 | 95 | 2 | ||||||||||||||||
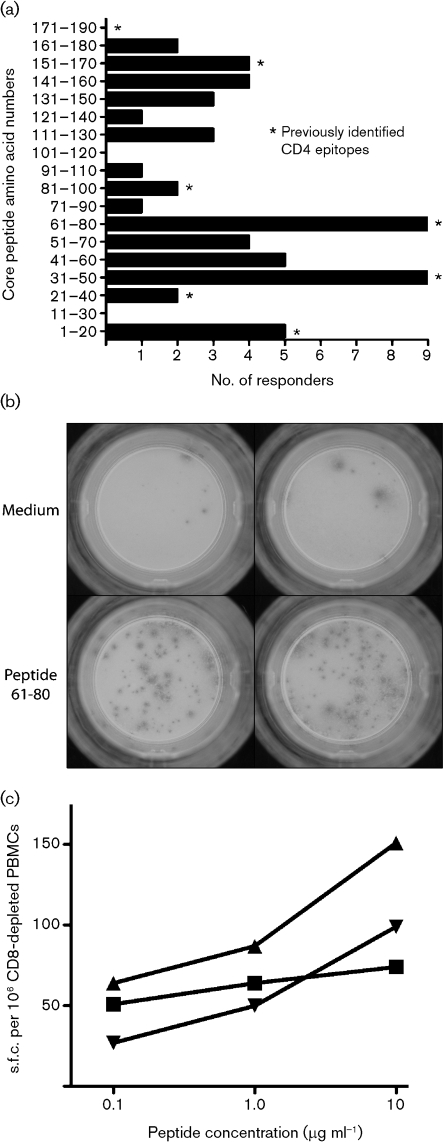
Fig. 1.
IFN-γ ELISPOT response to core peptides. (a) The frequency of CD4+ T-cell responses against the panel of core peptides is indicated. Data were derived from Table 2. The sequences of the individual peptides are available in Supplementary Table S1. All donors were tested against all peptides, and the total numbers of donors positive from the 61 tested is shown. (b) Typical ELISPOT assay from patient 306, showing a positive IFN-γ response to HCV core peptide 61–80 (s.f.u. per 106 CD8-depleted PBMCs=140). (c) Titration experiment using fresh ex vivo CD8-depleted PBMCs from donor 304. The peptide concentration used is displayed on the y-axis, with the background subtracted. Peptides: ▴, 31–50; ▪, 61–80; ▾, 151–170.
The group data are shown in Fig. 1(a). This revealed responses to the majority of peptides tested, with the commonest targets being peptide 31–50 (Lamonaca et al., 1999; MacDonald et al., 2002) and peptide 61–80 (Löhr et al., 1996). Both of these peptides were recognized in 9/32 individuals. The magnitude of the response varied from 50 to 732 s.f.c. per 106 CD8-depleted PBMCs (Table 2) and representative ELISPOT wells are shown in Fig. 1(b). These responses were reproduced in repeat assays, and titrations of the individual peptide, including epitope 61–80, showed that concentrations as low as 1–0.1 μg ml−1 were still able to stimulate 50 % maximal responses (Fig. 1c).
Impact of T-cell responses on viral sequence
We next addressed the relationship between the presence of CD4+ T-cell responses and viral sequence evolution. This is important for three reasons: (i) to define to what extent the detectable T-cell responses are targeting autologous sequences; (ii) to define the impact of potential T-cell-mediated selection pressure; and (iii) to analyse whether the non-responder group had accumulated significant mutations as a potential cause of T-cell failure. Sequences were therefore analysed as bulk sequence and, in addition, clonally derived sequences were obtained in a subset of patients.
Firstly, we analysed the relatedness of sequences to address whether responder and non-responder sequences clustered separately. Phylogenetic analysis of the core region sequences revealed a typical subdivision into genotype 1a and 1b subtypes (Fig. 2). No correlation was observed between HCV subgenotype and the CD4+ T-cell response. In one case, a CD4 responder and non-responder both shared identical core regions, placing them on the same branch of the tree.
Fig. 2.
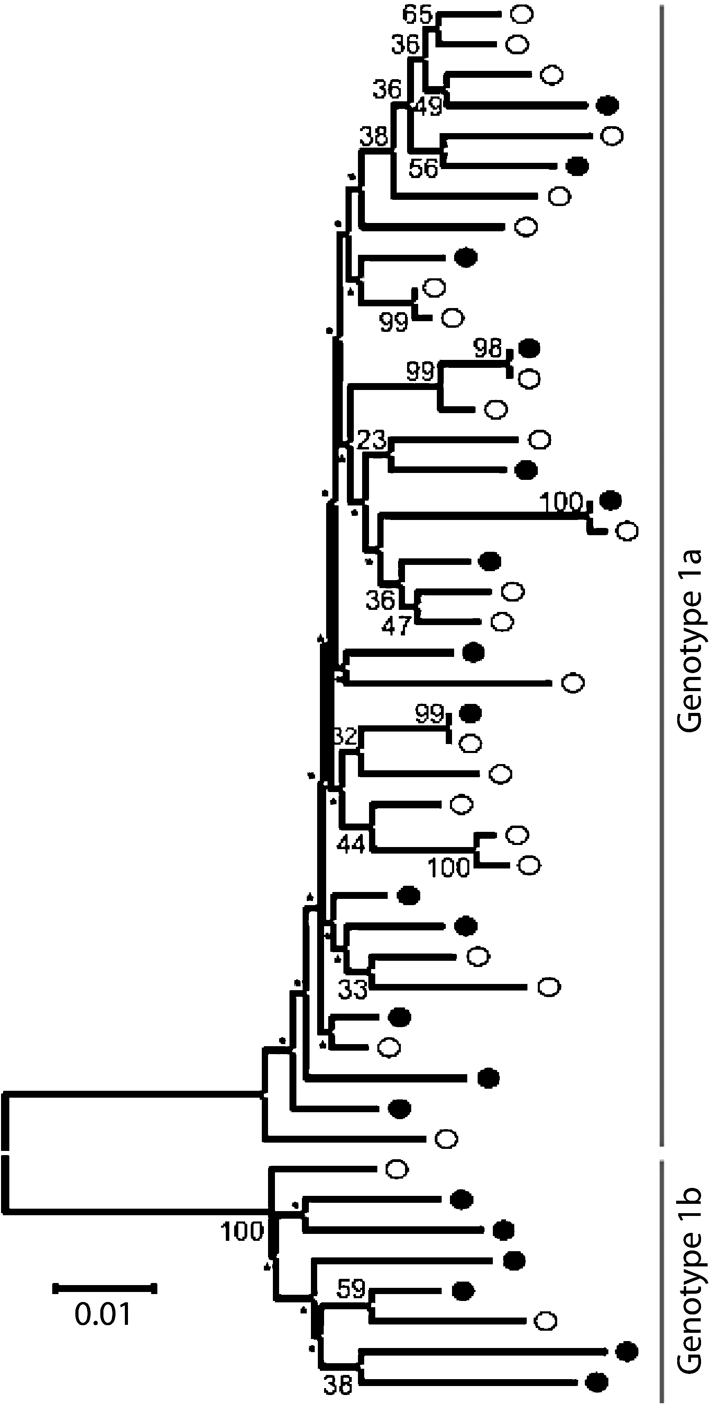
Relatedness of sequences amongst responders and non-responders. Phylogenetic tree of the CD4 responder (•) and non-responder (○) HCV core region based on the neighbour-joining method using 1000 bootstrap replicates (scores <30 are indicated by an asterisk). Bar, nucleotide substitutions per site.
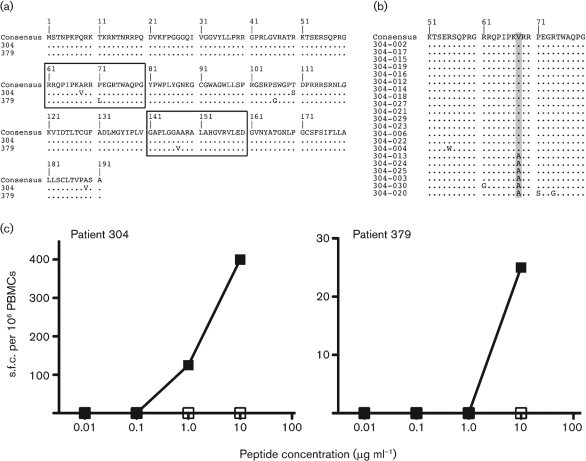
Forty-six sequences were obtained: 23 CD4 responder core sequences and 23 CD4 non-responder sequences. Amongst the responders, T-cell responses to previously defined peptides were observed in 12 donors and responses to novel peptides (i.e. not previously published as epitopes) in ten donors. From these, all but two donors (304 and 379) showed conserved sequences within the peptides targeted (i.e. identical to the peptides used in the assay). This indicated that the selection of escape mutants through immune pressure from CD4+ T cells targeting the HCV core region appears to be rare, at least in this group of individuals. However, single variants were observed within previously defined epitopes, peptide 61–80 (A68V) in one individual and peptide 141–160 (A147V) in another (Fig. 3a). Two additional mutations were observed in both individuals within peptide 101–120 (donor 304: T110S; donor 379: S106G); however, CD4 responses were not detected at this or neighbouring peptides.
Fig. 3.
Sequence mutants in targeted epitopes. (a) An alignment of the core region is shown. The upper line indicates the group consensus. The lower lines indicate donors 304 and 379 with mutations within targeted epitopes indicated. Dots indicate amino acids identical to the consensus sequence. (b) An alignment of the core region for cloned donor 304 is shown. Each clone was compared with the bulk sequencing product. The frequency of the variant within the epitope 61–80 is indicated by shading: A68V was observed in the majority of the sequenced population. (c) Peptide titrations using PBMCs from donors 304 and 379, using wild-type (▪) and mutant (□) peptide as indicated in Fig. 3(a). The assays were performed as in Fig. 1.
To determine the frequency at which these variants were observed within these two donors, the core–E1 region was cloned and sequenced (donor 304: n=21 sequences; donor 379: n=14 sequences). At aa 68 within donor 304, the mutation to valine was observed in the majority of the cloned population (Fig. 3b; 71.4 %). Within donor 379, the quasispecies population at amino acid position 147 remained identical to the mutant variant valine.
To establish whether T-cell reactivity was linked overall to positive selection within the core, the frequencies of synonymous (dS) and non-synonymous (dN) substitutions and mean pairwise diversity (π) were calculated for the CD4 responders and non-responders; these gave an indication of, respectively, the putative levels of selection and nucleotide diversity within each population (Table 3). Patients exhibiting a CD4+ T-cell response to the core peptides did not display a greater level of nucleotide diversity or level of selection (P>0.05). The values of dS/dN were substantially lower, as expected, in E1 compared with the core, but this did not differ between the two groups. Further analysis identified no evidence for significant positive selection at individual codons within the core region for both CD4 responders and non-responders. Cloning of the core–E1 region for patients 304 and 379 also identified no positively selected sites.
Table 3.
Genetic diversity of the HCV core and E1 in CD4+ T-cell responders and non-responders
| Core | E1 | |||
|---|---|---|---|---|
| dS/dN | π | dS/dN | π | |
| All individuals (n=46) | 26.86 | 0.048 | 8.13 | 0.149 |
| CD4+ responders (n=23) | 26.88 | 0.055 | 9.23 | 0.167 |
| CD4+ non-responders (n=23) | 25.83 | 0.041 | 6.93 | 0.132 |
Functional impact of epitope mutants
To test whether the donor CD4+ T cells were able to recognize autologous viral sequence in the two cases where variants were observed (donors 304 and 379), ex vivo ELISPOT assays were performed using wild-type and mutant peptides (Fig. 3c). The peptides were titrated to determine the functional avidity of the response. In both cases, there was a clear-cut lack of recognition of the autologous mutant peptide, despite consistent reactivity against the wild-type peptide, suggesting in both cases that CD4+ T cells were unable to respond to the endogenous variant.
DISCUSSION
Virus variation is a major feature of HCV infection, and immune escape from cellular and humoral immune responses is thought to play a significant role in the evolution of chronic infection (Bowen & Walker, 2005a, b; Cox et al., 2005b; Ray et al., 2005). During chronic infection, it is well recognized that there is a loss of CD8+ and CD4+ reactivity in the blood towards a range of peptides, compared with patients where infection has been resolved (Bowen & Walker, 2005a; Cox et al., 2005a; Day et al., 2003; Lauer et al., 2004; Lucas et al., 2007; Missale et al., 1996; Semmo et al., 2005, 2007). Although a link between immune pressure mediated by CD8+ T cells and selection of immune escape variants has been clearly shown (Bowen & Walker, 2005a, b; Cox et al., 2005a), few comparable data exist for CD4+ T cells. The impact of virus variation on T-cell function can be profound (Wang & Eckels, 1999), and CD4+ escape has been documented in other viral infections (Ciurea et al., 2001), as well as in specific cases in HCV (Puig et al., 2006). However, evaluation of such CD4+ T-cell responses in chronic hepatitis C is extremely difficult as they are typically described as ‘weak’ or ‘absent’ in persistent infection. However, the use of conventional assays of T-cell proliferation in these analyses may miss populations of CD4+ T cells with different functional profiles. We have recently shown using fresh PBMCs in both ELISPOT assays and a carboxyfluorescein succinimidyl ester proliferative evaluation (Semmo et al., 2005, 2007) that certain CD4 populations are maintained, but that they are low in both IL-2 production and proliferative capacity, features similar to HIV-specific CD4+ T-cell populations (Day & Walker, 2003). Assays may also be limited by substantial sequence mismatching between the viral antigens used in the assays and those present in the donor's circulating virus – this matters particularly in cases where superinfection has occurred (Harcourt et al., 2003; Schulze Zur Wiesch et al., 2007).
In this study, we first sought to examine HCV-specific CD4+ T-cell responses in a large, carefully selected cohort of chronic HCV patients in order to clearly map the frequency of the peptide-specific response. The HCV genotype 1 core was selected as an appropriate target due to previously identified reproducible responses to this antigen in studies of persistently infected patients (Barnes et al., 2002; Harcourt et al., 2006; Semmo et al., 2005, 2007; Weiner et al., 1995). In addition, the core is a relatively conserved protein and, as a result, sequence variation between the peptides used in the study and the viral strains present in donors should be limited. Peptides were tested individually in order to analyse the breadth of responses and to avoid competitive inhibition. The use of CD8-depleted PBMCs and a sensitive ex vivo ELISPOT assay for IFN-γ enabled us to detect relatively low-frequency cell populations and those with limited proliferative capacity, and thus provided a specific and accurate picture of the HCV CD4+ population in the cohort. Responses to non-structural antigens were very weak in this study, and similar results were also obtained using independently generated overlapping peptide sets spanning the entire genome (data not shown). Although such proteins would represent ideal targets for future studies, the lack of responses to these antigens in ex vivo assays severely limits the practicality and scale of such analyses.
HCV core-specific CD4+ T cells were readily detectable in just over 50 % of chronically infected patients using the IFN-γ ELISPOT assay, similar to results found in other studies (Semmo et al., 2005, 2007). The majority of core peptides were recognized, but responses focused particularly on aa 31–50 and 61–80. Within an individual, up to four different regions were recognized, with the majority of donors recognizing two or more distinct peptides even within this small region of the viral genome. Such data suggest that the level of multi-specificity of ex vivo responses in blood during chronic infection may previously have been underestimated. Even our analysis may be an underestimate, as the use of overlapping peptides as described might still fail to detect certain responses. The HLA class II types of those responding to individual peptides were not consistent in each case (data not shown), but data from other studies of virus-specific CD4+ T cells have shown that peptides typically bind and are presented by diverse HLA class II molecules (Gerlach et al., 2005; Lamonaca et al., 1999). However, the finding of highly targeted peptides within this conserved region, even though it is an underestimate of the total antiviral CD4+ T-cell response, may still provide an important focus in future for analyses of CD4+ T-cell responses, e.g. with MHC class II tetramers.
Sequencing of the core region of autologous virus revealed only two mutations within the recognized epitope, indicating that escape from recognition by specific CD4 cells is not a common occurrence. The CD4+ T-cell non-responder group acted as important controls in this respect, as it could be argued that the responses are most likely to be sustained in those with intact sequences; however, in the non-responder group, mutation was also rare. Phylogenetic and selection analysis did not resolve the two groups, and no signature of immune selection was detected within the region. Larger studies looking for HLA footprints may be required to define the impact of relatively infrequent selection events (Ray et al., 2005).
In the two cases where mutations did occur, these were novel mutations not found in sequence databases (Combet et al., 2007; Hraber et al., 2007) and both were associated with a functional impact on T-cell recognition. In the absence of the infecting strain, it is not possible to prove that the mutations occurred within the donors. However, in the case of donor 304, infection with HCV occurred as a result of a blood transfusion with no further apparent exposure. This makes it unlikely that superinfection with a mutant strain had occurred, but rather that this change has emerged within the host through immune selection pressures, with two-thirds of the viral clonal population possessing the A68V mutation.
As the core is a conserved viral gene, the failure to generate escape mutants may be related to the fitness cost to the virus of such changes. Interestingly, the A147V mutation lies at the beginning of a structural helix in the lipid droplet-binding domain (D2) of the core. Shavinskaya et al. (2007) showed that this amino acid substitution could enhance virus production through improved binding of the core to lipid droplets. Thus, loss of fitness does not seem to be a major argument for conservation of this residue. The impact of viral mutation may be greater on other sites within the core, although escape mutation sites in conserved regions of the core and NS3, which are targeted by CD8+ T cells, have been identified (Komatsu et al., 2006; Ray et al., 2005; Timm et al., 2007).
The impact of virus-specific CD4+ T cells on virus sequence and vice versa have been addressed in previous studies, although with more limited patient and hence responder numbers. In a single case report, induction of substantial CD4+ T-cell responses against HCV in a chimpanzee was associated temporally with the emergence of a mutation that eliminated recognition (Puig et al., 2006). In studies of T-cell lines derived from individuals with chronic infection, it has been observed that mutations can impact on the functional responses of T cells, including a switch away from a Th1 phenotype (Wang & Eckels, 1999). Despite these data, the overall role of virus escape in evasion of CD4+ T cells has not been fully addressed. Our data suggest that the phenomenon may occur, but that it is not common, at least among readily detectable responses. A recently performed longitudinal analysis of four infected chimpanzees concurs with this conclusion; these mutations occurred only rarely in this chronically infected HCV model (Fuller et al., 2008).
Exactly what the impact is of such persistent CD4+ T-cell responses with intact viral sequences on viral load and liver pathology is still not clear and may demand a larger study, incorporating larger numbers of antigenic targets. Studies to define the impact of target-specific CD8+ T-cell responses on HIV viral load (a setting where T-cell responses are much more readily detectable) have required cohorts in the order of 300–400 (Kiepiela et al., 2007). Alternatively, it may be that similar studies of liver-derived cells may be required to define the specificity and magnitude of the responses and their function at the site of infection (Penna et al., 2002; Schirren et al., 2000). Longitudinal studies may also reveal specific mutations, although these patients have been chronically infected for many years and acute infecting sequences are not available. For studies of CD8+ T-cell responses, cross-sectional approaches have revealed very obvious ‘footprints’ using a bulk sequencing approach, with similar or even smaller patient numbers, so although there are limitations in the approach used, it can be quite sensitive if selection pressure is strong (Dazert et al., 2009).
In conclusion, we observed sustained CD4+ T-cell responses in a large group of individuals persistently infected with HCV genotype 1, which targeted multiple peptides within the core protein. The responses were not associated with a clear virological footprint, and virologically responders and non-responders did not differ significantly, but in those cases where this was observed, mutation had an impact on T-cell recognition. This is inconsistent with a major role for immune escape in impairment of CD4+ T-cell responses against HCV, although it may occur in specific instances or in distinct antigenic targets. In this study, we focused on responses to the HCV core in genotype 1; it will be of importance to analyse whether the same process occurs in those responses targeting other viral proteins and/or in other genotypes, which may be under different functional constraints. Similarly, although cross-sectional studies do have significant power to detect escape, sequential studies of patients tracked through acute disease may provide an important alternative strategy. Defining the mechanisms underlying the failure of CD4+ T cells to contain HCV and the factors that determine their magnitude and function, longitudinally and especially in the liver, remain important goals in future studies.
METHODS
Study subjects.
Sixty-one subjects were included in the study from the Hepatitis Clinic at the John Radcliffe Hospital, Oxford, UK. All patients had persistent genotype 1 HCV infection and consented according to a locally approved protocol (COREC 04.OXA.010). Patients who had received treatment ending within the last 12 months were excluded. Of the 61 patients, 15 had received prior unsuccessful treatment with interferon and/or ribavirin (mean end 4.1 years previously, range 4–14 years). The clinical details of the patients are shown in Table 1.
T-cell assays
IFN-γ ELISPOT.
Fresh blood was obtained from the 61 individuals and PBMCs were separated on a density gradient. CD8+ cells were depleted using magnetic beads (Dynal) and the CD8− PBMCs assayed in an IFN-γ ELISPOT assay (MabTech) using 2×105 cells per well, against a pool of 18 core peptides (10 μg ml−1 final concentration for each peptide) to observe overall responses and a panel of 18 individual overlapping 20mer peptides covering the HCV genotype 1 core (aa 1–191; see Supplementary Table S1, available in JGV Online; 10 μg ml−1 final concentration) to provide fine detail. In addition, recombinant genotype 1 NS3–NS5 (Chiron; 2 μg ml−1), cytomegalovirus-infected cell lysate (Virusys; 2 μg ml−1) and phytohaemaglutinin were included as positive controls. Each antigen was tested in duplicate wells and the frequency of IFN-γ-producing cells was calculated by subtracting the mean number of s.f.c. per 106 CD8-depleted PBMCs in the negative-control wells (cells/medium alone) from the mean number of s.f.c. in the test wells (Semmo et al., 2005). A positive response was regarded as one in which the difference above the negative-control value of the s.f.c. per well was calculated to be significant (P<0.05; Excel binomdist). In specific experiments, a pool of genotype 1-derived NS5A peptides, consisting of 40 overlapping 18mers (BEI Resources) was used.
In further experiments to define the efficacy of the responses or examine the effect of mutations on T-cell recognition, wild-type and/or mutant peptides were tested in serial dilutions in RPMI 1640 in pairs, using the ELISPOT technique as above.
HCV sequence evaluation.
Viral RNA was extracted from plasma samples using a Viral RNA Extraction kit (Qiagen). Using a combined reverse transcription and first-round PCR step to amplify the core–E1 region, a 5063 bp external fragment was amplified using 10 pmol of primers utr-246 (5′-GACTGCTAGCCGAGTAGTGTTG-3′) and NS-5315 (5′-CGACCTCYARGTCNGCYCACATRC-3′) (Liu et al., 2004). A second-round PCR was performed with the inner primer utr-277 (5′-CCTTGTGGTACTGCCTGATAG-3′) and a modification of the outer primer C-E1 (5′-GTDGGNGACCARTTCATCATCAT-3′) (Corbet et al., 2003). Using a SuperScript III One-Step RT-PCR kit with Platinum Taq DNA polymerase (Invitrogen), RT-PCR cycling conditions were as follows: 55 °C for 30 min and 94 °C for 2 min, followed by 39 cycles of 15 s at 94 °C, 30 s at 57 °C and 5.5 min at 68 °C, with a final extension of 68 °C for 10 min. The inner PCR conditions were as follows: 94 °C for 2 min and ten cycles of 15 s at 94 °C, 30 s at 56 °C and 1 min at 72 °C, followed by 20 cycles of 15 s at 94 °C, 30 s at 56 °C and 1 min increasing by 5 s every cycle at 72 °C, with a final extension of 72 °C for 20 min using a high-fidelity Taq DNA polymerase (Expand High Fidelity PCR System; Roche). PCR fragments were gel purified (Qiagen) and the population was sequenced bidirectionally using Prism Big Dye (Applied Biosystems) on an ABI 3100 DNA automated sequencer.
Where necessary, PCR products were also cloned (TOPO TA; Invitrogen) and the DNA purified as above (Qiagen) and sequenced as above. Sequences were edited using Sequencher v4.8 (Gene Codes) and aligned with the Se-Al v2.0 sequence alignment editor (Rambaut, 2007). Kimura's two-parameter model was implemented using mega 4.0 to create neighbour-joining phylogenetic trees (Kimura, 1980). Bootstrap analyses were carried out with 1000 replicates. Levels of dS and dN mutations and π were calculated using mega 4.0. To detect evidence of selection at individual codons, single likelihood ancestor counting was used as implemented in DataMonkey (Kosakovsky Pond & Frost, 2005a, b). The analysis was conducted with both the HKY85 and general reversible models of nucleotide substitution with a cut-off P value of 0.1.
HCV load.
Based on the method outlined by Komurian-Pradel et al. (2001), HCV viral load quantification was carried out using real-time PCR with SYBR Green I detection (Roche). The 5′ HCV non-coding region was transcribed into cDNA using primer RC21 (5′-CTCCCGGGGCACTCGCAAGC-3′) (Besnard & Andre, 1994) and following the manufacturer's instructions for SuperScript II Reverse Transcriptase (Invitrogen). Real-time PCR was carried out with 1 μl cDNA with 10 pmol primer RC1 (5′-GTCTAGCCATGGCGTTAGTA-3′) and primer RC21 in a final volume of 25 μl. The reaction was performed in a LightCycler 480 (Roche). The PCR cycling conditions were as follows; an initial denaturation step at 95 °C for 15 min, followed by 45 cycles of 95 °C for 30 s, 60 °C for 30 s and 72 °C for 15 s. For each step, the temperature transition rate varied between 2.2 and 4.4 °C s−1, with fluorescence measurements taken after each elongation step. Conversion of copies ml−1 to IU ml−1 was performed using the HCV RNA genotype panel (National Institute for Biological Standards and Control, UK).
Statistical analysis.
Levels of ALT, viral load and T-cell responses were compared using the Mann–Whitney test. Pearson's chi-squared test was used to compare genetic diversity between groups for the core and E1. P<0.05 was considered statistically significant. Statistics were analysed using Prism V (Graphpad Software) and Excel (Microsoft).
Supplementary Material
Acknowledgments
This work was funded by the Wellcome Trust, the MRC, the James Martin 21st Century School, Oxford, UK, the EU (VIRGIL) programme and the NIHR Biomedical Research Centre Programme (Oxford, UK).
Footnotes
The GenBank/EMBL/DDBJ accession numbers for the sequences determined in this study are FN665803 and FN666283–FN666408.
A supplementary table showing the HCV core peptide amino acid sequences used in this study is available with the online version of this paper.
References
- Alter, M. J. (2007). Epidemiology of hepatitis C virus infection. World J Gastroenterol 13, 2436–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, E., Harcourt, G., Brown, D., Lucas, M., Phillips, R., Dusheiko, G. & Klenerman, P. (2002). The dynamics of T-lymphocyte responses during combination therapy for chronic hepatitis C virus infection. Hepatology 36, 743–754. [DOI] [PubMed] [Google Scholar]
- Besnard, N. C. & Andre, P. M. (1994). Automated quantitative determination of hepatitis C virus viremia by reverse transcription-PCR. J Clin Microbiol 32, 1887–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen, D. G. & Walker, C. M. (2005a). Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature 436, 946–952. [DOI] [PubMed] [Google Scholar]
- Bowen, D. G. & Walker, C. M. (2005b). Mutational escape from CD8+ T cell immunity: HCV evolution, from chimpanzees to man. J Exp Med 201, 1709–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciurea, A., Hunziker, L., Martinic, M. M., Oxenius, A., Hengartner, H. & Zinkernagel, R. M. (2001). CD4+ T-cell-epitope escape mutant virus selected in vivo. Nat Med 7, 795–800. [DOI] [PubMed] [Google Scholar]
- Combet, C., Garnier, N., Charavay, C., Grando, D., Crisan, D., Lopez, J., Dehne-Garcia, A., Geourjon, C., Bettler, E. & other authors (2007). euHCVdb: the European hepatitis C virus database. Nucleic Acids Res 35, D363–D366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbet, S., Bukh, J., Heinsen, A. & Fomsgaard, A. (2003). Hepatitis C virus subtyping by a core–envelope 1-based reverse transcriptase PCR assay with sequencing and its use in determining subtype distribution among Danish patients. J Clin Microbiol 41, 1091–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, A. L., Mosbruger, T., Lauer, G. M., Pardoll, D., Thomas, D. L. & Ray, S. C. (2005a). Comprehensive analyses of CD8+ T cell responses during longitudinal study of acute human hepatitis C. Hepatology 42, 104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, A. L., Mosbruger, T., Mao, Q., Liu, Z., Wang, X. H., Yang, H. C., Sidney, J., Sette, A., Pardoll, D. & other authors (2005b). Cellular immune selection with hepatitis C virus persistence in humans. J Exp Med 201, 1741–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day, C. L. & Walker, B. D. (2003). Progress in defining CD4 helper cell responses in chronic viral infections. J Exp Med 198, 1773–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day, C. L., Seth, N. P., Lucas, M., Appel, H., Gauthier, L., Lauer, G. M., Robbins, G. K., Szczepiorkowski, Z. M., Casson, D. R. & other authors (2003). Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers. J Clin Invest 112, 831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dazert, E., Neumann-Haefelin, C., Bressanelli, S., Fitzmaurice, K., Kort, J., Timm, J., McKiernan, S., Kelleher, D., Gruener, N. & other authors (2009). Loss of viral fitness and cross-recognition by CD8+ T cells limit HCV escape from a protective HLA-B27-restricted human immune response. J Clin Invest 119, 376–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepolder, H. M., Zachoval, R., Hoffmann, R. M., Wierenga, E. A., Santantonio, T., Jung, M. C., Eichenlaub, D. & Pape, G. R. (1995). Possible mechanism involving T-lymphocyte response to non-structural protein 3 in viral clearance in acute hepatitis C virus infection. Lancet 346, 1006–1007. [DOI] [PubMed] [Google Scholar]
- Eckels, D. D., Zhou, H., Bian, T. H. & Wang, H. (1999). Identification of antigenic escape variants in an immunodominant epitope of hepatitis C virus. Int Immunol 11, 577–583. [DOI] [PubMed] [Google Scholar]
- Eckels, D. D., Wang, H., Bian, T. H., Tabatabai, N. & Gill, J. C. (2000). Immunobiology of hepatitis C virus (HCV) infection: the role of CD4 T cells in HCV infection. Immunol Rev 174, 90–97. [DOI] [PubMed] [Google Scholar]
- Erickson, A. L., Kimura, Y., Igarashi, S., Eichelberger, J., Houghton, M., Sidney, J., McKinney, D., Sette, A., Hughes, A. L. & Walker, C. M. (2001). The outcome of hepatitis C virus infection is predicted by escape mutations in epitopes targeted by cytotoxic T lymphocytes. Immunity 15, 883–895. [DOI] [PubMed] [Google Scholar]
- Fuller, M. J., Bowen, D. G., Rutkiewicz, J. M., Shoukry, N. H., Hughes, A. L. & Walker, C. M. (2008). CD4 T cells do not exert selective pressure to promote persistent HCV infection. In 15th International Symposium on Hepatitis C Virus and Related Viruses, 5–9 October 2008, San Antonio, Texas, USA, p. 48.
- Gerlach, J. T., Ulsenheimer, A., Gruner, N. H., Jung, M. C., Schraut, W., Schirren, C. A., Heeg, M., Scholz, S., Witter, K. & other authors (2005). Minimal T-cell-stimulatory sequences and spectrum of HLA restriction of immunodominant CD4+ T-cell epitopes within hepatitis C virus NS3 and NS4 proteins. J Virol 79, 12425–12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui, A., Shoukry, N. H., Woollard, D. J., Han, J. H., Hanson, H. L., Ghrayeb, J., Murthy, K. K., Rice, C. M. & Walker, C. M. (2003). HCV persistence and immune evasion in the absence of memory T cell help. Science 302, 659–662. [DOI] [PubMed] [Google Scholar]
- Harcourt, G. C., Lucas, M., Godkin, A. J., Kantzanou, M., Phillips, R. E. & Klenerman, P. (2003). Evidence for lack of cross-genotype protection of CD4+ T cell responses during chronic hepatitis C virus infection. Clin Exp Immunol 131, 122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt, G., Gomperts, E., Donfield, S. & Klenerman, P. (2006). Diminished frequency of hepatitis C virus specific interferon gamma secreting CD4+ T cells in human immunodeficiency virus/hepatitis C virus coinfected patients. Gut 55, 1484–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hraber, P. T., Leach, R. W., Reilly, L. P., Thurmond, J., Yusim, K. & Kuiken, C. (2007). Los Alamos hepatitis C virus sequence and human immunology databases: an expanding resource for antiviral research. Antivir Chem Chemother 18, 113–123. [DOI] [PubMed] [Google Scholar]
- Kiepiela, P., Ngumbela, K., Thobakgale, C., Ramduth, D., Honeyborne, I., Moodley, E., Reddy, S., de Pierres, C., Mncube, Z. & other authors (2007). CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med 13, 46–53. [DOI] [PubMed] [Google Scholar]
- Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16, 111–120. [DOI] [PubMed] [Google Scholar]
- Klenerman, P. & Hill, A. (2005). T cells and viral persistence: lessons from diverse infections. Nat Immunol 6, 873–879. [DOI] [PubMed] [Google Scholar]
- Komatsu, H., Lauer, G., Pybus, O. G., Ouchi, K., Wong, D., Ward, S., Walker, B. & Klenerman, P. (2006). Do antiviral CD8+ T cells select hepatitis C virus escape mutants? Analysis in diverse epitopes targeted by human intrahepatic CD8+ T lymphocytes. J Viral Hepat 13, 121–130. [DOI] [PubMed] [Google Scholar]
- Komurian-Pradel, F., Paranhos-Baccala, G., Sodoyer, M., Chevallier, P., Mandrand, B., Lotteau, V. & Andre, P. (2001). Quantitation of HCV RNA using real-time PCR and fluorimetry. J Virol Methods 95, 111–119. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond, S. L. & Frost, S. D. W. (2005a). Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21, 2531–2533. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond, S. L. & Frost, S. D. W. (2005b). Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol 22, 1208–1222. [DOI] [PubMed] [Google Scholar]
- Lamonaca, V., Missale, G., Urbani, S., Pilli, M., Boni, C., Mori, C., Sette, A., Massari, M., Southwood, S. & other authors (1999). Conserved hepatitis C virus sequences are highly immunogenic for CD4+ T cells: implications for vaccine development. Hepatology 30, 1088–1098. [DOI] [PubMed] [Google Scholar]
- Lauer, G. M., Barnes, E., Lucas, M., Timm, J., Ouchi, K., Kim, A. Y., Day, C. L., Robbins, G. K., Casson, D. R. & other authors (2004). High resolution analysis of cellular immune responses in resolved and persistent hepatitis C virus infection. Gastroenterology 127, 924–936. [DOI] [PubMed] [Google Scholar]
- Lechner, F., Wong, D. K., Dunbar, P. R., Chapman, R., Chung, R. T., Dohrenwend, P., Robbins, G., Phillips, R., Klenerman, P. & Walker, B. D. (2000). Analysis of successful immune responses in persons infected with hepatitis C virus. J Exp Med 191, 1499–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., Netski, D. M., Mao, Q., Laeyendecker, O., Ticehurst, J. R., Wang, X. H., Thomas, D. L. & Ray, S. C. (2004). Accurate representation of the hepatitis C virus quasispecies in 5.2-kilobase amplicons. J Clin Microbiol 42, 4223–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löhr, H. F., Schlaak, J. F., Kollmannsperger, S., Dienes, H. P., Meyer zum Büschenfelde, K. H. & Gerken, G. (1996). Liver-infiltrating and circulating CD4+ T cells in chronic hepatitis C: immunodominant epitopes, HLA-restriction and functional significance. Liver 16, 174–182. [DOI] [PubMed] [Google Scholar]
- Lucas, M., Ulsenheimer, A., Pfafferot, K., Heeg, M. H., Gaudieri, S., Gruner, N., Rauch, A., Gerlach, J. T., Jung, M. C. & other authors (2007). Tracking virus-specific CD4+ T cells during and after acute hepatitis C virus infection. PLoS One 2, e649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald, A. J., Duffy, M., Brady, M. T., McKiernan, S., Hall, W., Hegarty, J., Curry, M. & Mills, K. H. (2002). CD4 T helper type 1 and regulatory T cells induced against the same epitopes on the core protein in hepatitis C virus-infected persons. J Infect Dis 185, 720–727. [DOI] [PubMed] [Google Scholar]
- McKiernan, S. M., Hagan, R., Curry, M., McDonald, G. S., Kelly, A., Nolan, N., Walsh, A., Hegarty, J., Lawlor, E. & Kelleher, D. (2004). Distinct MHC class I and II alleles are associated with hepatitis C viral clearance, originating from a single source. Hepatology 40, 108–114. [DOI] [PubMed] [Google Scholar]
- Missale, G., Bertoni, R., Lamonaca, V., Valli, A., Massari, M., Mori, C., Rumi, M. G., Houghton, M., Fiaccadori, F. & Ferrari, C. (1996). Different clinical behaviors of acute hepatitis C virus infection are associated with different vigor of the anti-viral cell-mediated immune response. J Clin Invest 98, 706–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann-Haefelin, C., McKiernan, S., Ward, S., Viazov, S., Spangenberg, H. C., Killinger, T., Baumert, T. F., Nazarova, N., Sheridan, I. & other authors (2006). Dominant influence of an HLA-B27 restricted CD8+ T cell response in mediating HCV clearance and evolution. Hepatology 43, 563–572. [DOI] [PubMed] [Google Scholar]
- Penna, A., Missale, G., Lamonaca, V., Pilli, M., Mori, C., Zanelli, P., Cavalli, A., Elia, G. & Ferrari, C. (2002). Intrahepatic and circulating HLA class II-restricted, hepatitis C virus-specific T cells: functional characterization in patients with chronic hepatitis C. Hepatology 35, 1225–1236. [DOI] [PubMed] [Google Scholar]
- Puig, M., Mihalik, K., Tilton, J. C., Williams, O., Merchlinsky, M., Connors, M., Feinstone, S. M. & Major, M. E. (2006). CD4+ immune escape and subsequent T-cell failure following chimpanzee immunization against hepatitis C virus. Hepatology 44, 736–745. [DOI] [PubMed] [Google Scholar]
- Rambaut A. (2007). Sequence alignment editor (version 2.0). University of Edinburgh, UK. Distributed by the author. http://tree.bio.ed.ac.uk/software/seal/.
- Ray, S. C., Fanning, L., Wang, X. H., Netski, D. M., Kenny-Walsh, E. & Thomas, D. L. (2005). Divergent and convergent evolution after a common-source outbreak of hepatitis C virus. J Exp Med 201, 1753–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehermann, B., Chang, K. M., McHutchison, J. G., Kokka, R., Houghton, M. & Chisari, F. V. (1996). Quantitative analysis of the peripheral blood cytotoxic T lymphocyte response in patients with chronic hepatitis C virus infection. J Clin Invest 98, 1432–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruys, T. A., Nanlohy, N. M., van den Berg, C. H., Hassink, E., Beld, M., van de Laar, T., Bruisten, S., Wit, F., Krol, A. & other authors (2008). HCV-specific T-cell responses in injecting drug users: evidence for previous exposure to HCV and a role for CD4+ T cells focussing on nonstructural proteins in viral clearance. J Viral Hepat 15, 409–420. [DOI] [PubMed] [Google Scholar]
- Schirren, C. A., Jung, M. C., Gerlach, J. T., Worzfeld, T., Baretton, G., Mamin, M., Hubert Gruener, N., Houghton, M. & Pape, G. R. (2000). Liver-derived hepatitis C virus (HCV)-specific CD4+ T cells recognize multiple HCV epitopes and produce interferon gamma. Hepatology 32, 597–603. [DOI] [PubMed] [Google Scholar]
- Schulze Zur Wiesch, J., Lauer, G. M., Timm, J., Kuntzen, T., Neukamm, M., Berical, A., Jones, A. M., Nolan, B. E., Longworth, S. A. & other authors (2007). Immunologic evidence for lack of heterologous protection following resolution of HCV in patients with non-genotype 1 infection. Blood 110, 1559–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semmo, N., Day, C. L., Ward, S. M., Lucas, M., Harcourt, G., Loughry, A. & Klenerman, P. (2005). Preferential loss of IL-2-secreting CD4+ T helper cells in chronic HCV infection. Hepatology 41, 1019–1028. [DOI] [PubMed] [Google Scholar]
- Semmo, N., Krashias, G., Willberg, C. & Klenerman, P. (2007). Analysis of the relationship between cytokine secretion and proliferative capacity in hepatitis C virus infection. J Viral Hepat 14, 492–502. [DOI] [PubMed] [Google Scholar]
- Shavinskaya, A., Boulant, S., Penin, F., McLauchlan, J. & Bartenschlager, R. (2007). The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J Biol Chem 282, 37158–37169. [DOI] [PubMed] [Google Scholar]
- Shoukry, N. H., Grakoui, A., Houghton, M., Chien, D. Y., Ghrayeb, J., Reimann, K. A. & Walker, C. M. (2003). Memory CD8+ T cells are required for protection from persistent hepatitis C virus infection. J Exp Med 197, 1645–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm, J., Li, B., Daniels, M. G., Bhattacharya, T., Reyor, L. L., Allgaier, R., Kuntzen, T., Fischer, W., Nolan, B. E. & other authors (2007). Human leukocyte antigen-associated sequence polymorphisms in hepatitis C virus reveal reproducible immune responses and constraints on viral evolution. Hepatology 46, 339–349. [DOI] [PubMed] [Google Scholar]
- Wang, H. & Eckels, D. D. (1999). Mutations in immunodominant T cell epitopes derived from the nonstructural 3 protein of hepatitis C virus have the potential for generating escape variants that may have important consequences for T cell recognition. J Immunol 162, 4177–4183. [PubMed] [Google Scholar]
- Wang, J. H., Layden, T. J. & Eckels, D. D. (2003). Modulation of the peripheral T-cell response by CD4 mutants of hepatitis C virus: transition from a Th1 to a Th2 response. Hum Immunol 64, 662–673. [DOI] [PubMed] [Google Scholar]
- Weiner, A., Erickson, A. L., Kansopon, J., Crawford, K., Muchmore, E., Hughes, A. L., Houghton, M. & Walker, C. M. (1995). Persistent hepatitis C virus infection in a chimpanzee is associated with emergence of a cytotoxic T lymphocyte escape variant. Proc Natl Acad Sci U S A 92, 2755–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.