Abstract
The redox proteomics technique normally combines two-dimensional gel electrophoresis, mass spectrometry and protein databases to analyze the cell proteome from different samples, thereby leading to the identification of specific targets of oxidative modification. Oxidative stress that occurs due to increased levels of reactive oxygen species and reactive nitrogen species can target most biomolecules, consequently leading to altered physiological function of the cells. Redox proteomics has identified oxidatively modified protein targets in different pathological conditions, consequently providing insight into the pathways involved in pathogenesis of these conditions. This approach also can be used to identify possible protective mechanisms to prevent or delay these disorders.
Keywords: Redox proteomics, two-dimensional gel electrophoresis, mass spectrometry, oxidative modification, protein carbonyls, protein-bound HNE, 3-nitrotyrosine
Introduction
Proteomics is the science involved to identify the proteome of a cell [1–2]. With advances in technology a number of approaches were developed to assess the cell proteome. Proteomics most commonly uses two-dimensional gel electrophoresis as the major mechanism of separation, though new methods of separation have been developed, such as 2D-high performance liquid chromatography (2D-HPLC) and isotopically coded affinity tags (ICAT) [3–4]. One of the oldest and most successful techniques that are employed in proteomics is 2D gel electrophoresis coupled to mass spectrometry [5]. Unlike the human genome that consists of about 30,000 genes (http://www.ncbi.nlm.nih.gov/genome/guide/Human/), the proteome contains several hundred thousand proteins. This diversity in the number of proteins could be explained based on the alternative splicing and post-translational modifications (PTMs). A large number of different PTMs, among which are reversible modification of proteins such as phosphorylation, carbonylation, nitration, acetylation, and glycosylation [6] have been identified. Some of the PTMs like oxidation of cysteine to sulfonic acid cause irreversible modification of proteins. Proteomics not only identifies proteins, but it also determines post-translational modifications, localization, interactions, function, and expression.
In the case of oxidatively modified proteins, redox proteomics [7] is used as a tool to identify such proteins, i.e., proteins that are modified by the reactive oxygen and reactive nitrogen species (ROS/RNS). Increased levels of ROS/RNS or decreased levels of antioxidant enzymes lead to a condition called oxidative/nitrosative stress. ROS and RNS can react with biomolecules such as proteins, lipids, carbohydrates and nucleic acids leading to oxidative modification [8–17]. Oxidative modification of proteins, for example, leads to modification of the structure and consequently either gain- or loss- of function of proteins. Oxidative modification of proteins thereby plays an important role in physiological and pathological conditions [18–19]. Oxidative stress has been shown to be one of the mechanisms involved in a number of diseases, including neurodegenerative disorders, ischemia, cancer, etc.
In this paper, we have discussed the approach that is most commonly used to identify the protein oxidation markers. Protein oxidation is most commonly indexed by the amount of protein carbonyls and protein-bound 3-nitrotyrosine (3-NT) [20]. Protein carbonyls are produced by backbone fragmentation, hydrogen atom abstraction at alpha carbons, oxidant attack on several amino acid side-chains (Lys, Arg, Pro, Thr, etc.), or by the formation of Michael adducts between His, Lys, and Cys residues and reactive alkenals (e.g., 4-hydroxy-2-trans-nonenal (HNE)) [19]. In addition, glycation/glycoxidation of Lys amino groups, forming advance glycation end products (AGEs) [7, 19–22], can also lead to formation of protein carbonyls. As noted, protein carbonyls are widely used as a marker to assess the extent of oxidation of proteins both in in vivo and in vitro conditions [7, 20, 22]. 3-NTs, ultimate products of ONOO−-mediated radical formation on tyrosine residues, is another protein oxidation marker [9, 23]. Protein nitration is a reversible and selective process that sometimes serves as a cellular signaling mechanism, similar to protein phosphorylation. In a neurodegenerative disease like Alzheimer disease (AD), mitochondrial abnormalities occur [24], associated with leakage of O2−•, which coupled to NO˙ leads to increased formation of highly reactive peroxynitrite. As noted above, ONOO− in the presence of CO2 can act on various amino acids such as cysteine, methionine, tryptophan, phenylalanine and tyrosine, which are particularly susceptible to nitration. A number of studies support the notion that nitrosative stress also contributes to disease, for example neurodegeneration in AD [7, 10, 14, 17, 25].
One of the products of lipid peroxidation, 4-hydroxy 2-trans nonenal (HNE) [26], can covalently modify cysteine, lysine, or histidine residues by Michael-addition [19, 26]. HNE causes membrane structural damage, changes conformation of proteins, produces diffusible secondary bioactive aldehydes, and induces cell death in many cell types [12, 27–34]. In AD subjects, the levels of free and protein-bound HNE were found to be significantly increased in brain, plasma, cerebrospinal fluid (CSF) etc., compared with control subjects [29, 35].
Our laboratory was the first to use redox proteomics to identify brain protein targets of oxidation in AD [36–37]. Using redox proteomics, our laboratory also identified the changes in brain protein carbonyls, HNE –adducts, glutathionylation, and the nitration of tyrosine residues of AD, mild cognitive impairment (MCI), and models of AD, Huntington disease (HD), amyotrophic lateral sclerosis (ALS) and Parkinson disease (PD) [10, 17, 36–42]. Table I shows carbonylated, HNE-bound, and 3-NT proteins that were identified in AD brain using redox proteomics approaches [10, 17, 35–37, 42–44]. The increase in the specific oxidation of proteins identified by proteomics agrees with previous studies that showed an increase in the total levels of oxidative stress in AD brain [19, 45–46], and the use of redox proteomics showed enolase as a common target of oxidative modification among protein carbonyls, 3-NT, and protein-bound HNE in AD, suggesting that the brain shows specific patterns of protein oxidative PTMs in AD. As seen in Table I, redox proteomics led to the identification of a number of brain proteins that regulate glucose metabolism as being oxidatively altered in AD, consistent with results from positron emission tomography (PET) studies showing decreased glucose utilization reported in AD brain [10, 16–17, 35–37, 42, 44]. Further, redox proteomics studies in AD brain led to identification of peptidyl prolyl cis/trans isomerase (Pin1), a protein that plays an important role in regulating the function of amyloid precursor protein (APP) and tau protein, and consequently potentially contributing to AD pathology [42, 47]. Other proteomic studies [21, 43, 48], in addition to those from our laboratory, have identified oxidatively modified proteins, consistent with reported oxidative stress in neurodegenerative diseases [14, 49–50].
Table 1.
Functional Categorization of Oxidatively Proteins Identified in AD.
| Protein Function | Carbonylationa | HNE-boundb | Nitrationc |
|---|---|---|---|
| Energy Dysfunction | Creatine kinase, α -enolase, triose phosphate isomerase, phosphoglyerate mutase 1 | α-enolase, | α-enolase, γ-enolase, lactate dehydrogenase, glyceraldehydes-3-phosphate dehydrogenase, triose phosphate isomerase |
| Mitochondrial dysfunction | ATP synthase Manganese superoxide dismutase | ATP synthase, Voltage dependent anion channel protein 1 | |
| Antioxidant defense/detoxification dysfunction | peroxiredoxin VI, Manganese superoxide dismutase | ||
| Cell cycle; Aβ production; Tau phosphorylation | Pin 1 | ||
| Lipid abnormalities and cholinergic dysfunction | Neuropolypeptide h3 | ||
| Neuritic abnormalities and structural dysfunction | DRP2, β-actin, gamma-SNAP | DRP2, Alpha tubulin | DRP2, β-actin |
| Excitotoxicity | Excitatory amino acid transporter, Glutamine synthetase | Glutamine synthetase | |
| pH dysfunction | Carbonic anhydrase II | Carbonic anhydrase II | |
| Protein degradation | UCHL 1, heat shock protein |
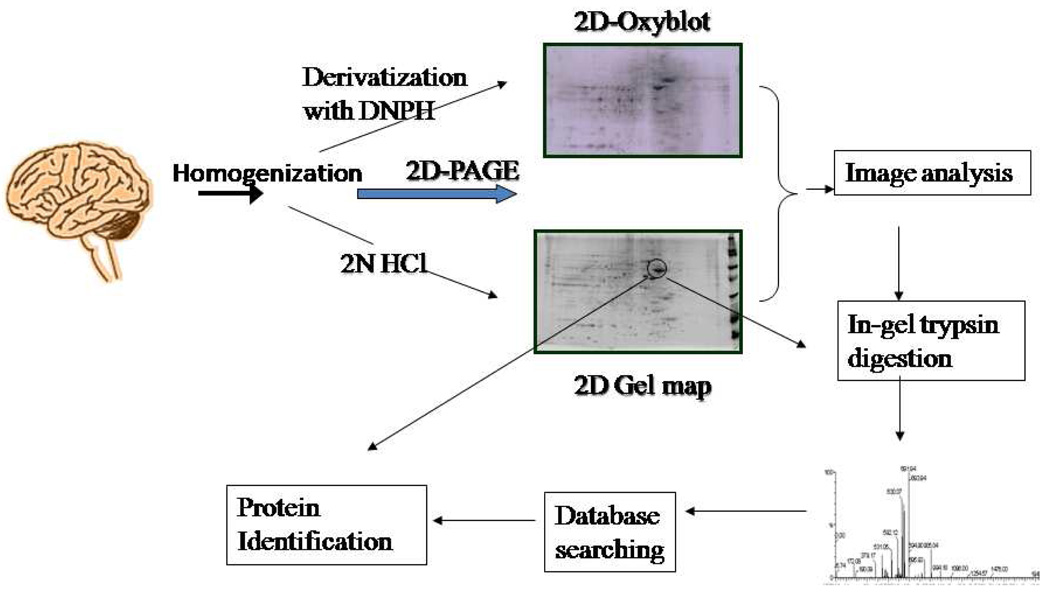
The combination of two-dimensional polyacrylamide gel electrophoresis (2D-PAGE), mass spectrometry (MS), and protein databases makes proteomics a powerful tool (Figure 1). However, this technique has a number of limitations including: (a) solubilization of membrane proteins, because the ionic detergents used for solubilization of such proteins can interfere with the isoelectric focusing process; (b) the mass range and the detection limits, which represent technical limitations of the method; and (c) proteins with high Lys/Arg content (which produce very low molecular weight tryptic peptides). Our laboratory and many others are trying to overcome these issues by using chaotropic agents, subcellular 2D gel electrophoresis, methods to concentrate the proteome being investigated, etc. High throughput proteomic techniques, such as HPLC, are also available to separate proteins without 2D electrophoresis [51]. However, the application of these techniques in redox proteomics is still relatively new, and more development in these techniques is needed.
Figure 1.
Outline of redox proteomics showing the incorporation of 2D-PAGE, MS, and protein database to identify oxidatively modified protein.
Principles
Protein carbonyls
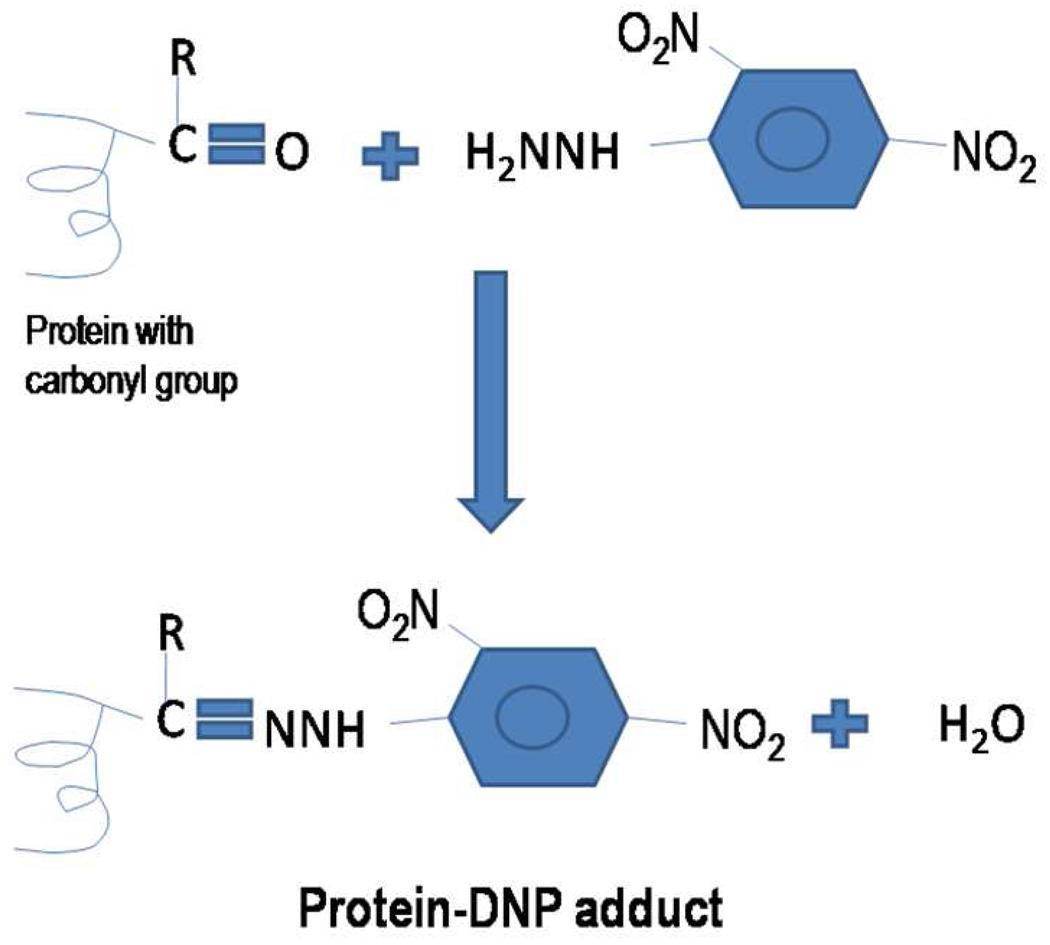
Protein carbonyls most often are detected by two methods, i.e., derivatization with 2,4-dinitrophenylhydrazine (DNPH), followed by immunochemical detection with an antibody against the resulting protein hydrazone, or formation of the Schiff base by biotin-hydrazide followed by detection of the protein-bound imine with enzyme- or fluorophore-linked avidin or streptavidin. Here we describe the DNPH-based detection method that is routinely used for detection of carbonylated proteins. In the DNPH method, samples are derivatized with dinitrophenyl hydrazine (DNPH). DNPH reacts with carbonyl groups to form protein resident 2,4-dinitrophenylhydrazone (DNP) (Figure 2), which can be detected using commercially available anti-DNP antibodies.
Figure 2.
Derivatization of the protein carbonyl group on a protein by 2,4 dinitrophenyl hydrazine to form a Schiff base product, 2,4-dinitrophenyl hydrazone.
Two dimensional gel electrophoresis
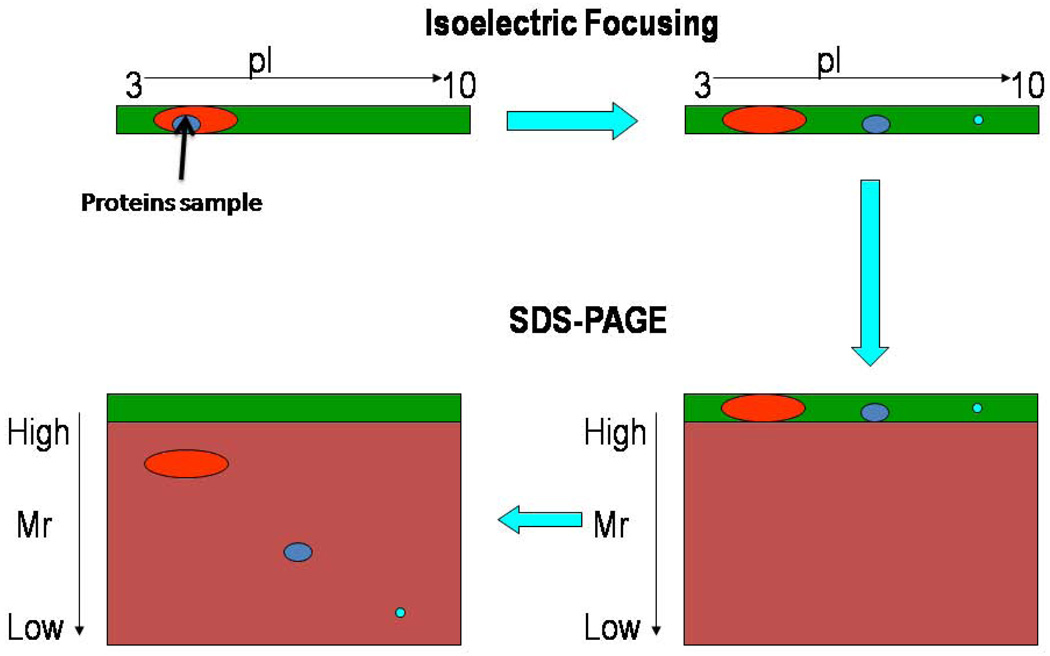
2D gel electrophoresis separates a mixture of proteins into single detectable protein spots in most cases. The separation of proteins is usually carried out in two steps, i.e., isoelectric focusing in the first dimension and molecular migration rate in the second dimension. In isoelectric focusing, proteins are separated based on their isoelectric point i.e, the pH at which the net charge on each protein is zero (Figure 3). In the second dimension, all the proteins are imparted a negative charge by addition of an anionic detergent such as sodium dodecyl sulfate (SDS) followed by separation of the proteins based on their relative mobility (Figure 3). Hence, on 2D gels proteins having smaller size to charge ratio will move faster than the proteins having larger size to charge ratio. These criteria allow separation of a large number of proteins, and each spot almost always represents a single protein.
Figure 3.
Two-dimensional gel electrophoresis allows separation of proteins based on the isoelectric point (pI) in the first dimension and relative mobility (Mr) in the second dimension.
PDQuest analysis
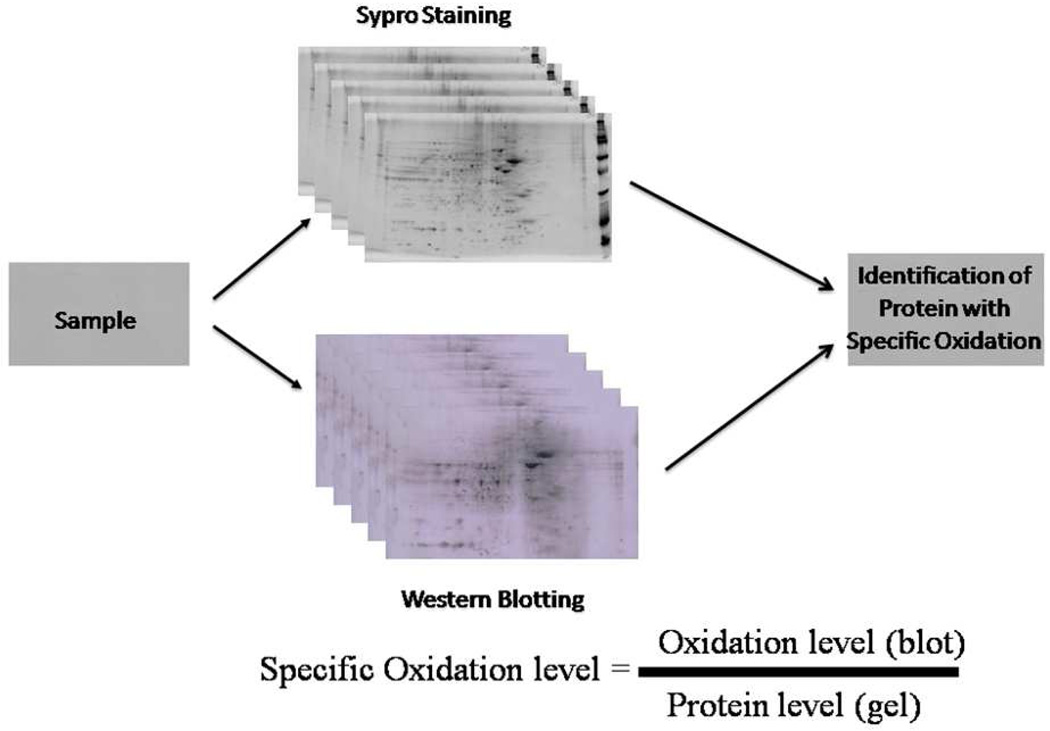
PDQuest image analysis allows gel-to-gel or blot to blot matching and analysis of visualized protein spots among different gels and blots (Figure 4). The principles of measuring intensity values by 2-D analysis software are similar to those of densitometric measurement. After completion of spot matching, the normalized intensity of each protein spot from individual gels (or blots) is compared between groups using statistical analysis. Although the software uses raw image-based alignment coupled with specific landmarks and neighboring spots to improve spot matching steps with significantly reduced analysis time, investigator-initiated hands-on processing is still necessary for higher accuracy of the matching. Normally, only spots that are statistically different between control and experimental groups are subjected to in-gel trypsin digestion for subsequent analysis by mass spectrometry. Each sample is run in triplicate to reduce variability and increase confidence of the resultant protein identification. In our experience, the percent CV is approximately 10 %. Hence, fold-change in levels or oxidative modification of at least 20–30 percent (often much more elevated oxidative modification is observed in AD brain proteins) over controls is necessary to pursue subsequent redox proteomics identification of specific protein spots.
Figure 4.
Determination of oxidized proteins by PDQuest software. PDQuest image analysis allows gel-to-gel or blot- to- blot matching and provides the intensity of each of the spots present on both gels and blots.
Mass Spectrometry (MS)
The primary MS methods used in our laboratory are peptide mass fingerprinting (PMF) following matrix assisted laser desorption ionization time of flight MS (MALDI-TOF) and electrospray ionization tandem MS (ESI-MS/MS). Full details of the methods of both techniques can be found elsewhere [52], and here a brief overview of each MS method will be given. In PMF, in-gel tryptic peptides of each spot of interest is mixed with an acidic solid matrix such as α-cyano-4-hyroxy cinnamic acid, which provides high sensitivity and negligible matrix adduction during the laser absorption [53], and subjected to laser radiation. The peptides are incorporated into the crystal lattice of the matrix during the condensation process. When the pulsed laser is applied to the matrix/analyte mixture, the peptides along with the matrix particles absorbs energy from the laser pulse and the matrix molecules containing the analyte are transferred into the gas phase. The positive ions of the peptides are formed in gas phase due to the acidic nature of the matrix in an as yet unknown process [54]. The ions are then accelerated into the mass analyzer where in the time of flight tube they are separated based on their m/z ratio. The peaks of the resulting mass spectrum represent the masses of the peptide ions of the sample of interest. Protein databases are available for theoretical digests of all known proteins which can be used for the identification of protein of interest. The protein database provides information such as molecular weight, pI and the probability of a random identification of a protein. The pI and molecular weight in the data base is compared to the experimental pI and molecular weight as a further indication of the correctness of the identification of the unknown protein. Many search engines perform this matching process with an output of a probability score for each theoretical digested protein indicating the correctness of the identification. The threshold score, which indicates if the experimental mass spectrum significantly matches the theoretical digested protein spectrum, is calculated by mathematical algorithms specific to each search engine and each experimental mass spectrum. Often this is manifested as a Mowse score (−10log10P), where P is the probability of a random identification. Given the high Mowse score required for significance, the p-value for PMF-identified proteins is often exceedingly small (often 10−8 or lower), giving confidence in the protein identification.
In the case of ESI MS/MS, tryptic peptide samples are loaded into a 96-well plate rack for nanoelectrospray infusion using an Advion Tri-Versa Nanomate (Ithaca, NY). Electrosprayed peptides are analyzed with an LTQ-Orbitrap XL (ThermoScientific, Waltham, MA) mass spectrometer. The Orbitrap normally is set to acquire a full MS scan at 60,000 resolution and in Data Dependent mode the eight most intense ions are selected for fragmentation and mass analyzed in the Orbitrap at 30,000 resolution. Conditions for fragmentation in the ion trap include a normalized collision energy of 35%, activation time of 30 ms, and selection of only +2 charge states or higher. Total acquisition time is approximately 5 minutes per sample. SEQUEST is used for database searching against the Uniprot SwissProt database. Filter criteria of returned protein lists included protein probabilities <0.01, peptide XCorr values >1.5 (for +1 charge state), 2.0 (+2 charge state), 2.5 (+3 charge state), and 3.0 (+4 charge state), peptide ΔCN values >0.1, and at least 2 peptides sequenced for each protein. As with MALDI, protein MW and pI information is used to assess individual protein identifications based on the location of the excised protein spot from the 2D gel. Only protein spots assigned to a single protein are further considered. As indicated above, the output of this MS method is the actual sequence of at least two tryptic peptides of the unknown protein, which, when searched against the SEQUEST database gives the identity of the protein.
In both PMF and ESI-MS/MS methods of protein identification, validation by independent means (often immunoprecipitation) is performed to ensure the correctness of the MS-identified proteins.
Materials
The chemicals used for redox proteomics and sources from whom they were purchased are largely from Sigma/Aldrich (St. Louis, MO, Milwaukee, WI, USA), unless stated otherwise: N-(2-Hydroxyethyl)piperazine-N'-2-ethanesulfonic acid buffer (HEPES), sodium chloride, potassium chloride (KCl), potassium phosphate monobasic (KH2PO4), MgSO4, leupeptin, pepstatin, type II S soybean trypsin inhibitor, phenylmethanesulfonylfluoride (PMSF), DNPH, hydrochloric acid, trichloroacetic acetic acid (TCA), ethanol, ethyl acetate, urea, thiourea, CHAPS, biolytes, dithiothreitol (DTT), bromophenol blue, Tris-HCl, SDS, glycerol, iodoacetamide (IA), running buffer (Bio-Rad, Hercules, CA), methanol, acetic acid, SYPRO Ruby Stain (Bio-Rad, Hercules, CA), phosphate buffered saline, Tween 20, sodium azide, bovine serum albumin (BSA), anti-dinitrophenyl hydrazone (anti-DNPH) antibody (Chemicon International, Temecula, CA), anti-rabbit conjugated to alkaline phoshatase antibody, SigmaFast tablet [5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (BCIP/NBT)], Criterion precast gels (8–16%; Bio-Rad, Hercules, CA), IPG strips (pH 3–10; Bio-Rad, Hercules, CA).
Instrumentation
The IEF instrument was purchased from Bio-Rad (Hercules, CA, USA), centrifuges were purchased from Beckman, UV transilluminator (λex=470 nm, λem=618 nm, Molecular Dynamics, Sunnyvale, CA, USA), µQuant Microtiter plate reader were purchased from Bio-Tek Instrument (Winooski, VT), vortex mixer was purchased from VWR International.
Protocol
Sample Preparation for Detection of Oxidatively Modified Proteins
Prepare 10% tissue homogenate in homogenization buffer {10 mM HEPES, 137 mM NaCl, 4.6 mM KCl, 1.1 mM KH2PO4, 0.6 mM MgSO4, 0.5 µg/mL leupeptin (stored as an aliquot at −20°C), 0.7 µg/mL pepstatin (stored as an aliquot at −20°C), 0.5 µg/mL type II S soybean trypsin inhibitor, 40 µg/mL PMSF dissolved in deionized water stored at 4°C). Centrifuge at 2,500g to remove intact cells and nuclei. Determine the protein content by using BCA reagent kit from Pierce (Rockford, IL).
For protein carbonyl detection: to 100–150 µg of the protein add DNPH (10 mM 2,4-dinitrophenyl hydrazine prepared in 2 N HCl solution (that can be stored at room temperature) four times the volume of sample, vortex the sample and incubate at room temperature for 20 min. Then, add a final concentration of 30% TCA and incubate the samples on ice for 10 min to precipitate the proteins. Centrifuge the sample at 10,000g for 5 min. Decant the supernatant and wash the pellet with ice cold ethanol and ethyl acetate solution (1:1 v/v). Suspend the final pellet in 200 µl of IEF rehydration buffer (8 M urea, 2 M thiourea, 2% CHAPS, 0.2% biolytes, 50 mM DTT, bromophenol blue dissolved in de-ionized water made fresh before use). Samples are incubated in rehydration buffer for a minimum of 1h at room temperature before loading in the IEF tray (samples can be incubated for a maximum of 2h).
For detection of HNE modified proteins, sample does not require any prederivatization. Samples are precipitated by TCA as described above, and the steps described above for protein carbonyls are followed for protein-bound HNE detection.
Detection of 3-NT modified proteins, like protein-bound HNE, does not require any prederivatization. Samples are precipitated by using 75% acetone, and the steps described above are followed for sample preparation.
First dimension or Isoelectric Focusing
Isoelectric focusing is performed with a Bio-Rad (Hercules, CA, USA) system using 110-mm, pH 3–10 or pH 4–7 immobilized pH gradients (IPG) strips. The selection of the IPG strips depends on nature of the samples and interest of the researcher.
Carefully load 180 µl of the samples into the bottom of the well in the isoelectric focusing (IEF) tray. Samples can be applied using either a (1) microliter syringe (washed between each sample by pipetting in deinonized water) or (2) disposable micropipette tips (discard each tip after a single use). While loading samples care should be taken to avoid bubbles which might interfere with current flow.
Remove the plastic sheet from the IPG strips (Bio-Rad, Hercules, CA) with forceps. Place the IPG strips on top of the sample with gel side facing down, making sure that the positive (+ ve) end of strip is towards the +ve end of the IEF tray. This is important for the correct connections.
Remove any air bubble trapped between the IPG strip and tray by gently tapping on the IPG strips with forceps.
Place the IEF tray in the IEF instrument and carry out an active rehydration overnight at 50 V (20°C). This step will enable the gel to swell and help in the penetration of samples into the IPG strip. Add 2mL of mineral oil (Bio-Rad, Hercules, CA) in each well after 1h, this will prevent both evaporation of rehydration buffer and drying of IPG strips. Carry out active rehydration step for about 16 h.
After 16 h, place wet paper wicks (Bio-Rad, Hercules, CA) on both the electrodes using a forceps to prevent burning of the IPG strips during isoelectric focusing. Carry out isoelectric focusing at 20 °C as follows: 300V for 2 h linear gradient, 500V 2 h linear gradient, 1000V 2 h linear gradient, 8000V 8 h linear gradient, 8000V 10 h rapid gradient.
After completion of IEF the IPG strips can be used directly for second dimension separation or stored in a −80 °C freezer until use.
Second Dimensional Electrophoresis
Incubate the IPG strips in 4mL of equilibration buffer containing DTT [50 mM Tris-HCl, 6 M urea, 1% (m/v) SDS, 30% (v/v) glycerol, 0.5% DTT dissolved in de-ionized water made fresh before use] in a disposable equilibration tray with lid (Bio-Rad, Hercules, CA, USA) with the gels side facing up for 10 min. Keep the equilibration tray in the dark. If the IPG strips have been stored in the −80 °C freezer, thaw the IPG strip at room temperature (thawed IPG strips change color from milky white to clear) before addition of the equilibration buffer.
Transfer the IPG strips into a new equilibration tray and add 4 mL of equilibration buffer containing iodoacetamide (IA) [50 mM Tris-HCl, 6 M urea, 1% (m/v) SDS, 30% (v/v) glycerol, 4.5% IA dissolved in de-ionized water made fresh before use], and incubate in the dark for another 10 min.
Wash the IPG strips in 1X running buffer (Bio-Rad, Hercules, CA, USA) [to remove excess equilibration buffer], and place the IPG strips with gel side facing up into criterion gels (Bio-Rad, Hercules, CA, USA).
Add warm agarose solution [Bio-Rad, Hercules, CA, USA] into the wells of criterion gels and then slowly push down the IPG strip until a contact is established between the gel and IPG strip. Remove any bubbles that are trapped in the agarose solution by gently pushing the IPG strip with forceps.
Load 2 µL of unstained (for gel only)/ stained molecular weight marker (for blot only) (Bio-Rad, Hercules, CA) into a standard well adjacent to the IPG strip. Allow the agarose to solidify for 10 min.
Place the gels in the electrophoresis tank filled with running buffer and then fill the upper tank with running buffer. Run the gels at 200V for 65 min at room temperature, until the dye front (bromophenol blue) just runs off the gel into the lower tank.
After the run is completed, disconnect the power supply (Bio-Rad, Hercules, CA) and disassemble the 2D-apparatus, and remove the gels.
Protein staining
Fix the gels containing non-derivatized proteins with unstained marker in 50mL of fixative solution [10% (v/v) methanol and 7% (v/v) acetic acid] for 1h at room temperature with gentle agitation [7, 42, 55]. Add 50mL of SYPRO Ruby gel stain (Bio-Rad, Hercules, CA, USA) and incubate overnight at room temperature on a rocking platform.
Oxyblot (For detection of Oxidatively modified proteins)
Gels containing DNPH derivatized proteins/nonderivatized proteins are transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA) using a semi-dry transfer unit (Bio-Rad, Hercules, CA) for detection of protein carbonyls and HNE/3-NT modified proteins, respectively. Prepare a transfer sandwich in the following order: first place one soaked filter paper on the transfer unit platform, followed by the nitrocellulose membrane, gel and one more wet filter paper.
Transfer the proteins at 15 V for 2h at room temperature. After transfer, block the nonspecific sites on the nitrocellulose membrane by incubating the membrane with 25 mL of blocking buffer (3% bovine serum albumin in wash blot) for 1 h at room temperature on a rocking platform.
After 1 h add 25 mL of the blocking buffer [3% bovine serum albumin (BSA) in PBST made fresh before use] containing (as appropriate for the analysis desired) either anti-DNPH /anti-HNE/ anti-3-NT antibody (1:100/1:1000/1:1000) (Chemicon International,Temecula, CA, USA) and incubate for 1 h at room temperature on a rocking platform.
Wash three times for 5 min each with 50 mL of wash blot [0.01% (w/v) sodium azide and 0.2% (v/v) Tween 20 dissolved in phosphate buffered saline (PBS) stored at room temperature].
Incubate the membrane in the secondary antibody (anti-Rabbit alkaline-phosphatase (ALP)-conjugated; 1:3000 in wash blot) for 1 h at room temperature on a rocking platform.
Wash three times for 5 min each with 50 mL of wash blot.
Develop the blot using SigmaFast tablets [5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (BCIP/NBT) [dissolved in 10mL DI water, prepared fresh]. After color development, drain out the developer and wash the membrane with deionized water and dry between filter papers.
Calculations and Expected Results
Scan the SYPRO Ruby stained and oxyblots using a UV transilluminator (λex=470 nm, λem=618 nm, Molecular Dynamics, Sunnyvale, CA, USA) and Adobe Photoshop on a Microtek Scanmaker 4900 (Cerritos, CA) respectively, and saved as ‘Tiff’ files.
Perform image analysis of oxyblots and 2D gel maps using PDQuest image analysis software (Bio-Rad, Hercules, CA, USA). This software gives the option of normalization of the actual protein content as measured by the intensity of a protein stain such as SYPRO ruby (Bio-Rad, Hercules, CA, USA and Molecular Probes, Eugene, OR, USA).
To determine the levels of specific oxidation divide the intensity of the spot on the blot by the intensity of spot on the gel (Figure 4). A minimum of n=6 independent samples (in triplicate) is run per group of samples. One of the best gels and one of the best blots showing the best resolution of spots is selected as master gel or blot to begin the analysis. PDQuest software allows the analysis of multiple gels and blots using powerful algorithms that accurately and quickly match protein spots in the “master” gel to the “master” blot. After completion of spot matching, to determine the levels of specific oxidation divide the intensity of the spot on the blot by the intensity of spot on the gel (Figure 4). Protein spots are considered significant if their p-value is less than 0.05.
Excise the spots showing a significant increase in specific oxidation compared to the control gel and perform an in gel-digestion with trypsin (Figure 1). However, first give consecutive treatments with 0.1 M ammonium bicarbonate (NH4HCO3), acetonitrile, 20 mM DTT in 0.1 M NH4HCO3, 55 mM iodoacetamide (IA) in 0.1 M NH4HCO3, 50 mM NH4HCO3 and acetonitrile. Then rehydrate the gel pieces with 20 ng/µL modified trypsin (Promega, Madison, WI) in 50 mM NH4HCO3 overnight at 37°C [56].
Salts and contaminants must be removed from tryptic peptide solutions using C18 ZipTip into a fresh tube. Step 1: To the remaining gel piece add ~20 µL of buffer A (5% acetonitrile, 0.1% formic acid), and sonicate it in water bath at 37°C for 15 min. Add ~30 µL of buffer B (95% acetonitrile, 0.1% formic acid, 1mM NH4HCO3) and sonicate it in water bath at 37°C for 15 min. Step 2: Transfer the supernatant obtained from step 1, to the trypic peptide solution aliquoted before. Concentrate the samples to ~10 µl using a speedVac. Step 3: Wash the ZipTip by aspirating 10 µL of solvent C (100% acetonitrile) 5 times through the same ZipTip, followed by equilibration of the ZipTip with 10 µL of buffer A. Step 4: Aspirate the samples for 4–5 times followed by washing of the ZipTip with buffer 10 µL of buffer A. Step 5: Elute the sample in 10 µL of buffer D (50% acetonitrile,, 0.1% formic acid). Submit the cleaned eluate obtained in step 5 for analysis by mass spectrometry, the output of which is used for protein identification.
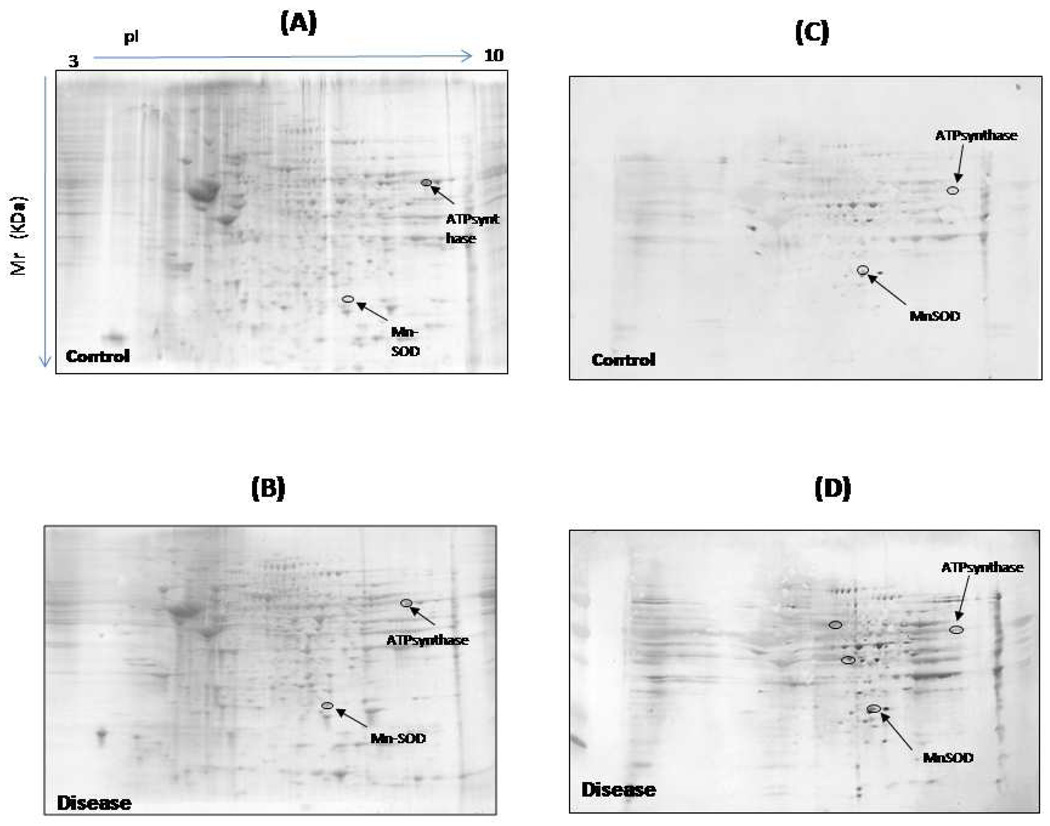
As described above, the results from the mass spectrometric analysis are searched against protein databases using automated search algorithms [57–58]. An example of the results produced is shown in Figure. 5.
Figure 5.
An example of gels from control (A) and disease samples (B) and their representative blots from control (C) and disease (D) samples.
Caveats
To minimize artifactual oxidation, brain samples should be obtained from subjects (or animals) at extremely short post mortem intervals (PMI, typically in our hands <3–4 h for human specimens; 1min or less for animal specimens), and all the processing of the samples should be done under identical conditions. If the sample handling leads to any oxidation this would be presumed to affect both the control and subject samples in a similar way.
It is important to include protease inhibitors in the sample homogenization buffer, since improper care of sample may lead to protein degradation that can have an effect on what is visualized and inaccurately reflect what is happening in the cell [59].
Proper washing of the pellet after derivatization is crucial to reduce the background signal on the blot due to excess DNPH which might interfere with image analysis, in addition to removal of lipids. Further, it is important to ensure that ethanol and ethylacetate solution are removed completely before adding rehydration buffer. The sample after derivatization usually has a high concentration of ions since an acidic buffer is used to optimize the reaction. The high level of ions in the buffer can cause variation of voltage and current during IEF, thereby preventing successful isoelectric separation. This phenomenon usually is manifested by horizontal smearing of the protein spot.
The sample should be incubated with IEF rehydration buffer for a minimum of 1 h to allow an efficient, quantitative solubilization of cellular proteins.
Too much of glycerol addition or too little DTT in the equilibration buffer may lead to smearing and streaking of the spots, which might interfere with the spot matching and final analysis.
When developing blots it is critical to maintain the same development time for all the blots, to prevent possible artifactual data generation. BCIP/NBT substrates generate an intense black purple precipitate at the site of enzyme binding. The reaction proceeds at a steady rate, thus allowing accurate control of the development of the reaction. This allows the relative sensitivity to be controlled by the length of incubation.
In almost all cases, 2D gel separation methods lead to one protein per spot. However, if a single protein spot from the 2D gel shows the presence of more than one protein in the same spot by MS, then immunoprecipitation techniques should be employed to identify the target of oxidation.
Acknowledgements
This research was supported by a NIH grant to D.A.B. [AG-05119].
List of Abbreviation
- 2D-HPLC
2D-high performance liquid chromatography
- 2D-PAGE
Two-dimensional polyacrylamide gel electrophoresis
- 3-NT
3-nitrotyrosine
- AD
Alzheimer’s disease
- AGEs
Advance glycation end products
- Anti-DNPH
Anti-dinitrophenyl hydrazone
- APP
Amyloid precursor protein
- BCIP
5-bromo-4-chloro-3-indolyl phosphate
- BSA
Bovine serum albumin
- DNP-
2,4-dinitrophenylhydrazone
- DNPH
Dinitrophenyl hydrazine
- DTT
Dithiothreitol
- HD
Huntington disease
- HEPES
N-(2-Hydroxyethyl)piperazine-N'-2-ethanesulfonic acid
- HNE
4-hydroxy-2-trans-nonenal
- HPLC
High pressure liquid chromatography
- IA
Iodoacetamide
- ICAT
Isotopically coded affinity tags
- IEF
Isoelectrofocusing
- KCl
Potassium chloride
- KH2PO4
Potassium phosphate monobasic
- MCI
Mild cognitive impairment
- MgSO4
Magnesium sulfate
- MS
Mass spectrometry
- NaCl
Sodium chloride
- NBT
Nitro blue tetrazolium
- NH4HCO3
Ammonium bicarbonate
- PBS
Phosphate buffered saline
- PD
Parkinson’s disease
- PET
Positron emission tomography
- pI
Isoelectric point
- Pin1
Peptidyl prolyl cis/trans isomerase
- PMI
Post mortem intervals
- PMSF
phenylmethanesulfonylfluoride
- PTMs
post-translational modifications
- RNS
Reactive nitrogen species
- ROS
Reactive oxygen species
- SDS
Sodium dodecyl sulfate
- TCA
Trichloric acetic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors have no conflict of interest.
References
- 1.O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 2.Rabilloud T. Two-dimensional gel electrophoresis in proteomics: old, old fashioned, but it still climbs up the mountains. Proteomics. 2002;2:3–10. [PubMed] [Google Scholar]
- 3.Smolka MB, Zhou H, Purkayastha S, Aebersold R. Optimization of the isotope-coded affinity tag-labeling procedure for quantitative proteome analysis. Anal Biochem. 2001;297:25–31. doi: 10.1006/abio.2001.5318. [DOI] [PubMed] [Google Scholar]
- 4.Wagner Y, Sickmann A, Meyer HE, Daum G. Multidimensional nano-HPLC for analysis of protein complexes. J Am Soc Mass Spectrom. 2003;14:1003–1011. doi: 10.1016/S1044-0305(03)00399-4. [DOI] [PubMed] [Google Scholar]
- 5.Anderson NL, Matheson AD, Steiner S. Proteomics: applications in basic and applied biology. Curr Opin Biotechnol. 2000;11:408–412. doi: 10.1016/s0958-1669(00)00118-x. [DOI] [PubMed] [Google Scholar]
- 6.Aebersold R, Goodlett DR. Mass spectrometry in proteomics. Chem Rev. 2001;101:269–295. doi: 10.1021/cr990076h. [DOI] [PubMed] [Google Scholar]
- 7.Dalle-Donne I, Scaloni A, Butterfield DA. Redox Proteomics: From protein modifications to cellular dysfunction and diseases. Hoboken, NJ: John Wiley and Sons; 2006. [DOI] [PubMed] [Google Scholar]
- 8.Butterfield DA. Amyloid beta-peptide (1-42)-induced oxidative stress and neurotoxicity: implications for neurodegeneration in Alzheimer's disease brain. A review. Free Radic Res. 2002;36:1307–1313. doi: 10.1080/1071576021000049890. [DOI] [PubMed] [Google Scholar]
- 9.Butterfield DA, Lauderback CM. Lipid peroxidation and protein oxidation in Alzheimer's disease brain: potential causes and consequences involving amyloid beta-peptide-associated free radical oxidative stress. Free Radic Biol Med. 2002;32:1050–1060. doi: 10.1016/s0891-5849(02)00794-3. [DOI] [PubMed] [Google Scholar]
- 10.Castegna A, Thongboonkerd V, Klein JB, Lynn B, Markesbery WR, Butterfield DA. Proteomic identification of nitrated proteins in Alzheimer's disease brain. J Neurochem. 2003;85:1394–1401. doi: 10.1046/j.1471-4159.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- 11.Lovell MA, Gabbita SP, Markesbery WR. Increased DNA oxidation and decreased levels of repair products in Alzheimer's disease ventricular CSF. J Neurochem. 1999;72:771–776. doi: 10.1046/j.1471-4159.1999.0720771.x. [DOI] [PubMed] [Google Scholar]
- 12.Lovell MA, Markesbery WR. Ratio of 8-hydroxyguanine in intact DNA to free 8-hydroxyguanine is increased in Alzheimer disease ventricular cerebrospinal fluid. Arch Neurol. 2001;58:392–396. doi: 10.1001/archneur.58.3.392. [DOI] [PubMed] [Google Scholar]
- 13.Markesbery WR, Kryscio RJ, Lovell MA, Morrow JD. Lipid peroxidation is an early event in the brain in amnestic mild cognitive impairment. Ann Neurol. 2005;58:730–735. doi: 10.1002/ana.20629. [DOI] [PubMed] [Google Scholar]
- 14.Smith MA, Richey Harris PL, Sayre LM, Beckman JS, Perry G. Widespread peroxynitrite-mediated damage in Alzheimer's disease. J Neurosci. 1997;17:2653–2657. doi: 10.1523/JNEUROSCI.17-08-02653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith MA, Richey PL, Taneda S, Kutty RK, Sayre LM, Monnier VM, et al. Advanced Maillard reaction end products, free radicals, and protein oxidation in Alzheimer's disease. Ann N Y Acad Sci. 1994;738:447–454. doi: 10.1111/j.1749-6632.1994.tb21836.x. [DOI] [PubMed] [Google Scholar]
- 16.Sultana R, Perluigi M, Butterfield DA. Redox proteomics identification of oxidatively modified proteins in Alzheimer's disease brain and in vivo and in vitro models of AD centered around Abeta(1-42) J Chromatogr B Analyt Technol Biomed Life Sci. 2006;833:3–11. doi: 10.1016/j.jchromb.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 17.Sultana R, Poon HF, Cai J, Pierce WM, Merchant M, Klein JB, et al. Identification of nitrated proteins in Alzheimer's disease brain using a redox proteomics approach. Neurobiol Dis. 2006;22:76–87. doi: 10.1016/j.nbd.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 18.Shen J, Yang X, Dong A, Petters RM, Peng YW, Wong F, et al. Oxidative damage is a potential cause of cone cell death in retinitis pigmentosa. J Cell Physiol. 2005;203:457–464. doi: 10.1002/jcp.20346. [DOI] [PubMed] [Google Scholar]
- 19.Butterfield DA, Stadtman ER. Protein oxidation processes in aging brain. Adv Cell Aging Gerontol. 1997:161–191. [Google Scholar]
- 20.Berlett BS, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 21.Dalle-Donne I, Scaloni A, Giustarini D, Cavarra E, Tell G, Lungarella G, et al. Proteins as biomarkers of oxidative/nitrosative stress in diseases: the contribution of redox proteomics. Mass Spectrom Rev. 2005;24:55–99. doi: 10.1002/mas.20006. [DOI] [PubMed] [Google Scholar]
- 22.Stadtman ER, Levine RL. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids. 2003;25:207–218. doi: 10.1007/s00726-003-0011-2. [DOI] [PubMed] [Google Scholar]
- 23.Beckman JS, Koppenol WH. Nitric oxide, superoxide, and peroxynitrite: the good, the bad, and ugly. Am J Physiol. 1996;271:C1424–C1437. doi: 10.1152/ajpcell.1996.271.5.C1424. [DOI] [PubMed] [Google Scholar]
- 24.Beal MF. Mitochondrial dysfunction in neurodegenerative diseases. Biochim Biophys Acta. 1998;1366:211–223. doi: 10.1016/s0005-2728(98)00114-5. [DOI] [PubMed] [Google Scholar]
- 25.Koeck T, Levison B, Hazen SL, Crabb JW, Stuehr DJ, Aulak KS. Tyrosine nitration impairs mammalian aldolase A activity. Mol Cell Proteomics. 2004;3:548–557. doi: 10.1074/mcp.M300141-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 27.Choudhary S, Zhang W, Zhou F, Campbell GA, Chan LL, Thompson EB, et al. Cellular lipid peroxidation end-products induce apoptosis in human lens epithelial cells. Free Radic Biol Med. 2002;32:360–369. doi: 10.1016/s0891-5849(01)00810-3. [DOI] [PubMed] [Google Scholar]
- 28.Hashimoto M, Sibata T, Wasada H, Toyokuni S, Uchida K. Structural basis of protein-bound endogenous aldehydes. Chemical and immunochemical characterizations of configurational isomers of a 4-hydroxy-2-nonenal-histidine adduct. J Biol Chem. 2003;278:5044–5051. doi: 10.1074/jbc.M210129200. [DOI] [PubMed] [Google Scholar]
- 29.Markesbery WR, Lovell MA. Four-hydroxynonenal, a product of lipid peroxidation, is increased in the brain in Alzheimer's disease. Neurobiol Aging. 1998;19:33–36. doi: 10.1016/s0197-4580(98)00009-8. [DOI] [PubMed] [Google Scholar]
- 30.Tamagno E, Robino G, Obbili A, Bardini P, Aragno M, Parola M, et al. H2O2 and 4-hydroxynonenal mediate amyloid beta-induced neuronal apoptosis by activating JNKs and p38MAPK. Exp Neurol. 2003;180:144–155. doi: 10.1016/s0014-4886(02)00059-6. [DOI] [PubMed] [Google Scholar]
- 31.Uchida K. 4-Hydroxy-2-nonenal: a product and mediator of oxidative stress. Prog Lipid Res. 2003;42:318–343. doi: 10.1016/s0163-7827(03)00014-6. [DOI] [PubMed] [Google Scholar]
- 32.Lauderback CM, Hackett JM, Huang FF, Keller JN, Szweda LI, Markesbery WR, et al. The glial glutamate transporter, GLT-1, is oxidatively modified by 4-hydroxy-2-nonenal in the Alzheimer's disease brain: the role of Abeta1-42. J Neurochem. 2001;78:413–416. doi: 10.1046/j.1471-4159.2001.00451.x. [DOI] [PubMed] [Google Scholar]
- 33.Subramaniam R, Roediger F, Jordan B, Mattson MP, Keller JN, Waeg G, et al. The lipid peroxidation product, 4-hydroxy-2-trans-nonenal, alters the conformation of cortical synaptosomal membrane proteins. J Neurochem. 1997;69:1161–1169. doi: 10.1046/j.1471-4159.1997.69031161.x. [DOI] [PubMed] [Google Scholar]
- 34.Owen JB, Sultana R, Aluise CD, Erickson MA, Price TO, Bu G, et al. Oxidative Modification to LDL-related Receptor Protein 1 (LRP1) in Hippocampus from Subjects with Alzheimer's Disease: Implications for Abeta Accumulation in AD Brain. Free Radic Biol Med. 2010 doi: 10.1016/j.freeradbiomed.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perluigi M, Sultana R, Cenini G, Di Domenico F, Memo M, Pierce WM, et al. Redox proteomics identification of 4-hydroxynonenal-modified brain proteins in Alzheimer's disease: Role of lipid peroxidation in Alzheimer's disease pathogenesis. Proteomics Clin Appl. 2009;3:682–693. doi: 10.1002/prca.200800161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castegna A, Aksenov M, Aksenova M, Thongboonkerd V, Klein JB, Pierce WM, et al. Proteomic identification of oxidatively modified proteins in Alzheimer's disease brain. Part I: creatine kinase BB, glutamine synthase, and ubiquitin carboxy-terminal hydrolase L-1. Free Radic Biol Med. 2002;33:562–571. doi: 10.1016/s0891-5849(02)00914-0. [DOI] [PubMed] [Google Scholar]
- 37.Castegna A, Aksenov M, Thongboonkerd V, Klein JB, Pierce WM, Booze R, et al. Proteomic identification of oxidatively modified proteins in Alzheimer's disease brain. Part II: dihydropyrimidinase-related protein 2, alpha-enolase and heat shock cognate 71. J Neurochem. 2002;82:1524–1532. doi: 10.1046/j.1471-4159.2002.01103.x. [DOI] [PubMed] [Google Scholar]
- 38.Butterfield DA, Poon HF, St Clair D, Keller JN, Pierce WM, Klein JB, et al. Redox proteomics identification of oxidatively modified hippocampal proteins in mild cognitive impairment: insights into the development of Alzheimer's disease. Neurobiol Dis. 2006;22:223–232. doi: 10.1016/j.nbd.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 39.Perluigi M, Poon HF, Maragos W, Pierce WM, Klein JB, Calabrese V, et al. Proteomic analysis of protein expression and oxidative modification in r6/2 transgenic mice: a model of Huntington disease. Mol Cell Proteomics. 2005;4:1849–1861. doi: 10.1074/mcp.M500090-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Poon HF, Frasier M, Shreve N, Calabrese V, Wolozin B, Butterfield DA. Mitochondrial associated metabolic proteins are selectively oxidized in A30P alpha-synuclein transgenic mice--a model of familial Parkinson's disease. Neurobiol Dis. 2005;18:492–498. doi: 10.1016/j.nbd.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 41.Poon HF, Hensley K, Thongboonkerd V, Merchant ML, Lynn BC, Pierce WM, et al. Redox proteomics analysis of oxidatively modified proteins in G93A-SOD1 transgenic mice--a model of familial amyotrophic lateral sclerosis. Free Radic Biol Med. 2005;39:453–462. doi: 10.1016/j.freeradbiomed.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 42.Sultana R, Boyd-Kimball D, Poon HF, Cai J, Pierce WM, Klein JB, et al. Redox proteomics identification of oxidized proteins in Alzheimer's disease hippocampus and cerebellum: An approach to understand pathological and biochemical alterations in AD. Neurobiol Aging. 2006;27:1564–1576. doi: 10.1016/j.neurobiolaging.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 43.Choi J, Levey AI, Weintraub ST, Rees HD, Gearing M, Chin LS, et al. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson's and Alzheimer's diseases. J Biol Chem. 2004;279:13256–13264. doi: 10.1074/jbc.M314124200. [DOI] [PubMed] [Google Scholar]
- 44.Newman SF, Sultana R, Perluigi M, Coccia R, Cai J, Pierce WM, et al. An increase in S-glutathionylated proteins in the Alzheimer's disease inferior parietal lobule, a proteomics approach. J Neurosci Res. 2007;85:1506–1514. doi: 10.1002/jnr.21275. [DOI] [PubMed] [Google Scholar]
- 45.Lovell MA, Ehmann WD, Butler SM, Markesbery WR. Elevated thiobarbituric acid-reactive substances and antioxidant enzyme activity in the brain in Alzheimer's disease. Neurology. 1995;45:1594–1601. doi: 10.1212/wnl.45.8.1594. [DOI] [PubMed] [Google Scholar]
- 46.Smith CD, Carney JM, Starke-Reed PE, Oliver CN, Stadtman ER, Floyd RA, et al. Excess brain protein oxidation and enzyme dysfunction in normal aging and in Alzheimer disease. Proc Natl Acad Sci U S A. 1991;88:10540–10543. doi: 10.1073/pnas.88.23.10540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Balastik M, Lim J, Pastorino L, Lu KP. Pin1 in Alzheimer's disease: multiple substrates, one regulatory mechanism? Biochim Biophys Acta. 2007;1772:422–429. doi: 10.1016/j.bbadis.2007.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korolainen MA, Nyman TA, Nyyssonen P, Hartikainen ES, Pirttila T. Multiplexed proteomic analysis of oxidation and concentrations of cerebrospinal fluid proteins in Alzheimer disease. Clin Chem. 2007;53:657–665. doi: 10.1373/clinchem.2006.078014. [DOI] [PubMed] [Google Scholar]
- 49.Smith MA, Taneda S, Richey PL, Miyata S, Yan SD, Stern D, et al. Advanced Maillard reaction end products are associated with Alzheimer disease pathology. Proc Natl Acad Sci U S A. 1994;91:5710–5714. doi: 10.1073/pnas.91.12.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Q, Woltjer RL, Cimino PJ, Pan C, Montine KS, Zhang J, et al. Proteomic analysis of neurofibrillary tangles in Alzheimer disease identifies GAPDH as a detergent-insoluble paired helical filament tau binding protein. Faseb J. 2005;19:869–871. doi: 10.1096/fj.04-3210fje. [DOI] [PubMed] [Google Scholar]
- 51.Soreghan BA, Yang F, Thomas SN, Hsu J, Yang AJ. High-throughput proteomic-based identification of oxidatively induced protein carbonylation in mouse brain. Pharm Res. 2003;20:1713–1720. doi: 10.1023/b:pham.0000003366.25263.78. [DOI] [PubMed] [Google Scholar]
- 52.MacCoss MJ, Yates JR., 3rd Proteomics: analytical tools and techniques. Curr Opin Clin Nutr Metab Care. 2001;4:369–375. doi: 10.1097/00075197-200109000-00006. [DOI] [PubMed] [Google Scholar]
- 53.Beavis RC, Chait BT. Cinnamic acid derivatives as matrices for ultraviolet laser desorption mass spectrometry of proteins. Rapid Commun Mass Spectrom. 1989;3:432–435. doi: 10.1002/rcm.1290031207. [DOI] [PubMed] [Google Scholar]
- 54.Butterfield DA, Boyd-Kimball D, Castegna A. Proteomics in Alzheimer's disease: insights into potential mechanisms of neurodegeneration. J Neurochem. 2003;86:1313–1327. doi: 10.1046/j.1471-4159.2003.01948.x. [DOI] [PubMed] [Google Scholar]
- 55.Poon HF, Castegna A, Farr SA, Thongboonkerd V, Lynn BC, Banks WA, et al. Quantitative proteomics analysis of specific protein expression and oxidative modification in aged senescence-accelerated-prone 8 mice brain. Neuroscience. 2004;126:915–926. doi: 10.1016/j.neuroscience.2004.04.046. [DOI] [PubMed] [Google Scholar]
- 56.Thongboonkerd V, Luengpailin J, Cao J, Pierce WM, Cai J, Klein JB, et al. Fluoride exposure attenuates expression of Streptococcus pyogenes virulence factors. J Biol Chem. 2002;277:16599–16605. doi: 10.1074/jbc.M200746200. [DOI] [PubMed] [Google Scholar]
- 57.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 58.Yates JR, 3rd, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 59.Fountoulakis M, Takacs B. Effect of strong detergents and chaotropes on the detection of proteins in two-dimensional gels. Electrophoresis. 2001;22:1593–1602. doi: 10.1002/1522-2683(200105)22:9<1593::AID-ELPS1593>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]